| Original Article | ||
Open Vet J. 2023; 13(1): 48-63 Open Veterinary Journal, (2023), Vol. 13(1): 48–63 Original Research Methanol extract of Black soldier fly (Hermetia illucens) prepupae against Aeromonas and Staphylococcus aureus bacteria in vitro and in silicoDahliatul Qosimah1,2*, Sanarto Santoso3, Maftuch Maftuch4, Husnul Khotimah5, Loeki Enggar Fitri6, Aulanni’am Aulanni’am7 and Lucia Tri Suwanti81Doctoral Study Program in Medical Science, Faculty of Medicine, Universitas Brawijaya, Malang, Indonesia 2Laboratory Microbiology and Immunology, Faculty of Veterinary Medicine, Universitas Brawijaya, Malang, Indonesia 3Laboratory of Microbiology, Faculty of Medicine, Universitas Brawijaya, Malang, Indonesia 4Laboratory of Fish Diseases, Faculty of Fisheries and Marine Science, Universitas Brawijaya, Malang, Indonesia 5Laboratory of Pharmacology, Faculty of Medicine, Universitas Brawijaya, Malang, Indonesia 6Laboratory of Parasitology, Faculty of Medicine, Universitas Brawijaya, Malang, Indonesia 7Department of Chemistry, Faculty of Sciences, Universitas Brawijaya, Malang, Indonesia 8Veterinary Parasitology Department, Faculty of Veterinary Medicine, Airlangga University, Surabaya, Indonesia Submitted: 05/09/2022 Accepted: 12/12/2022 Published: 11/01/2023 *Corresponding Author: Dahliatul Qosimah. Doctoral Study Program in Medical Science, Faculty of Medicine, Universitas Brawijaya, Malang, Indonesia. Email: dahlia_qosimah [at] ub.ac.id © 2023 Open Veterinary Journal
AbstractBackground: Staphylococcus and Aeromonas bacteria are pathogens in humans and animals. The therapy disrupts the virulence structure of the bacteria, resulting in bacterial death. Currently, chemical drugs have resulted in many resistant bacteria, so it is necessary to find alternative natural materials that are not toxic and do not quickly induce resistance. Aims: This study aimed to analyze the potential of methanol extract from Black soldier fly (BSF) prepupae as an antibacterial agent against Staphylococcus aureus and Aeromonas through in silico and in vitro tests. Methods: The BSF prepupae methanol extract was analyzed for protein and fatty acid contents. Disc diffusion method, minimal inhibitory concentration, and minimum bactericidal concentration test were used for in vitro tests against Staphylococcis and Aeromonas. Molecular docking of the active ingredients (defensin, chitin, and chitosan as well as fatty acids) in BSF was downloaded from the NCBI database and docked by the Hex Cuda version 8.0 program with Correlation type parameters Shape + Electro and Grid Dimension version 0.6. Docking results were analyzed using the Discovery Studio program version 21.1.1. Results: The highest fatty acid contents in the extract were palmitic acid and myristic acid. Methanol extract from BSF prepupae acted as a bactericidal agent against S. aureus at a concentration of 320 mg/ml, in contrast to Aeromonas, which still showed bacterial growth. The results of the in silico test showed that defensin–aerolysin and defensin–hemolysin was bound to the same active site area. However, the amount of binding energy produced by 69-Defensin-83-aerolysin was higher than all defensin types in BSF against Aeromonas. Chitin and chitosan showed a bond on the active site of aerolysin and hemolysin, but chitosan had a stronger bond than chitin. In silico study also showed the strongest binding affinity of BSF fatty acids to isoleucyl-tRNA synthetase of S. aureus. Conclusion: The study showed that methanol extract from BSF prepupae had potential capability as an antibacterial agent against S. aureus than Aeromonas in vitro and in silico. Keywords: Aeromonas, Staphylococcus aureus, Prepupae, Antimicrobe, Methanol extract. IntroductionAeromonas spp. bacteria are opportunistic pathogens and can cause disease in fish under immunosuppressive conditions or act as secondary infections. Motile Aeromonas septicemia is chronic, so the mortality rate is high (AlYahya et al., 2018), and it is resistant to multidrug antibiotics, which allows the bacteria to survive. Aeromonas salmonicida) is one of five quarantine fish diseases in Indonesia that requires special attention (Kementrian Kelautan dan Perikanan (KKP) RI, 2021). Staphylococcus aureus is a typical flora on the skin, mucous membranes, and skin glands in warm-blooded animals, and about 30% colonize humans (Tadesse et al., 2018). Staphylococcus aureus has zoonotic potential as it can be transmitted from dogs to humans (Nadăs et al., 2021). Staphylococcus aureus is a crucial bacterium that causes infection in humans worldwide. This bacterial infection requires prompt treatment because it can be fatal in immunosuppressed patients susceptible to bacterial resistance (Cortes et al., 2020). The main targets of antibiotics in Staphylococcus sp. are cell walls, ribosomes, and nucleic acids (Foster, 2017). β-lactam antibiotics as the drug of choice against S. aureus infection have become ineffective since the emergence of methicillin-resistant S. aureus (MRSA) strains (Qureshi and Chaudhari, 2019). To overcome this problem, we need novel antibacterial agents from natural ingredients that are effective, non-toxic, and have no risk of causing resistance to bacteria. Black soldier fly larva (BSFL) is widely used in the community because it can produce fatty acids and bioactive ingredients such as antimicrobial peptides (AMPs; Nardiello et al., 2022). BSF prepupae contains 32% proteins, 37% lipids, 19% minerals, and 9% chitin (Caligiani et al., 2018). Antimicrobial peptides act against broad-spectrum and resistant bacteria (Alvarez et al., 2019). The fatty acid content in BSF prepupa extract can act as an antibacterial, anti-inflammatory, and wound-healing agent (Casillas-Vargas et al., 2021). There are various BSF active ingredients, so this study aims to analyze prepupa BSF as an antibacterial against Aeromonas and S. aureus. Materials and MethodsPreparation of S. aureus and MRSA bacteria and antibiotic sensitivity testBacteria were cultured in the Faculty of Medicine, Universitas Brawijaya. Bacteria were grown in BPA (Baird Parker Agar) (Merck™) media and incubated at a temperature of 35 C ± 1°C, which showed presumptive growth of S. aureus, namely black colonies with a clear zone around it (halo-shaped), afterward Gram staining, catalase, oxidase, and coagulase tests were performed (Javid et al., 2018). The bacteria were then grown on brain heart infusion broth (BHIB) media (Merck™) to make a suspension that would be used for disk dilution and diffusion tests. The antibiotics used for disk diffusion were penicillin (P), methicillin (MET), ampicillin (AMP10), bacitracin (B), and amoxicillin (AMX) (Oxoid™). Preparation of A. hydrophila and A. salmonicida bacteria and antibiotic sensitivity testBacteria from BHIB media were grown on Trypticase Soy Agar media (Merck™) and incubated at 27°C for 24 hours, and then biochemical tests were carried out (Al Laham and Al Fadel, 2014). The several antibiotic discs include oxytetracycline (OXY), doxycycline (DOXY), tetracycline (TE), erythromycin (ERY), cefadroxil (CFR), bacitracin (B), colistin sulfate (CS), gentamicin (CN), and enrofloxacin (ENR) (Oxoid™). The results of the antibiotic sensitivity test were adjusted to the tables of the Clinical and Laboratory Standards Institute M45 (CLSI, 2015) and M100 (CLSI, 2021). Preparation of BSFL growth mediaThe study used 7-day-old BSFL. The BSFL were grown on organic substrates (fermented fruit and tofu waste, with a ratio of 1:1 as much as 300 g each). The substrates were put through a fermentation process for 48 hours. The fermentation process occurred in anaerobic conditions with a mixture of EM4, molasses, water, and media in a ratio of 2:1:50:500 (ml). Larvae were harvested during the prepupae period at the age of 25 days. Larvae were reared at a temperature of 33°C–40°C, with a relative humidity of 60%–70% (Lin et al., 2021). BSF prepupae were washed (Fig. 1), euthanized with hot water, and dried using a microwave at 1,000 watts for 5 minutes. This research used prepupae than an adult fly of BSF because flies don’t have fat content (Caligiani et al., 2018). Extraction procedures and extract fatty acids testing using the gas chromatography-mass spectrometer detector (GC-MSD) method The dry maggot powder was ground, soaked in absolute methanol, and filtered. The filtrate was taken and evaporated with a rotatory evaporator at 45°C–60°C. Five hundred grams of dry BSF prepupa were extracted to produce 10.08 g of brown extract (Choi et al., 2012). The extract was dissolved with 1% dimethyl sulfoxide (DMSO) to obtain concentrations of 40, 80, 160, and 320 mg/ml. The extract results were analyzed for fatty acid content (%) using the GC/MSD method (Mazurek et al., 2017) and for protein using Kjeldahl methods. Disc diffusion methodEach test bacterium was grown into BHIB media according to the growth phase (incubation time and length), then the turbidity was adjusted to the McFarland standard 0.5 (1.5 × 108 CFU/ml). It was then resuspended with phosphate buffer saline to a concentration of 105 CFU/ml. A loopful of bacteria was swabbed on top of Muller Hinton agar (MHA). Sterile blank disks were dipped into each concentration of the tested extract and then pasted on the MHA medium (Abuelsaad et al., 2013).
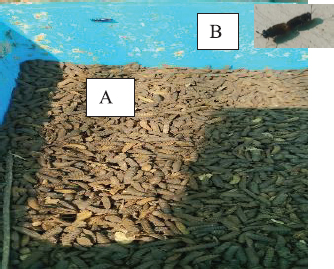
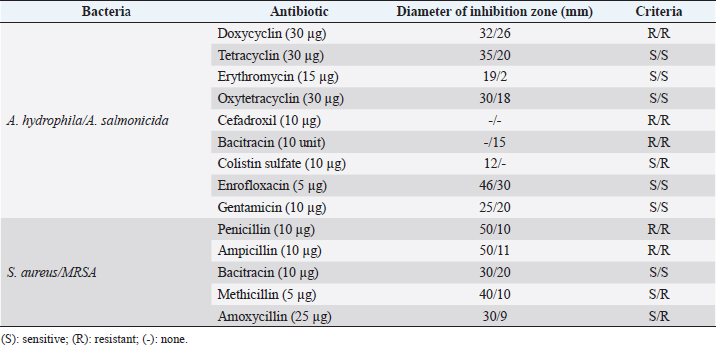
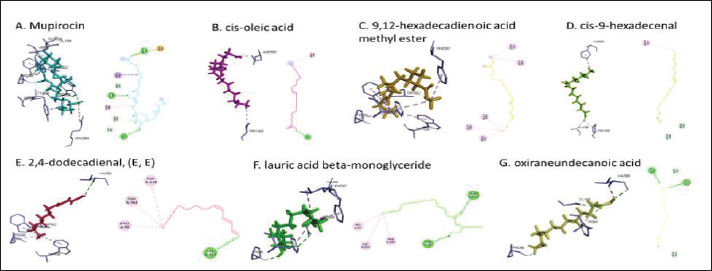
Fig. 1. Dry prepupae BSF (A) and BSF (B). Determination of minimal inhibitory concentration (MIC)One ml of test bacteria grown on BHIB media with a McFarland standard concentration of 0.5 was mixed with 1 ml of an extract with tested concentration. Similar things were repeated for each testing concentration. The tubes were then incubated and observed for turbidity. MIC is the lowest extract concentration capable of inhibiting bacterial growth (Abuelsaad et al., 2013). Determination of minimum bactericidal concentrations (MBC)Tubes of MIC that did not show turbidity were swabbed onto MHA media and incubated. MBC is the lowest extract concentration that kills bacterial growth (Abuelsaad et al., 2013). Molecular docking and analysis of fatty acids to S. aureusThe structure of the fatty acids contained in BSF was downloaded from the PubChem NCBI database cis-oleic acid (CID 445639), 9,12-hexadecadienoic acid methyl ester (CID 5365666), cis-9-hexadecenal (CID 5364643), 2,4-dodecadienal (CID 5367530), lauric acid beta-monoglyceride (CID 74297), oxiraneundecanoic acid (CID 86131368), and control penicillin (CID 2349). The 3D structure of targeting ligand is imported into the Molegro virtual docker version 5.0 program for fatty acid docking without preparation (Filimonov et al., 2014). The target proteins used as antibacterial targets for S. aureus are penicillin-binding protein [PBP; Protein Data Bank (PDB) ID 4CJN] (Bouley et al., 2015), isoleucyl-tRNA synthetase (IARS; PDB ID: 1QU2) (Cortes et al., 2020), and DNA gyrase (GDP ID: 2XCQ) (Bax et al., 2010). The 3D structure of the protein was downloaded from the PDB database. Comparative controls for docking validation were quinazolinone for PBP (PDB ID 4CJN) (Bouley et al., 2015), Mupirocin for IARS (PDB ID: 1QU2), and penicillin (CID 2349). Each protein was prepared by removing the binding ligand and water molecule. Furthermore, the active site is predicted with binding cavities parameters based on the van der Waals force, with a maximum of five active sites. This active site will be used for the docking area. Predictions were made with the Molegro Virtual Docker version 5.0 program (Bitencourt-Ferreira and De Azevedo, 2019). Docking between the ligand and the target protein interacts in the special area. The grid of PBP’s (PDB ID 4CJN) is X=11.43; Y=−6.86; Z=−69.69; radius 13, IARS (GDP ID: 1QU2) X=29.40; Y=74.79; Z=71.53; radius 15. Meanwhile, the DNA gyrase target protein grid (GDP ID: 2XCQ) is X=−24.02; Y=105.01; Z=21.81; radius 15. Other docking parameters are MolGrid 0.3A, 10× docking repetitions, and an RMSD threshold of less than 1. The docking results were analyzed with PyMol version 2.2 and Discovery Studio version 21.1.0 program. Molecular docking and analysis of defensin to AeromonasProtein defensin-like BSF was downloaded from the NCBI database with ID XP_037915109.1. Defensin proteins predicted bioactive peptide sequences with the expasy peptide cutter webserver (https://web.expasy.org/cgi-bin/peptide_cutter/peptidecutter.pl). Defensin bioactive peptides with amino acid residues of 6–53 were modeled using http://swissmodel.expasy.org/ and PEP-FOLD3 (https://bioserv.rpbs.univ-parisdiderot.fr/services/PEP-FOLD3/) (Bienert et al., 2017). Defensin, hemolysin, and aerolysin bioactive peptides were docked with the Hex Cuda version 8.0 program with Correlation type Shape + Electro and Grid Dimension version 0.6 parameters (Macindoe et al., 2010). The docking results were analyzed using the Discovery Studio program version 21.1.1. Molecular docking and analysis of docking chitin dan chitosan to AeromonasChitin Octamer (CID 24978517) and Chitosan (CID 71853) downloaded canonical smiles from NCBI’s PubChem database and modeled them with MolView (https://molview.org/). Aeromonas hydrophilla target proteins are aerolysin and hemolysin. The 3D structure of aerolysin was downloaded from the PDB database (PDB ID 1PRE), while the 3D structure of hemolysin was downloaded from the UNIPROT database with ID P55870 (Poux et al., 2017). The docking of chitin and chitosan compounds with aerolysin and hemolysin proteins was carried out using the Hex Cuda version 8.0 program with Correlation type Shape + Electro and Grid Dimension parameters 0.6 (Macindoe et al., 2010; Ritchie et al., 2008). The docking results were analyzed using the Discovery Studio program version 21.1.1. Statistical analysisData from the disk diffusion test were analyzed statistically using the one-way analysis of variance test with a 95% confidence level. Ethical approvalThis research received an ethical approval certificate from Universitas Brawijaya, Malang, Indonesia (Certified no. 035-KEP-UB-2022). ResultsAntibiotic sensitivity The results of the antibiotic sensitivity test showed that Aeromonas hydrophila bacteria were resistant against CFR, DOXY, AMX, and B, but sensitive against TE, ERY, OXY, ENR, CS, and CN; A. salmonicida showed resistance against CS, CFR, B, AMX, and DOXY, but sensitive against TE, ERY, OXY, ENR, CN. Staphylococcus aureus was resistant against P, AMP10, but sensitive against B, MET, and AML; and MRSA bacteria showed sensitivity only to the antibiotic B (Table 1). Table 1. Sensitivity of antibiotics against A. hydrophila, A. salmonicida, S. aureus, MRSA bacteria (CLSI, 2015).
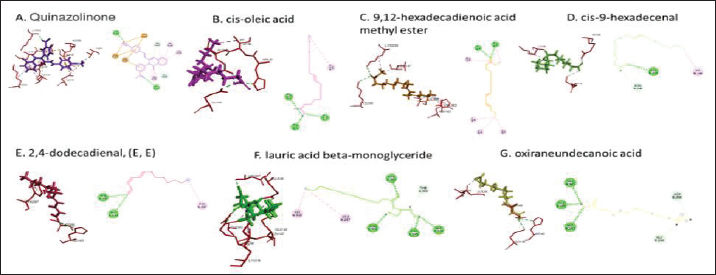
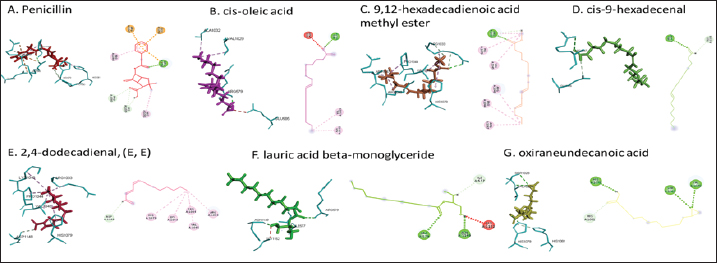
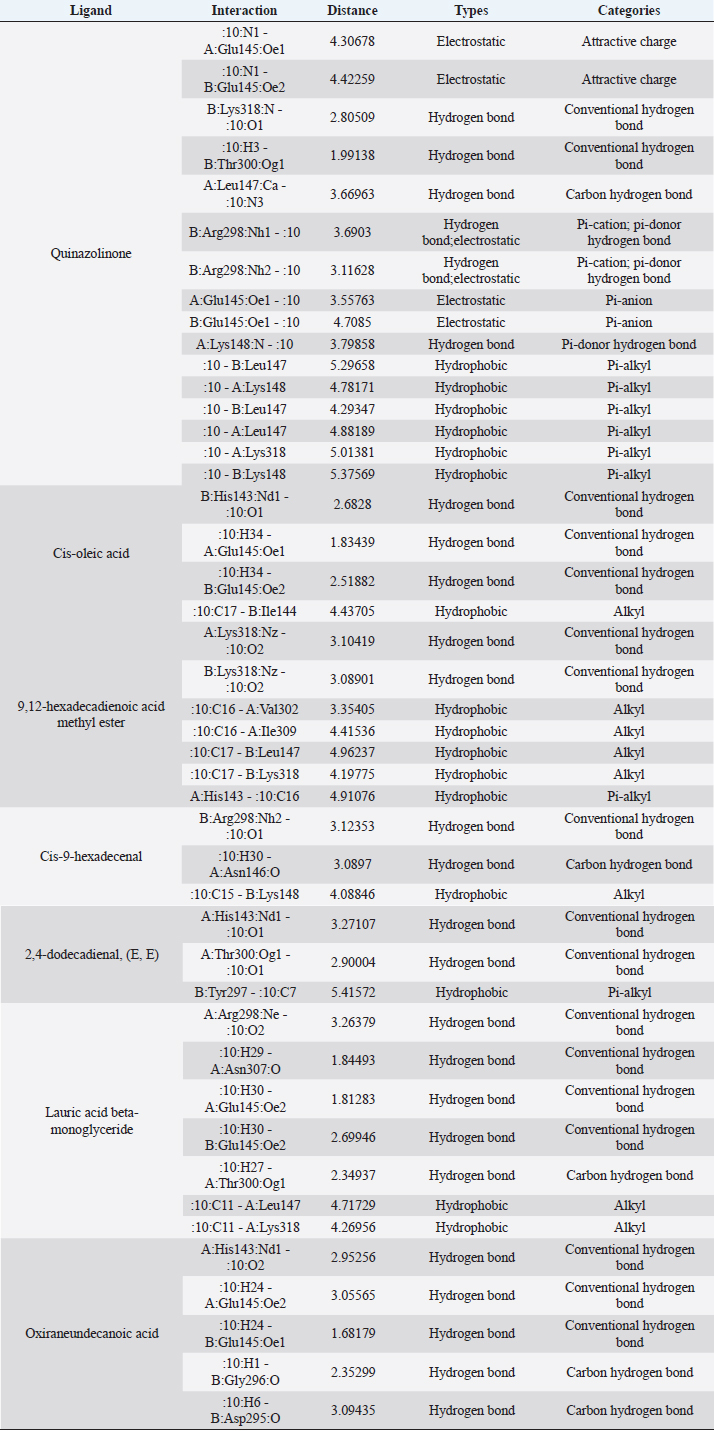
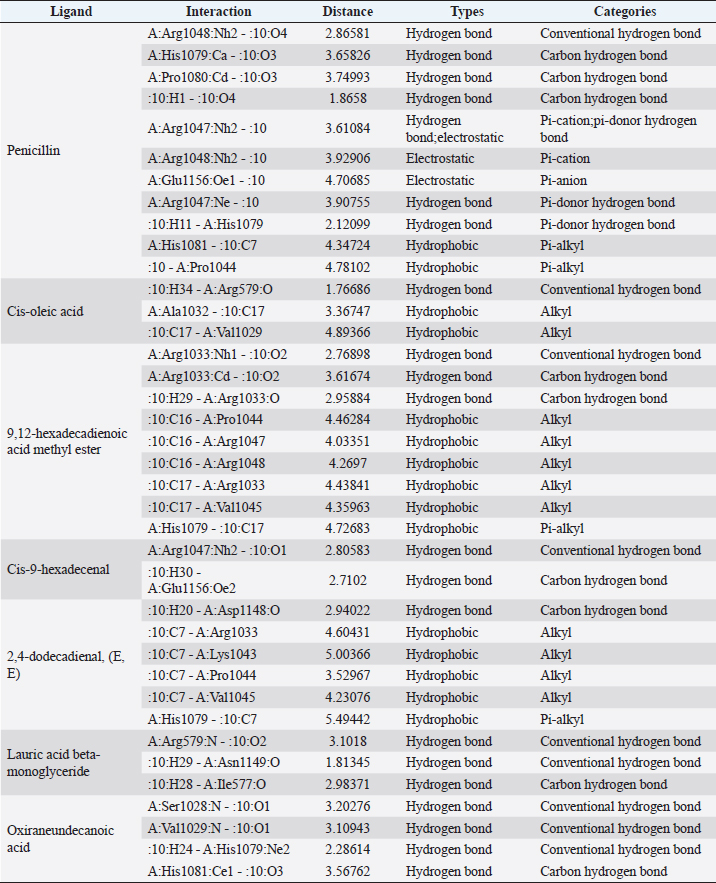
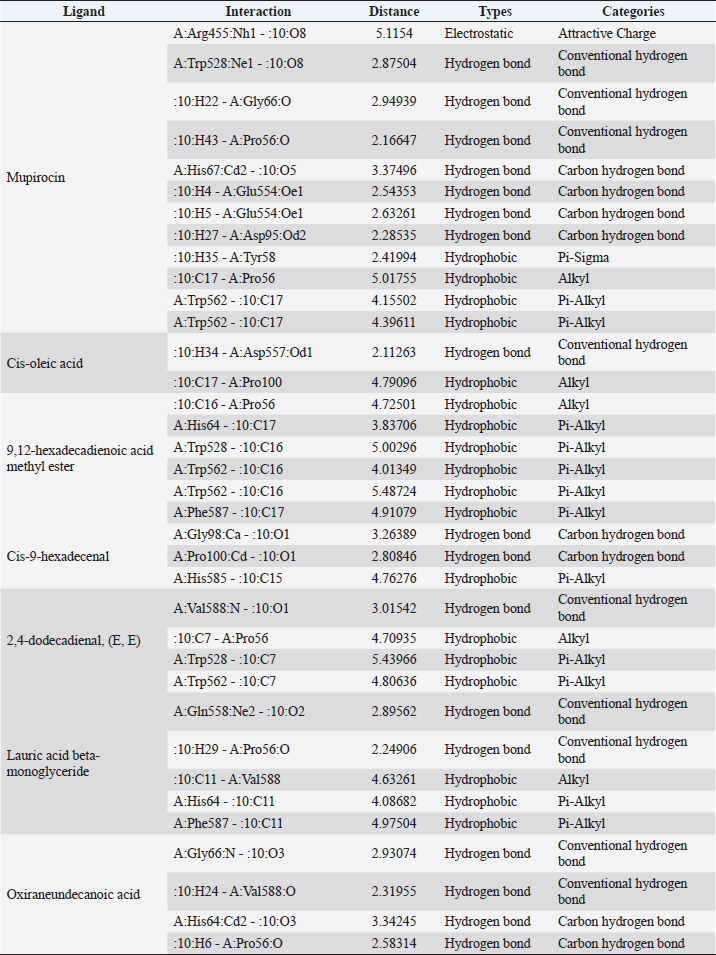
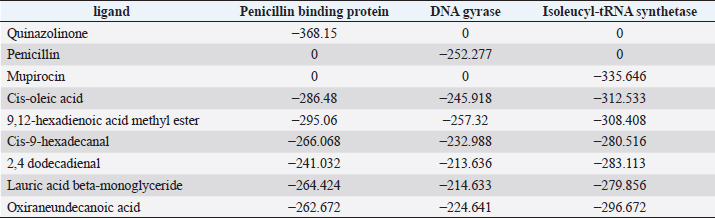
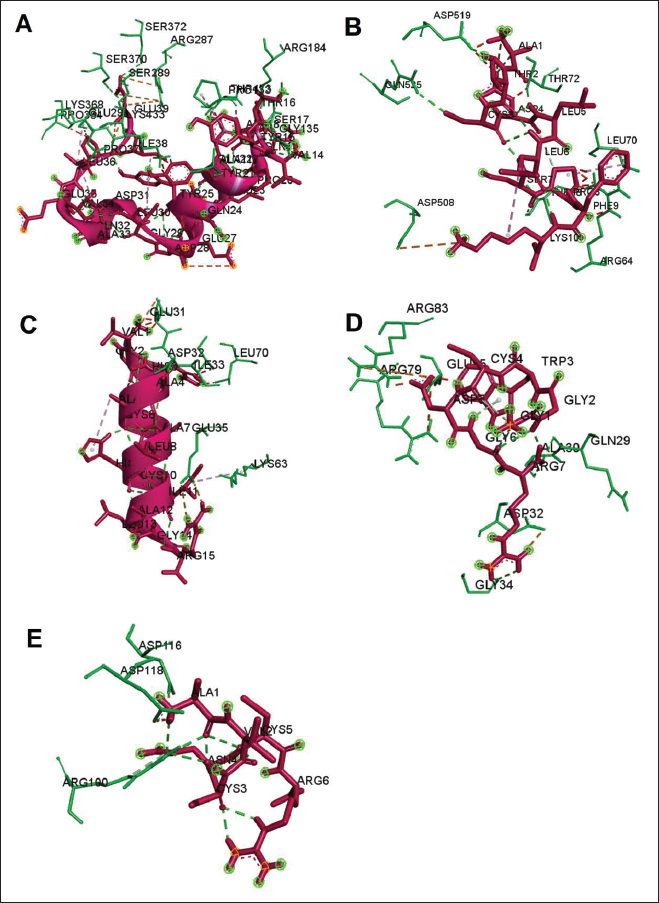
Active ingredients in methanol extract from BSF prepupaeThe study showed that fatty acids in the following percentage were found in the methanol extract of BSF prepupae: myristic acid (21.04), stearic acid (4.62), palmitic acid (25.85), pentadecanoic acid (0.40), oleic acid (18.30), palmitoleic acid (8.58), arachidic acid (0.37), lauric acid (0.03), and linoleic acid (17.6). The highest fatty acids in this study were palmitic acid and myristic acid, while the lowest were arachidic acid and oleic acid. Protein content in the methanol extract of BSF prepupae was 7.44%. Disk diffusion test, MIC, and MBC testInhibition of methanolic extract of BSF prepupae against the growth of S. aureus and MRSA was found at a concentration of 160 mg/ml, indicated by the formation of an inhibition zone (Table 2). The MIC test was not able to determine the ability of the extract to inhibit bacterial growth because the suspension was dark in color at all test concentrations, so the results were read directly from the MBC test, which indicated the killing power or no bacterial growth was found at a concentration of 320 mg/ml. Methanol extract of BSF prepupa could not inhibit growth or kill A. hydrophila and A. salmonicida bacteria at all test concentrations. Molecular docking and analysis of fatty acids to S. aureusThe fatty acids found in BSF are cis-oleic acid, 9,12-hexadecadienoic acid methyl ester, cis-9-hexadecenal, 2,4-dodecadienal, lauric acid beta-monoglyceride, and oxiraneundecanoic acid. The 3D views of those six fatty acid compounds and quinazolinone showed the same binding area as the PBP of S. aureus. These fatty acids and quinazolinone bind to the amino groups of residues Glu 145, Lys 318, Arg 298, Lys 148, Leu 147, His 143, Leu 147, Arg 298, and Thr 300 (Table 3). The bonds between the fatty acid and quinazolinone are electrostatic, hydrogen, and hydrophobic, with the highest affinity of bonds being quinazolinone (Fig. 2). Fatty acid in BSF and penicillin showed inhibition in the same area against DNA gyrase in Figure 2. The same amino acid active site was residues Arg 1048, His 1079, Arg 1047, Glu 1156, His 1079, His 1081, Pro 1044, Arg 579, and Val 1029 (Table 4). The highest interaction of all fatty acids is with IARS consecutively are Cis-oleic acid (−312.533 kcal/mol); 9,12-hexadienoic acid methyl ester (−308.408 kcal/mol) and Lauric acid beta-monoglyceride (−279.856 kcal/mol). This interaction was higher than PBP and DNA gyrase (Fig. 3, Supplementary data in Table 6). There is a favorable bond between Mupirocin and fatty acids in IARS. Mupirocin has a higher binding activity than fatty acids of BSF to IARS. There are beneficial interactions at the same active site between Mupirocin and fatty acids on IARS, namely residues Trp 528, Pro 56, Trp 562, Pro 100, Val 588, and His 64 (Table 5). The highest ligand complex binding energy (fatty acid of BSF, penicillin, quinazolinone, and Mupirocin) to the protein of S. aureus is PBP’s on Quinazolinone, to DNA gyrase is penicillin and to IARS is Mupirocin (Fig. 4; Supplymentary data in Table 6). Molecular docking of peptide defensin to AeromonasProtein defensins like BSF were predicted with peptide cutter and trypsin enzyme to produce 10 peptides with amino acid sequence lengths of 1–53 (unpublished data). Five of ten sequences of bioactive defensin peptides were selected (AVCNCR; GGWCDGR; ATCDLLSPFK; VGHAACALHCIALGR; SVLVLGLIVAAFAVYTSAQPYQLQYEEDGLDQAVELPIEEEQLPSQVVEQHYR). Table 2. Antibacterial sensitivity test of methanol extract of BSF prepupae.
Fig. 2. The 3D and 2D views of interactions between PBP and target ligands.
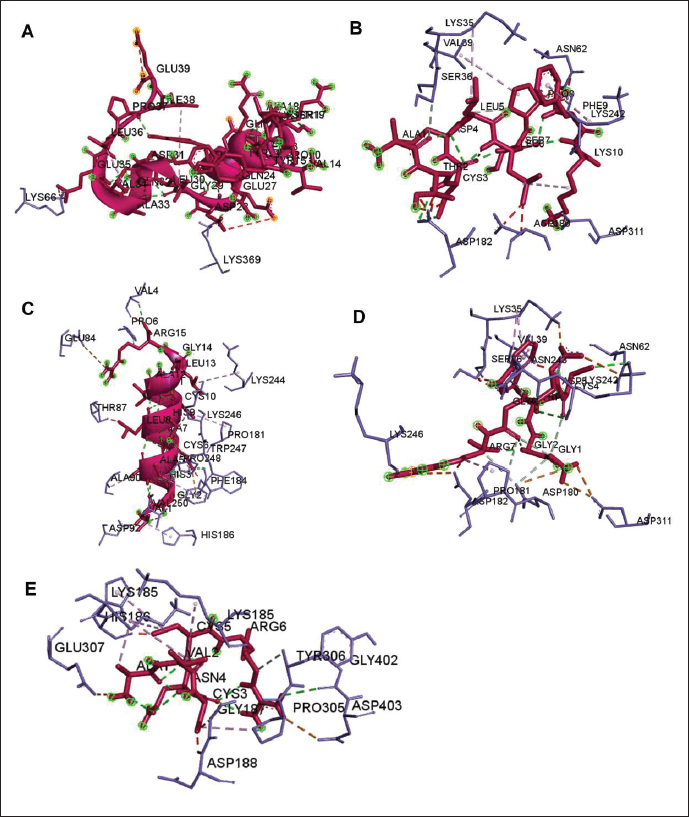
Fig. 3. The 3D and 2D views of interactions between DNA gyrase and target ligands. Defensin peptides from BSF (3-defensin-53, 59-defensin-68, 69-defensin-83, 85-Defensin-91-Aerolysin, and 92-Defensin-97-Aerolysin) bind to aerolysin from A. hydrophila on the active site. The amino acid residues of the five defensin peptides were Ser 36, Lys 242, Lys 35, Ala 1, and Cys 3. Bond types of all defensin peptides are hydrogen bond, electrostatic, hydrophobic, and Unfavorable (Fig. 5). Defensin peptides (3-defensin-53, 59-defensin-68, 69-defensin-83, 85-Defensin-91-Aerolysin, and 92-Defensin-97-Aerolysin) bind to hemolysin at the active site. The amino acid residues of the four defensin peptides were Ala 1, Cys 3, Asn 62, Asp 180, Val 39, Asp 182, and Asp 311 (Fig. 5). Bond types 3-Defensin-53–hemolysin, 59-Defensin-68-hemolysin, and 69-Defensin-83-hemolysin are hydrogen bonds, electrostatic, hydrophobic, and unfavorable. The bond types of 85-Defensin-91-hemolysin are hydrogen bond, electrostatic and Unfavorable. The bond types of 92-Defensin-97-hemolysin are hydrogen bond and electrostatic. The amino acid residues are Glu 39 and Ala 1 (Fig. 6). Table 3. Interaction between penicillin binding protein and target ligand.
The lower binding energy, the stronger interaction, protein 59-defensin-68; 69-defensin-83; 85-defensin-91, and 92-defensin-97 have stronger interactions with aerolysin protein than hemolysin. Molecular docking of chitin and chitosan to AeromonasChitin and chitosan show interaction with aerolysin protein of Aeromonas. The residue of the active site bound by the two compounds is Asn 79, with the type of bond that is hydrogen bonds. Chitin produces five hydrogen bonds and lower bond energies from the interaction of chitosan with aerolysin. While chitosan shows four hydrogen bonds in Asn 79. Chitin binds to aerolysin on the inactive site. Chitin and chitosan have the same residue, namely Asn 79, because of the same structure. Both do not bind to aerolysin. The lower the binding energy, the stronger the interaction; the bond between chitin and Aerolysin (−271.2 kJ/mol) is more potent than chitosan—Aerolysin (−259.8 kJ/mol). In contrast to aerolysin, in the complex interaction of chitin and chitosan with hemolysin, chitosan shows more active site residues than chitin. Although the 3D view of the two complexes—hemolysin shows chitin and chitosan bound to hemolysin in the same area, the binding energy produced by chitin is lower than that of chitosan. The lower the binding energy, the stronger the interaction. Chitin—Hemolysin (−516.8 kJ/mol) has a more robust interaction than chitosan—Hemolysin (−455.7 kJ/mol). The strongest are ionic bonds (hydrogen and hydrophobic) because they can donate and accept hydrogen atoms. The calculation of the amount of energy depends on the number of bonds–any binding effect (hydrogen, hydrophobic, or van der wall). Unfavorable includes covalent bonds. The amino acid residues of chitin-hemolysin and chitosan-hemolysin are Asn 483, Gln 539, Ala 509, and Asp 519. Chitin-hemolysin amino acid residues form 4 hydrogen bonds and 23 Unfavorable bonds on chitin-hemolysin bonds, forming Conventional Hydrogen Bonds, Carbon Hydrogen Bonds, and Unfavorable Bumps. In comparison, chitosan-hemolysin amino acid residues form 13 hydrogen bonds and 19 Unfavorable bonds and form Conventional Hydrogen Bonds, Pi-Alkyl, Carbon Hydrogen Bonds, Unfavorable Bumps, Unfavorable Donor-Donor, and Unfavorable Acceptor-Acceptor (Data unpublished). Table 4. Interaction between DNA gyrase and target ligand.
Table 5. Interaction between isoleucyl-tRNA synthetase and target ligand.
Table 6. The binding energy of the ligand–protein complex.
Fig. 4. 3D and 2D views of interactions between IARS and target ligands. DiscussionAntibiotic sensitivity, disk diffusion test, and dilution testThe four bacteria used in this study were multidrug-resistant (MDRs). Resistant bacteria will adversely affect the environment and hosts, especially humans (Nadăs et al., 2021). Aeromonas is currently showing global MDR on strains that infect ornamental fish and food fish (Dhanapala et al., 2021). The results showed that A. hydrophila bacteria resisted the same antibiotics as A. salmonicida. MRSA bacteria are more resistant to antibiotics than S. aureus. Staphylococcus aureus and MRSA bacteria showed they were still sensitive to bacitracin antibiotics. Bacitracin use for treating systemic infections is limited because it shows high nephrotoxicity (Bessa et al., 2016). The ability of the extract to kill bacteria (bactericidal) observed in the MBC test required concentration two times higher than the concentration needed to inhibit bacterial growth (bacteriostatic) through the MBC test. The effect of the extract as bactericidal or bacteriostatic in vitro depends on the growth medium used (optimal MHA medium for antibacterial sensitivity test) and the concentration of the bacteria being tested. Bactericidal is the direct ability of antibacterial agent to kill target bacteria cells. At the same time, bacteriostatic requires phagocytic cells to prevent bacterial growth, so it is less effective to use without good immunity from host cells. Thus, for unhealthy individuals, especially under immunosuppressive conditions, it is more recommended to use an antibacterial with a bactericidal effect than a bacteriostatic one (Nemeth et al., 2015). The content of BSF prepupae methanol extractThe fatty acid contents in the methanol extract of BSF prepupae were myristic acid, stearic acid, palmitic acid, pentadecanoic acid, oleic acid, palmitoleic acid, arachidic acid, lauric acid, and linoleic acid. The highest fatty acid contents in the extract were palmitic acid and myristic acid. Saturated fatty acids (SFAs) such as lauric acid, stearic acid, palmitic acid, and capric acid can Fight broad-spectrum bacteria (Casillas-Vargas et al., 2021). This study showed that methanol extract affected Gram-positive bacteria such as S. aureus more than Gram-negative bacteria such as Aeromonas. The methanol extract from the BSF prepupa contains long and medium-chain fatty acids. Being hydrophobic, it is difficult for them to penetrate the hydrophilic bacterial cell membrane to kill the bacteria (Agoramoorthy et al., 2007). Fatty acids can prevent adhesion and biofilm formation and maintain the pH of the body's surface to be acidic, allowing it to kill certain bacteria. The most potential SFA as an antibacterial agent is lauric acid, while the un SFA is oleic acid, which kills S. aureus bacteria (Jumina et al., 2019). Research conducted by Casillas-Vargas et al. (2021) showed that lauric acid has more potential as an anti-bacterial agent against Gram-positive bacteria such as Staphylococcus and Streptococcus than against Gram-negative ones such as Escherichia coli. Monolaurin and its derivatives work by binding directly to the lipid bilayer of S. aureus bacteria to cause bacterial cell membrane damage and death.
Fig. 5. Interaction between defensin peptide and aerolysin protein. A. 3-defensin-53, B. 59-defensin-68, C. 69-defensin-83, D. 85-Defensin-91, and E. 92-Defensin-97.
Fig. 6. Interaction between defensin peptide and hemolysin protein. A. 3-defensin-53, B. 59-defensin-68, C. 69-defensin-83, D. 85-Defensin-91, and E. 92-Defensin-97. Molecular docking and analysis of fatty acids against S. aureusMolecular docking was used to evaluate the potential of BSF fatty acids, quinazolinone, penicillin, and mupirocin antibiotics in inhibiting the cell wall of S. aureus bacteria through the action of PBP, IARS, and DNA gyrase. Molecular docking was carried out to see the bond between fatty acids from BSF and other antibacterial materials such as quinazolinone, Mupirocin, and penicillin to enzymes produced by S. aureus. Quinazolinone and its derivatives can fight Gram-positive bacteria through cell wall inhibition and disruption of DNA structure (Jafari et al., 2016). Mupirocin is an antibiotic against Gram-positive bacteria which works through the inhibition of IARS thereby preventing the amino acid isoleucine from combining into peptides and thus failing protein synthesis (Hetem and Bonten, 2013). Penicillin is a β-lactam group that acts in the final phase of bacterial cell wall formation for transpeptidation, transglucosylation, and carboxypeptidation reactions (Alves et al., 2014). Fatty acids have a broad spectrum as antibacterial agents and do not induce resistance. Long-chain polyunsaturated fatty acids have antibacterial activity against Gram-positive bacteria (Desbois et al., 2009). In silico study showed the strongest interaction of BSF fatty acids to IARS via hydrogen, electrostatic, and hydrophobicity interaction with amino acid residues present in the cell membranes of S. aureus. According to a study by Shaaban et al. (2021), clove alcoholic extract contains hexadecanoic acid methyl ester, which can inhibit the growth of MDR bacteria such as S. aureus W35, Pseudomonas aeruginosa D31, and Klebsiella pneumoniae DF30 through mechanisms including interfering with membrane activity of bacterial cells, cellular energy, and enzymatic activity to cause bacterial death further. Oleic acid acts as an antibacterial agent against broad-spectrum bacteria (Ali et al., 2017) by changing their membrane profile, causing bacterial death (Nagendra Prasad et al., 2019). The in silico results showed that the strongest interaction of BSF fatty acids was oleic acid for aerolysin and linoleic acid for hemolysin and aerolysin bacteria Aeromonas (data unpublished). Based on the results of in silico research, it can be concluded that oleic acid affects the growth of S. aureus and Aeromonas bacteria. Molecular docking was used to evaluate the potential of BSF fatty acids, quinazolinone, penicillin, and mupirocin antibiotics in inhibiting the cell wall of S. aureus bacteria through the action of PBP, IARS, and DNA gyrase. Molecular docking of peptide defensin to AeromonasWhen Aeromonas bacteria enter the body of fish, amphibians, or humans, the bacteria will travel through the bloodstream to the target organs by secreting cytotoxic enterotoxins, which are one of the main virulence factors. The toxin binds to high-affinity receptors to form a heptameric pore-forming complex that allows the passage of small particles into the plasma membrane, which results in the death of the host tissue (Al Laham and Al Fadel, 2014). According to Ahmed et al. (2018), the most common virulent gene molecules found in fish infected with A. hydrophila are hemolysin and aerolysin. BSFL can eat organic waste substances more efficiently than other insects, such as Drosophila melanogaster, Apis mellifera, and Bombyx mori. BSFL are also able to survive in extreme environments with many microbes because they have an innate immune system that can produce various substances, such as peptides (Xia et al., 2021) defensins in their bodies, which are protein components rich in cysteine (Irawan et al., 2020), According to Li et al. (2017), the defensin peptides DLP2 and DLP4 from BSF at 873.5 and 801.3 mg/l were expressed in Pichia pastoris, showing faster antimicrobial activity against an MRSA, and a longer post-antibiotic effect than vancomycin. Antimicrobial peptide defensins bind to the lipid-peptide interface when interacting with membranes that have strong electrostatic bonds with the bacterial plasma membrane (Bhat et al., 2020), inhibiting or even stopping the enzyme catalytic activity (Jia et al., 2019), and damaging the cell wall of bacteria thus resulting in bacterial death. In this research, an in silico study to examine the bond between defensins from BSF to Aeromonas showed that AMP defensins from BSF have strong hydrogen and hydrophobic bonds to aerolysin and hemolysin. These results are consistent with the molecular docking of the antimicrobial peptide RY12WY, which has a strong affinity for the target protein Aeromonas sobria; aerolysin, and outer membrane protein. Antimicrobial activity of RY12WY at low concentrations was also reported against A. hydrophila, Edwardsiella tarda, S. aureus, Vibrio parahaemolyticus, Pseudomonas aeruginosa, and E. coli at low concentrations (Bhat et al., 2020). However, AMP also has weaknesses, namely stability and toxicity. Low serum levels can induce cross-resistance upon prolonged exposure to bacteria (Nepal et al., 2021), so it is necessary to explore further the suitability of peptide properties with its target. Molecular docking of chitin to AeromonasUntil now, there is no scientific data on using chitin from BSF as an antibacterial agent. Based on the in silico test, it was shown that both chitin and chitosan had a bond on the aerolysin and hemolysin active sites of Aeromonas, but chitosan had a stronger bond than chitin. Chitin is an unbranched long-chain polysaccharide formed from N-acetylglucosamine units linked by −1.4 covalent bonds. Chitin is deacetylated into chitosan, which can be used for various biomedical applications (Tan et al., 2021). Chitosan, chitin, and their derivatives function as bacteriostatic and bactericidal agents. Polycationic chitosan can interfere with bacteria metabolism through electrostatic accumulation on the surface of bacterial cells and block bacterial transcription through adsorption of chitosan that penetrates DNA molecules. The antimicrobial activity of chitosan depends on its physical properties, especially its molecular weight and degree of deacetylation. The higher the degree of deacetylation of chitosan, the higher the antibacterial ability (Benhabiles et al., 2012). Chitosan extract from white shrimp (Penaeus indicus) can inhibit the growth of S. aureus and E. coli bacteria (Kusnadi et al., 2022) Methanol extract of BSF prepupa functions as an antibacterial agent against S. aureus and MRSA bacteria in vitro and silico. The content of BSF prepupa (antimicrobial peptide, chitosan, and fatty acids) can act as an antibacterial agent against Aeromonas. Still, this study found no inhibitory activity against bacterial growth. Hence, it is necessary to explore further the extraction method and solvent used so that the bioactive contents in BSF prepupa can be optimally produced as antibacterial agents. According to Zarrinmehr et al. (2022), mixtures of solvents with higher ratio of non-polar solvents such as chloroform and hexane can be applied at higher temperatures to produce more fat and fatty acid methyl esters profile and have higher oxidative stability. AcknowledgmentMany thanks to the Doctoral Study Program in Medical Science, Faculty of Medicine, Universitas Brawijaya, and Faculty of Veterinary Medicine, Universitas Brawijaya, for their support. Conflict of interestThe authors declare that there is no conflict of interest. Author contributionsD.Q.S., S.S., M.M., and H.K.: making research designs, analyzing and writing manuscripts; L.E.K., A.A., and L.T.: writing manuscript. ReferencesAbuelsaad, A.S.A., Mohamed, I., Allam, G. and Al-Solumani, A.A. 2013. Antimicrobial and immunomodulating activities of hesperidin and ellagic acid against diarrheic Aeromonas hydrophila in a murine model. Life Sci. 93, 714–722. Agoramoorthy, G., Chandrasekaran, M., Venkatesalu, V. and Hsu, M.J. 2007. Antibacterial and antifungal activities of fatty acid methyl esters of the blind-your-eye mangrove from India. Brazilian. J. Microbiol. 38, 739–742. Ahmed, H.A., Mohamed, M.E.M., Rezk, M.M., Gharieb, R.M.A. and Abdel-Maksoud, S.A. 2018. Aeromonas hydrophila in fish and humans; prevalence, virulotyping and antimicrobial resistance. Slov. Vet. Res. 55, 113–124. Al Laham, S. and Al Fadel, F.M. 2014. Antibacterial activity of various plants extracts against antibiotic-resistant Aeromonas hydrophila. Jundishapur J. Microbiol. 7, 1–7. Ali, S.E., Chehri, K., Karimi, N. and Karimi, I. 2017. Computational approaches to the in vitro antibacterial activity of Allium hirtifolium Boiss against gentamicin-resistant Escherichia coli: focus on ribosome recycling factor. Silico Pharmacol. 5, 1–9. Alvarez, D., Wilkinson, K.A., Treilhou, M., Téné, N., Castillo, D. and Sauvain, M. 2019. Prospecting peptides isolated from black soldier fly (Diptera: Stratiomyidae) with antimicrobial activity against Helicobacter pylori (Campylobacterales: Helicobacteraceae). J. Insect Sci. 19, 1–5. Alves, M.., Froufe, H.J.C., Costa, A.F.T., Santos, A.F., Oliveira, L.G., Osório, S.R.M., Abreu, R.M.V. and Pintado, M., Ferreira, I.C.F.R. 2014. Docking studies in target proteins involved in antibacterial action mechanisms: Extending the knowledge on standard antibiotics to antimicrobial mushroom compounds. Molecules 19, 1672–1684. AlYahya, S.A., Ameen, F., Al-Niaeem, K.S., Al-Sa’adi, B.A., Hadi, S. and Mostafa, A.A. 2018. Histopathological studies of experimental Aeromonas hydrophila infection in blue tilapia, Oreochromis aureus. Saudi J. Biol. Sci. 25, 182–185. Bax, B.D., Chan, P.F., Eggleston, DS., Fosberry, A., Gentry, D.R., Gorrec, F., Giordano, I., Hann, M.M., Hennessy, A., Hibbs, M., Huang, J., Jones, E., Jones, J., Brown, K.K., Lewis, C.J., May, E.W., Saunders, M.R., Singh, O., Spitzfaden, C.E., Shen, C., Shillings, A., Theobald, A.J., Wohlkonig, A., Pearson, ND. and Gwynn, MN. 2010. Type IIA topoisomerase inhibition by a new class of antibacterial agents. Nature 466, 935–940. Benhabiles, M.S., Salah, R., Lounici, H., Drouiche, N., Goosen, M.F.A. and Mameri, N. 2012. Antibacterial activity of chitin, chitosan and its oligomers prepared from shrimp shell waste. Food Hydrocoll. 29, 48–56. Bessa, G.R., Quinto, V.P., Machado D.C., Lipnharski, C., Weber, M.B., Bonamigo, RR. and D'Azevedo, PA. 2016. Staphylococcus aureus resistance to topical antimicrobials in atopic dermatitis. An. Bras. Dermatol. 91, 604–610. Bhat, R.A., Thakuria, D., Pant, V., Khangembam, V.C., Tandel, R.S., Shahi, N., Sarma, D., Tripathi, G., Krishnani, K.K. and Krishna, G. 2020. Antibacterial and antioomycete activities of a novel designed RY12WY peptide against fish pathogens. Microb. Pathog. 149, 104591. Bienert, S., Waterhouse, A., De Beer, T.A.P., Tauriello, G., Studer, G., Bordoli, L. and Schwede, T. 2017. The SWISS-MODEL Repository-new features and functionality. Nucleic. Acids. Res. 45, D313–D319. Bitencourt-Ferreira, G. and De Azevedo, W.F.Jr. 2019. Molegro Virtual Docker for Docking. Methods in Molecular Biology (Clifton, N.J.). Methods. Mol. Biol. 2053, 149–167. Bouley, R., Kumarasiri, M., Peng, Z., Otero, L., Song, W., Suckow, M., Schroeder, V., Wolter, W., Lastochkin, E., Antunes, N., Pi, H., Vakulenko, S., Hermoso, J., Chang, M. and Mobashery, S. 2015. Discovery of antibiotic (E)-3-(3-CarboxyphenyI)-2-(4-cyanostyryl)quinazolin-4(3H)-one. J. Am. Chem. Soc. 137, 1738–1741. Caligiani, A., Marseglia, A., Leni, G., Baldassarre, S., Maistrello, L., Dossena, A. and Sforza, S. 2018. Composition of black soldier fly prepupae and systematic approaches for extraction and fractionation of proteins, lipids and chitin. Food Res. Int. 105, 812–820. Casillas-Vargas, G., Ocasio-Malavé, C., Medina, S., Morales-Guzmán, C., Del Valle, R.G., Carballeira, N.M. and Sanabria-Ríos, D.J. 2021. Antibacterial fatty acids: An update of possible mechanisms of action and implications in the development of the next-generation of antibacterial agents. Prog. Lipid Res. 82, 101093. Choi, W.H., Yun, J.H., Chu, J.P. and Chu, K.B. 2012. Antibacterial effect of extracts of Hermetia illucens (Diptera: Stratiomyidae) larvae against Gram-negative bacteria. Entomol. Res. 42, 219–226. CLSI. 2021. Performance standards for antimicrobial susceptibility testing. CLSI supplement M100. Informational supplement M100-S31. CLSI. CLSI. 2015. Methods for antimicrobial dilution and disk susceptibility testing of infrequently isolated or fastidious bacteria. CLSI. Cortes, E., Mora, J. and Márquez, E. 2020. Modelling the anti-methicillin-resistant Staphylococcus aureus (MRSA) activity of qsar, cannabinoids a study, docking. Crystals 10, 1–20. Desbois, A.P., Mearns-Spragg, A. and Smith, V.J. 2009. A fatty acid from the diatom Phaeodactylum tricornutum is antibacterial against diverse bacteria including multi-resistant Staphylococcus aureus (MRSA). Mar. Biotechnol. 11, 45–52. Dhanapala, P.M., Kalupahana, R.S., Kalupahana, A.W., Wijesekera, D.P.H., Kottawatta, S.A., Jayasekera, N.K., Silva-Fletcher, A. and Jagoda, S.S.S.S. 2021. Characterization and antimicrobial resistance of environmental and clinical Aeromonas species isolated from fresh water ornamental fish and associated farming environment in Sri Lanka. Microorg. 9, (10), 2106 Filimonov, D.A., Lagunin, A.A., Gloriozova, T.A., Rudik, A.V., Druzhilovskii, D.S., Pogodin, P.V. and Poroikov, V.V. 2014. Prediction of the biological activity spectra of organic compounds using the pass online web resource. Chem. Heterocycl. Compd. 50, 444–457. Foster, T.J. 2017. Antibiotic resistance in Staphylococcus aureus. Current status and future prospects. FEMS Microbiol. Rev. 41, 430–449. Hetem, D.J. and Bonten, M.J.M. 2013. Clinical relevance of mupirocin resistance in Staphylococcus aureus. J. Hosp. Infect. 85, 249–256. Irawan, A.C., Astuti, D.A., Wibawan, I.W.T., Hermana, W. and dan Makanan, D.T. 2020. Supplementation of black soldier fly (Hermetia illucens) on productivity and blood hematology. J. Ilmu-Ilmu Peternak. 30, 50–68. Jafari, E., Khajouei, M.R., Hassanzadeh, F., Hakimelahi, G.H. and Khodarahmi, G.A. 2016. Quinazolinone and quinazoline derivatives: Recent structures with potent antimicrobial and cytotoxic activities. Res. Pharm. Sci. 11, 1–14. Javid, F., Taku, A., Bhat, M., Badroo, G., Mudasir, M. and Sofi, T. 2018. Molecular typing of Staphylococcus aureus based on coagulase gene. Vet. World. 11, 423–430. Jia, B., Ma, Y., Liu, B., Chen, P., Hu, Y. and Zhang, R. 2019. Synthesis, antimicrobial activity, structure-activity relationship, and molecular docking studies of indole diketopiperazine alkaloids. Front. Chem. 7, 1–13. Jumina, J., Lavendi, W., Singgih, T., Triono, S., Steven, Y. and Koketsu, M. 2019. Preparation of Monoacylglycerol Derivatives from Indonesian Edible Oil and Their Antimicrobial Assay against Staphylococcus aureus and Escherichia coli. Sci. Rep. 9, 1–8. Kementrian kelautan dan perikanan (KKP) RI. 2021. Keputusan menteri kelautan dan perikanan republik Indonesia Nomor 17 tahun 2021 tentang penetapan jenis penyakit ikan karantina, organisme penyakit, golongan, dan media pembawa. Kusnadi, Purgiyanti, Kumoro, A. and Legowo, A.M. 2022. The antioxidant and antibacterial activities of chitosan extract from white shrimp shell (Penaeus indicus) in the waters north of Brebes, Indonesia. Biodiversitas 23, 1267–1272. Li, Z., Mao, R., Teng, D., Hao, Y., Chen, H., Wang, Xiumin, Wang, Xiao, Yang, N. and Wang, J. 2017. Antibacterial and immunomodulatory activities of insect defensins-DLP2 and DLP4 against multidrug-resistant Staphylococcus aureus. Sci. Rep. 7, 1–17. Lin, Y., Rong, J., Wei, X., Sui, Z., Xiao, J. and Huang, D. 2021. Proteomics and ultrastructural analysis of Hermetia illucens (Diptera: Stratiomyidae) larval peritrophic matrix. Proteome Sci. 19, 1–14. Macindoe, G., Mavridis, L., Venkatraman, V., Devignes, M.D. and Ritchie, D.W. 2010. HexServer: an FFT-based protein docking server powered by graphics processors. Nucleic Acids Res. 38, 445–449. Mazurek, B., Chmiel, M. and Górecka, B. 2017. Fatty acids analysis using gas chromatography-mass spectrometer detector (GC/MSD)—method validation based on berry seed extract samples. Food Anal Methods, 10(8),2868–2880. Nadăs, G., Novac, C., Matei, I., Bouari, C., Gal, Z., Tamas-Krumpe, O., Macri, A. and Fit, N. 2021. Prevalence of antimicrobial resistant bacteria from conjunctival flora in an eye infection prone breed (Saint bernard). Molecules, 26(8), 2219. Nagendra Prasad, H.S., Karthik, C.S., Manukumar, H.M., Mallesha, L. and Mallu, P. 2019. New approach to address antibiotic resistance: miss loading of functional membrane microdomains (FMM) of methicillin-resistant Staphylococcus aureus (MRSA). Microb. Pathog. 127, 106–115. Nardiello, M., Scieuzo, C., Salvia, R., Farina, D., Franco, A., Cammack, J., Tomberlin, J., Falabella, P. and Persaud, K. 2022. Odorant binding proteins from Hermetia illucens: potential sensing elements for detecting volatile aldehydes involved in early stages of organic decomposition. Nanotechnology 33, 205501. Nemeth, J., Oesch, G. and Kuster, S.P. 2015. Bacteriostatic versus bactericidal antibiotics for patients with serious bacterial infections: systematic review and meta-analysis. J. Antimicrob. Chemother. 70, 382–395. Nepal, A., Ræder, S., Søgaard, C., Haugan, M. and Otterlei, M. 2021. Broad-spectrum antibacterial peptide kills extracellular and intracellular bacteria without affecting epithelialization. Front. Microbiol. 12, 1–12. Poux, S., Arighi, C.N., Magrane, M., Bateman, A., Wei, C., Lu, Z., Boutet, E., Bye-A-Jee, H., Famiglietti, M., Roechert, B. and Consortium, U. 2017. On expert curation and scalability: UniProtKB/Swiss-Prot as a case study. Bioinformatics 33, 3454–3460. Qureshi, S.I. and Chaudhari, H.K. 2019. Design, synthesis, in-silico studies and biological screening of quinazolinone analogues as potential antibacterial agents against MRSA. Bioorg. Med. Chem. 27, 2676–2688. Ritchie, D.W., Kozakov, D. and Vajda, S. 2008. Accelerating and focusing protein-protein docking correlations using multi-dimensional rotational FFT generating functions. Bioinformatics 24, 1865–1873. Shaaban, M.T., Ghaly, M.F. and Fahmi, S.M. 2021. Antibacterial activities of hexadecanoic acid methyl ester and green-synthesized silver nanoparticles against multidrug-resistant bacteria. J. Basic Microbiol. 61, 557–568. Tadesse, S., Alemayehu, H., Tenna, A., Tadesse, G., Tessema, T., Shibeshi, W. and Eguale, T. 2018. Antimicrobial resistance profile of Staphylococcus aureus isolated from patients with infection at tikur anbessa Specialized Hospital, addis ababa, Ethiopia. BMC Pharmacol. Toxicol. 19, 1–8. Tan, Y., Chin, Y. and Chen, W. 2021. Comparison of sustainable lipid and protein removal methods for the isolation of insect chitin from black soldier fly exoskeleton. ACS Food. Sci. Technol. 1, 698–706. Xia, J., Ge, C. and Yao, H. 2021. Antimicrobial peptides from black soldier fly (Hermetia illucens) as potential antimicrobial factors representing an alternative to antibiotics in livestock farming. Animals 11, (7):1937. Zarrinmehr, M., Daneshvar, E., Nigam, S., Gopinath, K., Biswas, J., Kwon, E., Wang, H., Farhadian, O. and Bhatnagar, A. 2022. The effect of solvents polarity and extraction conditions on the microalgal lipids yield, fatty acids profile, and biodiesel properties. Bioresour. Technol. 344. | ||
| How to Cite this Article |
| Pubmed Style Qosimah D, Santosa S, Maftuch M, Khotimah H, LEF, AA, Suwanti LT. Methanol extract of Black soldier fly (Hermetia illucens) prepupae against Aeromonas and Staphylococcus aureus bacteria in vitro and in silico. Open Vet J. 2023; 13(1): 48-63. doi:10.5455/OVJ.2023.v13.i1.6 Web Style Qosimah D, Santosa S, Maftuch M, Khotimah H, LEF, AA, Suwanti LT. Methanol extract of Black soldier fly (Hermetia illucens) prepupae against Aeromonas and Staphylococcus aureus bacteria in vitro and in silico. https://www.openveterinaryjournal.com/?mno=111732 [Access: July 27, 2024]. doi:10.5455/OVJ.2023.v13.i1.6 AMA (American Medical Association) Style Qosimah D, Santosa S, Maftuch M, Khotimah H, LEF, AA, Suwanti LT. Methanol extract of Black soldier fly (Hermetia illucens) prepupae against Aeromonas and Staphylococcus aureus bacteria in vitro and in silico. Open Vet J. 2023; 13(1): 48-63. doi:10.5455/OVJ.2023.v13.i1.6 Vancouver/ICMJE Style Qosimah D, Santosa S, Maftuch M, Khotimah H, LEF, AA, Suwanti LT. Methanol extract of Black soldier fly (Hermetia illucens) prepupae against Aeromonas and Staphylococcus aureus bacteria in vitro and in silico. Open Vet J. (2023), [cited July 27, 2024]; 13(1): 48-63. doi:10.5455/OVJ.2023.v13.i1.6 Harvard Style Qosimah, D., Santosa, . S., Maftuch, . M., Khotimah, . H., , . L. E. F., , . A. A. & Suwanti, . L. T. (2023) Methanol extract of Black soldier fly (Hermetia illucens) prepupae against Aeromonas and Staphylococcus aureus bacteria in vitro and in silico. Open Vet J, 13 (1), 48-63. doi:10.5455/OVJ.2023.v13.i1.6 Turabian Style Qosimah, Dahliatul, Sanarto Santosa, Maftuch Maftuch, Husnul Khotimah, Loeki Enggar Fitri, Aulanni'am Aulanni'am, and Lucia Tri Suwanti. 2023. Methanol extract of Black soldier fly (Hermetia illucens) prepupae against Aeromonas and Staphylococcus aureus bacteria in vitro and in silico. Open Veterinary Journal, 13 (1), 48-63. doi:10.5455/OVJ.2023.v13.i1.6 Chicago Style Qosimah, Dahliatul, Sanarto Santosa, Maftuch Maftuch, Husnul Khotimah, Loeki Enggar Fitri, Aulanni'am Aulanni'am, and Lucia Tri Suwanti. "Methanol extract of Black soldier fly (Hermetia illucens) prepupae against Aeromonas and Staphylococcus aureus bacteria in vitro and in silico." Open Veterinary Journal 13 (2023), 48-63. doi:10.5455/OVJ.2023.v13.i1.6 MLA (The Modern Language Association) Style Qosimah, Dahliatul, Sanarto Santosa, Maftuch Maftuch, Husnul Khotimah, Loeki Enggar Fitri, Aulanni'am Aulanni'am, and Lucia Tri Suwanti. "Methanol extract of Black soldier fly (Hermetia illucens) prepupae against Aeromonas and Staphylococcus aureus bacteria in vitro and in silico." Open Veterinary Journal 13.1 (2023), 48-63. Print. doi:10.5455/OVJ.2023.v13.i1.6 APA (American Psychological Association) Style Qosimah, D., Santosa, . S., Maftuch, . M., Khotimah, . H., , . L. E. F., , . A. A. & Suwanti, . L. T. (2023) Methanol extract of Black soldier fly (Hermetia illucens) prepupae against Aeromonas and Staphylococcus aureus bacteria in vitro and in silico. Open Veterinary Journal, 13 (1), 48-63. doi:10.5455/OVJ.2023.v13.i1.6 |