| Original Article | ||
Open Vet J. 2021; 11(4): 771-779 Open Veterinary Journal, (2021), Vol. 11(4): 771–779 Original Research Study of the effects of Escherichia coli lipopolysaccharide on innate immunity: The expression profile of TLR4 and CD14 genes in rat liverAmmar M. Al-Aalim1*, Ali A. Al-iedani2 and Mohammad A. Hamad11Department of Microbiology, College of Veterinary Medicine, University of Mosul, Mosul, Iraq 2Department of Microbiology, College of Veterinary Medicine University of Basrah, Basrah, Iraq *Corresponding Author: Ammar M. Al-Aalim. Department of Microbiology, College of Veterinary Medicine, University of Mosul, Mosul, Iraq. Email: ammarmahmmod [at] uomosul.edu.iq Submitted: 11/09/2021 Accepted: 01/12/2021 Published: 26/12/2021 © 2021 Open Veterinary Journal
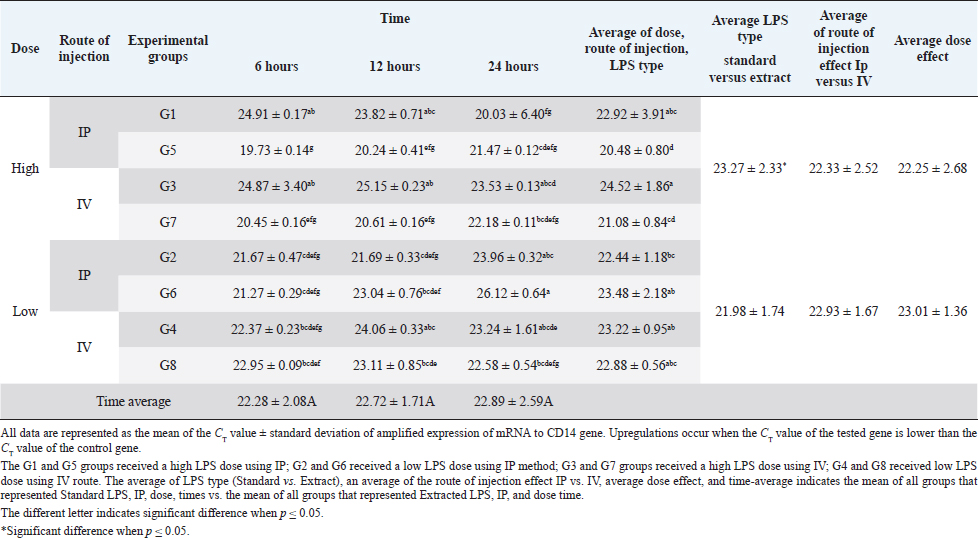
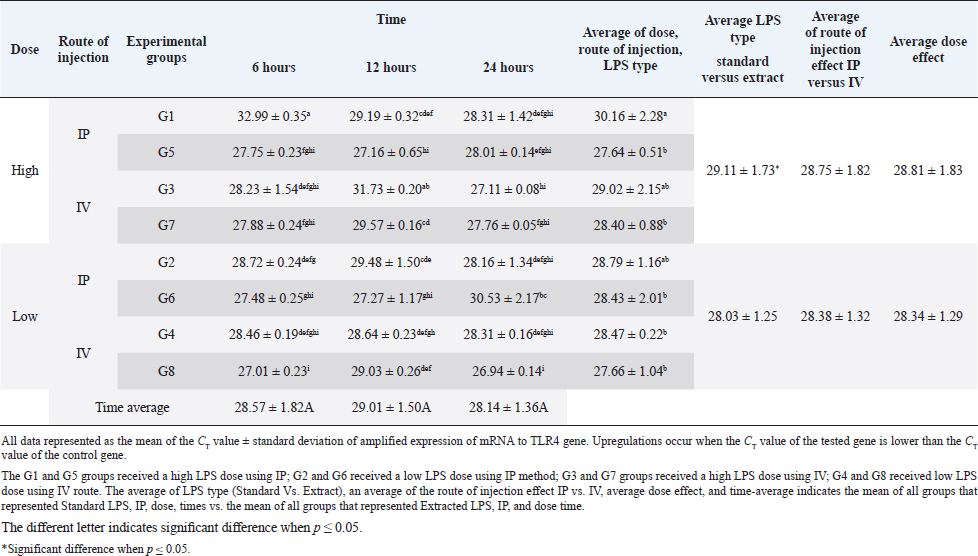
AbstractBackground: Bacterial endotoxin [lipopolysaccharide (LPS)] is essential for bacterial virulence as it has a biphasic effect which is either harmful and leads to aseptic shock and death or assists the body defense mechanisms as it stimulates B-cells activation. Many studies have noted that LPS do their action through activation of CD14/ TLR4 pathways, which occur mainly in liver cells, including Kupffer cells, hepatocytes, and liver sinusoidal endothelial cells, which are responsible for cytokines releases and shows the good or bad LPS effect. Aim: The current study aimed to disclose the expression changes in the profile of innate immunological receptors TLR4 and CD14 in rats’ livers after stimulation with LPS. Methods: Ten groups of male Wistar albino rats were used to study the effects of two types of LPS [extracted LPS from the local strain of Escherichia coli (ELPS) and standard E. coli (SLPS)]; these were given by using different doses (5 mg/kg and 100 μg/kg); the LPS were injected either intravenously or intraperitoneally. The TLR4 and CD14 mRNA expression patterns were estimated using qPCR after 6, 12, and 24 hours postinjection. Results: The results show that there is a negative effect of ELPS on liver CD14 and TLR4, regardless of the dose and route of administration. On the other hand, the SLPS has an upregulatory impact on the liver gene expression. Also, different times show no effect on the gene expression of the two genes. Conclusion: This study concludes that both LPS types used were able to stimulate the CD14 and TLR4 gene expression in the liver in different doses and routes of injection. Also, this study showed the possibility of using ELPS as an immunomodulator in rats. Keywords: Escherichia coli LPS, Rat liver, Innate immunity, TLR4, CD14. IntroductionThe lipopolysaccharide (LPS) chemical structure is conserved in all Gram-negative bacteria (Ebbensgaard et al., 2018), which consist mainly of three-part lipid A molecules, oligosaccharide core, and the O-antigen (Erridge et al., 2002; Mazgaeen and Gurung, 2020). The LPS associates with bacterial virulence, resists the phagocytic effect, plays a role in antigenic variation (Sampath, 2018), and also plays a role in bacterial resistance to complement action and bacteriophage (Putker et al., 2015; Bertani and Ruiz, 2018), as well as antimicrobial substances (May and Grabowicz, 2018), especially cationic antimicrobial peptides (Ebbensgaard et al., 2018). Finally, LPS is capable of inhibiting the immune host response (Bertani and Ruiz, 2018). The researchers noted that the exact effect of LPS is dose-dependent. In high doses, the LPSs have a wide range of bad effects such as coagulation (Stief, 2009), inflammation, capillary leak, tissue toxicity aseptic shock, and lethality (Sampath, 2018) in low doses. LPSs act as a pathogen-associated molecular patterns (PAMP), which is a potent stimulator of innate and cellular immunity through the activation of TLR4/CD14/LPS complex pathway (Erridge et al., 2002; Steimle et al., 2016; Tsukamoto et al., 2018; Mazgaeen and Gurung, 2020). The activation of TLR4 pathways leads to the activation to more than 1,000 gene transcriptions (Steimle et al., 2016). Especially, interleukin 6 (IL6), tumor necrosis factor α (TNFα), and IL1 and IL12 during an inflammatory cytokines storm are responsible for septic shock (Mazgaeen and Gurung, 2020), but they also have some beneficial effects such as anti-tumor (Gonçalves et al., 2016), radioprotection, activation of homoeotic (Schuettpelz and Link. 2013), activation of complement and B-cells (Kaca et al., 2009), and, finally, as an adjuvant to assist the immunity system’s defense (Kuznetsova et al., 2020). The immune system’s responses against LPS occur mainly in the liver, which regulates the cytokines released from Kupffer cells (liver resides microphage), hepatocytes, and liver sinusoidal endothelial cells (antigen-presenting cells resemble) after recognition and modulates of LPS through TLR4 and CD14 (Jirillo et al., 2002). This study aimed to disclose the expression changes in the profile of innate immunological receptors TLR4 and CD14 in rats’ livers after stimulation with LPS. Materials and MethodsOne hundred and twenty male Wistar albino rats, with weights ranging from 220 to 280 g, were divided into 10 groups, each containing 12 rats randomly (total n=120). The animals were injected with SLPS or ELPS using two different doses of LPS 5 mg/kg and 100 μg/kg. Animals in the groups G1–G4 were used for ELPS. However, the groups G5–G8 were used for SLPS, and the last two groups G9 and G10 were used as control. G1 and G5 groups received a high LPS dose (5 mg/kg) using the intraperitoneal route (IP), G2 and G6 received a low LPS dose (100 μg/kg) using IP; G3 and G7 groups received a high LPS dose using the intravenous route (IV); G4 and G8 received a low LPS dose by IV. Finally, G9 and G10 served as a control and received distal water. Rats were euthanized humanely by cervical dislocation at 6, 12, and 24 hours, respectively; the livers were collected from different animal groups. All liver samples were stored at −80°C in the deep freezer until used for detection of TLR4 and CD14 gene expressions using two steps qPCR using Promega system/USA. Isolation of RNA was carried out by using extraction and purification SV total RNA isolation system (Promega/USA). The GoScript™ Reverse Transcription System (Promega/USA) with Oligo(dT) 15 primer was used to synthesize the first strand of cDNA. Five microliters of cDNA were mixed with 20 μl reaction mixture, GoTag ® qPCR master (Promega/USA) 10 μl, forward primer 2.5 μl, reverse primer 2.5 μl, and nuclease-free water 5 μl, for detecting all gene expressions (Table 1). The qPCR system (PCRmax/UK) was used to amplify all genes, the reaction conditions were initial denaturation at 95°C, 2 minutes, 40 cycles with the second denaturation at 95°C, 15 seconds, finally, annealing and extension at 60°C, 1 minute. The gene expression was normalized with the GAPDH housekeeping gene, and then the fold changes were calculated using ΔΔCT (Livak and Schmittgen, 2001). Finally, all qPCR products were migrated in 1.5% agarose gel electrophoresis and visualized using a transilluminator and photographic using a digital camera. The result data were statistically analyzed using IBM SPSS program statistics version 24. All data were tested using the analysis of variance, Duncan’s test , and t-test, which recorded significant differences in experimental parameters. Ethical approvalAll animals were treated with the ethical rule of animal care and sample collection (University of Melbourne Animal Care and Use Standards Committee, 2019). The study design and the animal experiments were approved by the Mosul University/Local College of veterinary medicine/Institutional Animal Care and Use Committee under approved ID: UN.VET.2021.008. ResultsThe CD14 mRNA gene expression showed an upregulation after the animals were injected with either LPS types, which differed between groups compared to the control group. The existing study results revealed an increase in gene expression in G5 with a rise in fold change variation from other groups (Fig. 1). Statistical analysis showed a similar expression of CD14 mRNA, which was observed at different experiment times. The effects of ELPS on CD14 gene expression were low regardless of the dose and route of administration compared with SLPS; however, the differences regarding doses and routes of injection were not significant statistically (Table 2). The TLR4 mRNA gene expression showed upregulation after the animals were injected with either LPS type, which differed between groups compared to the control group. The current study (Fig. 2) explains the fold changes of TLR4 mRNA gene expression Overall, the injection route and type of LPS show the reduction of TLR4 gene expression in G1 rats compared to other groups, excluding G2 and G3 animals. The statistical analysis of TLR4 mRNA exhibits a similar expression at different experimental times. SLPS demonstrates more elevation on the regulatory effect of the TLR4 gene on the rats’ model than ELPS, which exhibits a lower effect on the same model. At the same time, different routes and doses appear to have little impact on TLR4 mRNA expression (Table 3). Table 1. The specific primers used for gene expression.
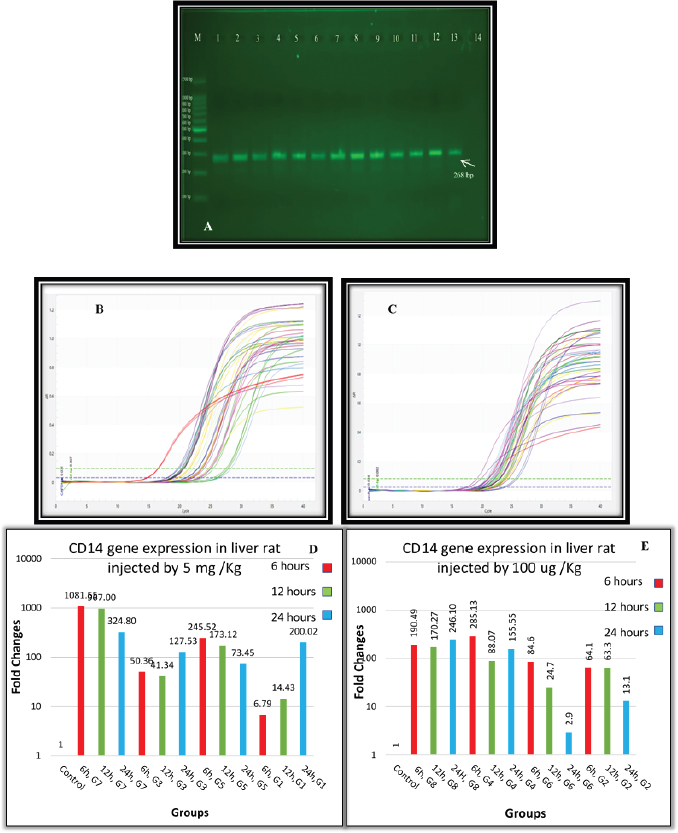
Fig. 1. (A) Gel electrophoresis of CD14 (M: kilobase marker; well 1, 2, 3: G7 at 6, 12, 24 hours; well 4, 5, 6: G3 at 6, 12, 24 hours; well 7, 8, 9: G5 at 6, 12, 24 hours; well 10, 11, 12: G1 at 6, 12, 24 hours; 13: control positive; and 14: control negative). (B) Amplification curve of CD14 using qPCR (5 mg/kg). (C) Amplification curve of CD14 using qPCR (100 μg/kg). (D) Gene expression of CD14 of rat groups that received high doses (5 mg/kg) and different routes of injections. (E) Gene expression of CD14 of rat groups that received high doses (100 μg/kg) and different routes of injections. Table 2. Analysis of CD14 gene expression induced by ELPS and SLPS.
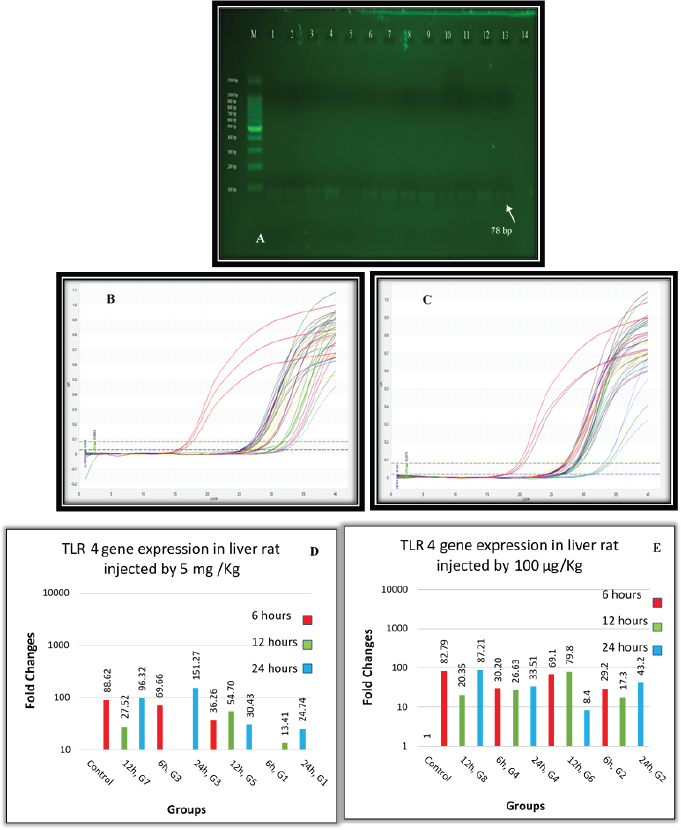
Fig. 2. (A) Gel electrophoresis of TLR4 (M: kilobase marker; well 1, 2, 3: G7 at 6, 12, 24 hours; well 4, 5, 6: G3 at 6, 12, 24 hours; 7, 8, 9: G5 at 6, 12, 24 hours; well 10, 11, 12: G1 at 6, 12, 24 hours; 13: control positive; and 14: control negative). (B) Amplification curve of TLR4 using qPCR (5 mg/kg). (C) Amplification curve of TLR4 using qPCR (100 μg/kg). (D) Gene expression of TLR4 of rat groups that received high doses (5 mg/kg) and different routes of injections. (E) Gene expression of TLR4 of rat groups that received high doses (100 μg/kg) and different routes of injections. Table 3. Analysis of TLR4 gene expression induced by ELPS and SLPS.
DiscussionThe innate immune responses are responsible for rapid detection, removal of foreign materials, and induction of inflammatory response by recognition of PAMP found in many bacterial infections. LPSs are capable of inducing innate immune inflammatory response associated with inflammatory cytokines (Mazgaeen and Gurung, 2020). So, the study of this cytokines gives a good indication about the health and ability of innate immunity to invading microbes. The most important receptors and cytokines are CD14, TLR4, IL1, and TNFα, which increase during LPS recognition and immune responses against it. The liver plays a pivotal role in the responses of the immune system against LPS, Kupffer cells (liver resides microphage), hepatocytes, and liver sinusoidal endothelial cells (antigen-presenting cells resemble) and can recognize and modulate the response to LPS through TLR4, CD14, and regulation of cytokines releases (Jirillo et al., 2002). The studies of the gene expression of CD14 with LPS showed an upregulation of the CD14 gene in all liver cells to reach a peak at 3–6 hours after using different doses with ELPS or SLPS injected by different methods; this result agrees with Li et al. (2003), who recorded an increase in hepatic CD14 protein level at 3 hours to reach a peak at 12 hours. After 24 hours of LPS administration, the expression of CD14 in the liver was decreased in some groups. This is due to gradually downregulating CD14 mRNA to baseline level in liver cells after 24 hours. The animals in the G3 and G1 groups that were injected with high doses of LPS intraperitoneally revealed a decrease in CD14 mRNA gene expression at 6 hours and a reincrease at 24 hours; this may be due to the delay in response of liver cells in those suffering from intensive injury with low CD14 expression, and this result agrees with Hozumi et al. (2013). In this research, no significant difference was noted between the overall time used, and this may be related partially to the time of samples collection. The collection of samples started after 6 hours of LPS injection. This may have led to missing the early expression which normally occurs within minutes or hours. The ELPS causes less elevation in CD14 mRNA gene expression level contrary to SLPS. This may result from differences in the chemical structure which reflected different ligands that reacted differently with the binding site of CD14 (Cunningham et al., 2000). Both routes of injection show a similar effect because the LPS concentration in the liver raised gradually after LPS injection, especially the smooth form of LPS (Jacque et al., 2006). Furthermore, a similar effect was seen in both high and low doses on CD14 mRNA expression, and this may result partially from the presence of sCD14 in plasma, which expresses as acute phase protein and may compete with mCD14 to bind and neutralize the LPS-induced response in vitro or in vivo (Bas et al., 2004). The assay of TLR4 mRNA expression shows biphasic upregulation after 6 and 24 hours in most rat groups. A similar pattern was reported by Huang et al. (2017), who noted that the mRNA of TLR4 expression was “increased, decreased, then increased again” the significant increases occurred at 6 and 12 hours in chicken liver stimulated with Salmonella LPS. The rapid increase in mRNA of TLR4 in the early phase reveals the early immune stimulation against the LPS. Under normal conditions liver, Kupffer cells process a low baseline of TLR4 mRNA, and under the LPS stimulation, these cells respond by upregulation of TLR4 mRNA. Hepatocytes also produce remarkable TLR4 mRNA and protein under the same condition (Huang et al., 2017). Increase in mRNA of TLR4 occurs to overcome the LPS effect after activation of TLR4 downstream signaling to produce pro-inflammatory cytokine (IL1, TNFα, and IFNδ) through NF-kB pathways (Mazgaeen and Gurung, 2020; Ciesielska et al., 2021). The second phase of increase in TLR4 mRNA expression occurs after 24 hours is interesting and result from LPS acute injury to liver cells results in TLR4 response through NLRP3 (Gong et al., 2019) or by damage-associated molecular pattern to initiate tissue repair (Ciesielska et al., 2021). The ELPS produces lower induction on TLR4 mRNA gene expression in contrast to SLPS; this reflects different LPS structures, which affect the binding site of CD14 (Cunningham et al., 2000), and expand the effect on TLR4 mRNA expression; meanwhile, different effects on TLR4 mRNA expression by different Escherichia coli LPS strains and/or organs investigated were found (Grasa et al., 2017). The time of TLR4 expression shows no difference in TLR4 mRNA expression pattern. Studies have showed that the expression time pattern was different, depending on tissue affected, for example, the heart increased TLR4 mRNA expression at 3 hours, followed by a decrease at 6 hours and increased at 24 hours which reflects tissue macrophage/monocyte effect on TLR4 mRNA expression (Matsumura et al., 2000). TLR4 expression showed a similar effect on both routes or doses used, and this may relate to the effect of both soluble and membrane-bound CD14 molecules, which mediated delivery and internalization of TLR in both low and high doses and stimulate downstream signaling for production of TNFα (Schwabe et al., 2006; Ciesielska et al., 2021). Liu et al. (2018) showed a positive correlation between TLR4 mRNA expression and TNFα and IL12 concentration. This led us to propose that CD14 may partially regulate TLR4 expression through TNFα production by its LPS delivery to TLR4 receptors. ConclusionThe existing study concluded that both LPS types used were able to stimulate the CD14 and TLR4 gene expression in the liver in low and high doses with different injection routes. The statistical analysis reveals that ELPS produces less elevations of CD14 and TLR4 gene expression levels in contrast to SLPS. Furthermore, no effect of doses or time was noted on the expression of the two genes, so this study shows the possibility of using E. coli (local strain) as an immunomodulator in rats. AcknowledgmentsThe authors are grateful for the support provided by the head of the Department of Microbiology and the deanship of the College of Veterinary Medicine, Mosul University, to accomplish this study. Conflict of interestThe authors declare that there is no conflict of interest. Authors’ contributionsAll authors have designed, written, reviewed, discussed, agreed, and contributed to this article before and during the submission of their article. ReferencesBas, S., Gauthier, B.R., Spenato, U., Stingelin, S. and Gabay, C. 2004. CD14 is an acute-phase protein. J. Immunol. 172, 4470–4479. Bertani, B. and Ruiz, N. 2018. Function and biogenesis of lipopolysaccharides. EcoSal. Plus. 8, 1–33. Ciesielska, A., Matyjek, M. and Kwiatkowska, K. 2021. TLR4 and CD14 trafficking and its influence on LPS-induced pro-inflammatory signaling. Cell. Mol. Life Sci. 78, 1233–1261. Cunningham, M.D., Shapiro, R.A., Seachord, C., Ratcliffe, K., Cassiano, L. and Darveau, R.P. 2000. CD14 employs hydrophilic regions to “capture” lipopolysaccharides. J. Immunol. 164, 3255–3263. Ebbensgaard, A., Mordhorst, H., Aarestrup, F.M. and Hansen, E.B. 2018. The role of outer membrane proteins and lipopolysaccharides for the sensitivity of Escherichia coli to antimicrobial peptides. Front. Microbiol. 9,1–13. Erridge, C., Bennett-Guerrero, E. and Poxton, I.R. 2002. Structure and function of lipopolysaccharides. Microb. Infect. 4, 837–851. Gonçalves, M., Cappellari, Á.R., Santos, A.A.D., Marchi, F.O.D., Macchi, F.S., Antunes, K.H., de Souza A.P.D. and Morrone, F.B. 2016. Effect of LPS on the viability and proliferation of human oral and esophageal cancer cell lines. Bra. Arch. Biol. Techol. 59, 1–10. Gong, Q., He, L., Wang, M., Zuo, S., Gao, H., Feng, Y. and Li, J. 2019. Comparison of the TLR4/NFκB and NLRP3 signalling pathways in major organs of the mouse after intravenous injection of lipopolysaccharide. Pharma. Biol. 57, 555–563. Grasa, L., Gonzalo, S., de Martino, A. and Murillo, M.D. 2017. The lipopolysaccharide from Escherichia coli O127: B8 induces inflammation and motility disturbances in rabbit ileum. World Rabb. Sci. 25, 185–191. Hozumi, H., Tada, R., Murakami, T., Adachi, Y. and Ohno, N. 2013. Comparative analysis of hepatic CD14 expression between two different endotoxin shock model mice: relation between hepatic injury and CD14 expression. PloS. One. 8, 1–10. Huang, X.Y., Ansari, A.R., Huang, H.B., Zhao, X., Li, N.Y., Sun, Z.J., Peng K., Zhong J. and Liu, H.Z. 2017. Lipopolysaccharide mediates immuno-pathological alterations in young chicken liver through TLR4 signaling. BMC. Immune. 18, 1–9. Jacque, B., Stephan, K., Smirnova, I., Kim, B., Gilling, D. and Poltorak, A. 2006. Mice expressing high levels of soluble CD14 retain LPS in the circulation and are resistant to LPS-induced lethality. Europ. J. Immunol.36, 3007–3016. Jirillo, E., Caccavo, D., Magrone, T., Piccigallo, E., Amati, L., Lembo, A., Kalis, C. and Gumenscheimer, M. 2002. The role of the liver in the response to LPS: experimental and clinical findings. J. Endot. Res. 8, 319–327. Kaca, W., Arabski, M., Fudała, R., Holmström, E., Sjöholm, A., Weintraub, A., Futoma−Kołoch, B., Bugla−Płoskońska, G. and Doroszkiewicz, W. 2009. Human complement activation by smooth and rough Proteus mirabilis lipopolysaccharides. Archi. Immunol. Therap. Exp. 57, 383–391. Kocak, H., Tokcaer-Keskin, Z., Insal, B., Gursel, I. and Akcali, K.C. 2019. Administration of bone marrow derived mesenchymal stem cells modulate TLR expression during Liver regeneration. Trakya Uni. J. Natu. Sci. 20, 1–10. Kuznetsova, V.S., Ivaschenko, S.V. and Domnitsky, I.Y. 2020. Polyazolidinammonium as an adjuvant in immunization with lipopolysaccharide of Yersinia pseudotuberculosis. In IOP Earth and Environmental Science Conference Series, pp: 1–9. Li, X.H., Gong, J.P., Shi, Y.J., Liu, C.A. and Peng, Y. 2003. In vitro expression of CD14 protein and its gene in Kupffer cells induced by lipopolysaccharide. Hepatobiliary Pancreat. Dis. Int. 2, 571–575. Liu, J.Y., Liu, H.H., Kou, J., Zhao, Y.M., Wang, J.W., Li, L. and Du, X.H. 2018. The influence of intravenous lipopolysaccharide injection on TLR4 transcription levels in duck (Anas domestica) liver, spleen and bursa of fabricius. Pak. J. Zoo. 50, 1237–1244. Livak, K.J. and Schmittgen, T.D. 2001. Analysis of relative gene expression data using real-time quantitative PCR and the 2-ΔΔCT method. Meth. 25, 402–408. Matsumura, T., Ito, A., Takii, T., Hayashi, H. and Onozaki, K. 2000. Endotoxin and cytokine regulation of toll-like receptor (TLR) 2 and TLR4 gene expression in murine liver and hepatocytes. J. Interfer. Cytok. Res. 20, 915–921. Mazgaeen, L. and Gurung, P. 2020. Recent advances in lipopolysaccharide recognition systems. Int. J. Mol. Sci. 21, 379–397. May, K.L. and Grabowicz, M. 2018. The bacterial outer membrane is an evolving antibiotic barrier. Proc. Natl. Acad. Sci. U. S. A. 115(36), 8852–8854. Pal, S., Nath, P., Biswas, S., Mukherjee, U. and Maitra S. 2019. Nonylphenol attenuates SOCS3 expression and M1 polarization in lipopolysaccharide-treated rat splenic macrophages. Ecotoxico. Enviro. Saf. 174, 574–583. Putker, F., Bos, M.P. and Tommassen, J. 2015. Transport of lipopolysaccharide to the Gram-negative bacterial cell surface. FEMS. Microb. Rev. 39, 985–1002. Sampath, V. 2018. Bacterial endotoxin-lipopolysaccharide; structure, function and its role in immunity in vertebrates and invertebrates. Agricul. Natu. Res. 52, 115–120. Schuettpelz, L. and Link, D. 2013. Regulation of hematopoietic stem cell activity by inflammation. Front. Immunol. 4, 1–9. Schwabe, R.F., Seki, E. and Brenner, D.A. 2006. Toll-like receptor signaling in the liver. Gastroent. 130, 1886–1900. Steimle, A., Autenrieth, I.B. and Frick, J.S. 2016. Structure and function: lipid a modifications in commensals and pathogens. Inter. J. Med. Microbiol. 306, 290–301. Stief, T.W. 2009. Coagulation activation by lipopolysaccharides. Clin. Appl. Thromb. Hemost. 15, 209–219. Tsukamoto, H., Takeuchi, S., Kubota, K., Kobayashi, Y., Kozakai S., Ukai, I., Shichiku, A., Okubo, M., Numasaki, M., Kanemitsu, Y., Matsumoto, Y., Nochi, T., Watanabe, K., Aso, H. and Tomioka, Y. 2018. Lipopolysaccharide (LPS)-binding protein stimulates CD14-dependent toll-like receptor 4 internalization and LPS-induced TBK1–IKK_–IRF3 axis activation. J. Biol. Chem. 293, 10186–10201. University of Melbourne Animal Care and Use Standards Committee. 2019. Human killing of mice and rats. Office for research ethics & integrity, animal care & use standard. Melbourne University, Melbourne, Australia. Zhishang C., Ying L., Ai, S., Wencheng W., Hongwei X., Yushan J. and Hui, L. 2020. Protective effect and underlying mechanism of aplysin on ethanol-induced liver injury in Rats. Nut. Hyg. Food Sci. 41, 131–139. | ||
| How to Cite this Article |
| Pubmed Style AL-Aalim AM, Al-iedani AA, Hamad MA. Study The Effects Of Systemic Injection Of Escherichia coli Lipopolysaccharide On The Expression Profile Of TLR4 And CD14 Genes Of Rat's Liver. Open Vet J. 2021; 11(4): 771-779. doi:10.5455/OVJ.2021.v11.i4.30 Web Style AL-Aalim AM, Al-iedani AA, Hamad MA. Study The Effects Of Systemic Injection Of Escherichia coli Lipopolysaccharide On The Expression Profile Of TLR4 And CD14 Genes Of Rat's Liver. https://www.openveterinaryjournal.com/?mno=123489 [Access: July 27, 2024]. doi:10.5455/OVJ.2021.v11.i4.30 AMA (American Medical Association) Style AL-Aalim AM, Al-iedani AA, Hamad MA. Study The Effects Of Systemic Injection Of Escherichia coli Lipopolysaccharide On The Expression Profile Of TLR4 And CD14 Genes Of Rat's Liver. Open Vet J. 2021; 11(4): 771-779. doi:10.5455/OVJ.2021.v11.i4.30 Vancouver/ICMJE Style AL-Aalim AM, Al-iedani AA, Hamad MA. Study The Effects Of Systemic Injection Of Escherichia coli Lipopolysaccharide On The Expression Profile Of TLR4 And CD14 Genes Of Rat's Liver. Open Vet J. (2021), [cited July 27, 2024]; 11(4): 771-779. doi:10.5455/OVJ.2021.v11.i4.30 Harvard Style AL-Aalim, A. M., Al-iedani, . A. A. & Hamad, . M. A. (2021) Study The Effects Of Systemic Injection Of Escherichia coli Lipopolysaccharide On The Expression Profile Of TLR4 And CD14 Genes Of Rat's Liver. Open Vet J, 11 (4), 771-779. doi:10.5455/OVJ.2021.v11.i4.30 Turabian Style AL-Aalim, Ammar M, Ali A Al-iedani, and Mohammad A Hamad. 2021. Study The Effects Of Systemic Injection Of Escherichia coli Lipopolysaccharide On The Expression Profile Of TLR4 And CD14 Genes Of Rat's Liver. Open Veterinary Journal, 11 (4), 771-779. doi:10.5455/OVJ.2021.v11.i4.30 Chicago Style AL-Aalim, Ammar M, Ali A Al-iedani, and Mohammad A Hamad. "Study The Effects Of Systemic Injection Of Escherichia coli Lipopolysaccharide On The Expression Profile Of TLR4 And CD14 Genes Of Rat's Liver." Open Veterinary Journal 11 (2021), 771-779. doi:10.5455/OVJ.2021.v11.i4.30 MLA (The Modern Language Association) Style AL-Aalim, Ammar M, Ali A Al-iedani, and Mohammad A Hamad. "Study The Effects Of Systemic Injection Of Escherichia coli Lipopolysaccharide On The Expression Profile Of TLR4 And CD14 Genes Of Rat's Liver." Open Veterinary Journal 11.4 (2021), 771-779. Print. doi:10.5455/OVJ.2021.v11.i4.30 APA (American Psychological Association) Style AL-Aalim, A. M., Al-iedani, . A. A. & Hamad, . M. A. (2021) Study The Effects Of Systemic Injection Of Escherichia coli Lipopolysaccharide On The Expression Profile Of TLR4 And CD14 Genes Of Rat's Liver. Open Veterinary Journal, 11 (4), 771-779. doi:10.5455/OVJ.2021.v11.i4.30 |