| Original Article | ||
Open Vet J. 2021; 11(1): 42-51 doi: 10.4314/ovj.v11i1.8 Open Veterinary Journal, (2021), Vol. 11(1): 42–51 Original Research http://dx.doi.org/10.4314/ovj.v11i1.8 Molecular characterization of duck plague virus from selected Haor areas of BangladeshKamrul Ahmed Khan1,2, Md. Alimul Islam1, Abdullah Al Momen Sabuj1, Md. Abul Bashar1, Md. Saiful Islam1, Md. Golzar Hossain1, Muhammed Tofazzal Hossain1 and Sukumar Saha1*1Department of Microbiology and Hygiene, Bangladesh Agricultural University, Mymensingh, Bangladesh 2Department of Livestock Services, Ministry of Fisheries and Livestock, Dhaka, Bangladesh *Corresponding Author: Sukumar Saha. Department of Microbiology and Hygiene, Bangladesh Agricultural University, Mymensingh-2202, Bangladesh. Email: sukumar.saha [at] bau.edu.bd Submitted: 26/08/2020 Accepted: 31/12/2020 Published: 23/01/2021 © 2021 Open Veterinary Journal
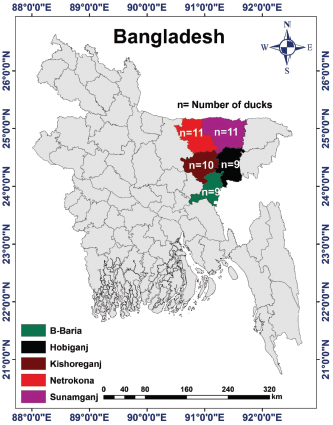
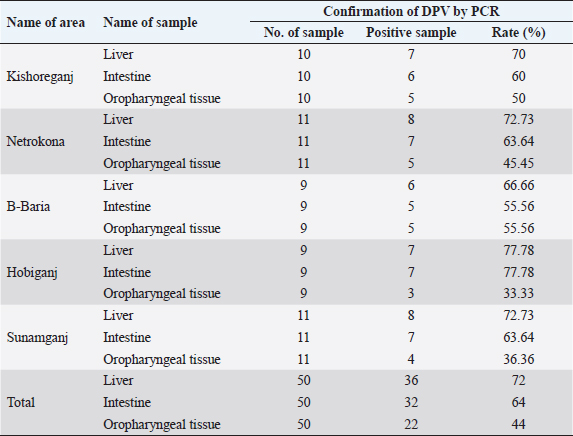
AbstractBackground: Duck viral enteritis, commonly known as duck plague (DP), is an acute and contagious fatal disease in ducks, geese, and swans caused by the DP virus (DPV). It poses a serious threat to the growth of duck farming in the Haor (wetland) areas of Bangladesh. Aim: This study aimed to detect the circulating DPV by molecular characterization, followed by phylogenetic analysis, targeting the UL30 gene in infected ducks from five Haor districts in Bangladesh and to observe the variation in the genome sequence between the field virus and vaccine strain of DPV. Methods: A total of 150 samples (liver, 50; intestine, 50; and oropharyngeal tissue, 50) were collected from DP-suspected sick/dead ducks from 50 affected farms in Kishoreganj, Netrokona, B. Baria, Habiganj, and Sunamganj districts in Bangladesh. For the identification of DPV in collected samples, polymerase chain reaction (PCR) was utilized. Nucleotide sequences of the amplified UL30 gene were compared with those of other DPV strains available in GenBank. Results: Of the 150 samples, 90 (60%) were found to be positive for DPV, as confirmed by PCR. Organ-wise prevalence was higher in the liver (72%), followed by the intestine (64%) and oropharyngeal tissue (44%). Regarding areas, the highest and lowest prevalence in the liver and intestine was observed in Habiganj and B. Baria, respectively, whereas the highest and lowest prevalence in the oropharyngeal tissue was observed in B. Baria and Habiganj, respectively. Two isolates, BAU/KA/DPV(B1)/2014 from Kishoreganj and BAU/KA/DPV(B4)/2014 from Sunamganj were sequenced, and phylogenetic analysis revealed that these isolates are evolutionarily closely related to Chinese isolates of DPV. Additionally, the isolates of DPV BAU/KA/DPV(B1)/2014 and BAU/KA/DPV(B4)/2014 showed the highest (98%) similarity to each other. The nucleotide sequence of the isolate BAU/KA/DPV(B1)/2014 exhibited higher nucleotide variability (246 nucleotides) than that of the vaccine strain (accession no. EU082088), which may affect protein function and additional drug sensitivity. Conclusion: Based on the findings of the molecular study, it can be assumed that the Bangladeshi isolates and all Chinese isolates of DPV may have a common ancestry. Keywords: Duck plague virus, Polymerase chain reaction, Duck embryo, Nucleotide sequence. IntroductionFlood-prone wetland areas of northeast Bangladesh, commonly known as “Haor” areas, are suitable for duck rearing because of their geographical advantages, such as natural feed availability, abundant water for swimming, and tolerable temperature (Huque et al., 2011). Although duck rearing is highly profitable in Bangladesh (Ahmed and Huque, 1994), the most important constraint is the existence of infectious diseases, among which duck plague (DP) is the most common disease. DP, caused by DP virus (DPV), is an acute, sometimes chronic, contagious, and fatal viral infection that occurs naturally in ducks, geese, and swans (Kaleta et al., 2007; Hanan et al., 2014) and is characterized by digestive mucosal eruptions, tissue hemorrhages, vascular damage, changes in lymphoid organs, and degenerative lesions in parenchymatous organs (Davison et al., 1993; Shawky et al., 2000; Campagnolo et al., 2001). The causal agent is also known as Anatid herpesvirus-1, a member of the subfamily Alphaherpesvirinae of the family Herpesviridae (Li et al., 2006; Fadly et al., 2008; ICTV, 2017). Similar to other alphaherpesviruses, DPV contains four structural components: an amorphous tegument, an icosahedral capsid, a bilayer lipid envelope, and a double-stranded DNA comprising 64.3% guanine and cytosine content (Yuan et al., 2005; Cai et al., 2013). The genome length of DPV is approximately 158 kbp and encodes 78 putative proteins. The genomic organization of DPV comprises four regions: a unique long (UL) region, a unique short region, a unique short internal repeat region, and a unique short terminal repeat region (Chen et al., 2013). Sequence analysis has shown that the viral genome contains 67 genes with homology within most members of the Alphaherpesvirinae (Li et al., 2006). Among these regions, the genes in the UL region of its genome are well conserved. Therefore, the viral DNA polymerase encoded by the UL30 gene might be used for molecular detection by polymerase chain reaction (PCR) amplification of this gene (Li et al., 2006). The disease was first identified in the Netherlands in 1923 (Baudet, 1923) and was termed DP (Bos, 1942). Subsequently, the disease has been reported worldwide (Calnek et al., 1997). Ducks of all age groups (from 1–-week-old to adults) are susceptible to DP, and morbidity and mortality rates vary from 5% to 100% (Calnek et al., 1997; Hossain et al., 2004). Domestic ducks (Anas platyrhynchos), including Indian Runner, White Pekin, and Khaki Campbell, as well as mixed breeds and Muscovy ducks, are commonly infected by DPV (Campagnolo et al., 2001; Akter et al., 2004; Konch et al., 2009; Woźniakowski and Samorek-Salamonowicz, 2014). Within 3–7 days after infection, clinical signs appear in domestic ducks, and following clinical manifestation, death usually occurs within 1–5 days. The susceptible duck population may be infected via both direct and indirect routes (Sandhu and Shawky, 2003; Kaleta et al., 2007; Wang et al., 2013). Migratory waterfowls play a vital role in spreading the disease (Gough, 1984; Kathryn et al., 2001; Wang et al., 2011). The major characteristic symptoms of DP include sudden high and persistent flock mortality, photophobia, half-closed pasted eyelids, drowsiness, ataxia, ruffled feathers, ocular and nasal discharge, inappetence, extreme thirst, distress, soiled vents, greenish watery diarrhea, and maintaining an upright stance using wings for support, indicating weakness and depression (Baki et al., 1993; King et al., 2011). Necropsy findings indicate hemorrhage in the liver, intestine, spleen, heart, body cavity, and pericardium; plaques in the intestine and esophagus, and lesions in the bursa and thymus (Davison et al., 1993; Sandhu and Shawky, 2003; Konch et al., 2009). Clinical signs and gross pathology associated with a DP outbreak vary with the species, age, sex, and immune status of the affected birds and viral virulence (OIE, 2012). Reactivation of the dormant virus has a higher probability of transmitting DP to a healthy duck population. The birds that recover from the disease commonly become a carrier of DPV and shed virulent virus in the feces or on the surface of the eggs for years (Richter and Horzinek, 1993; Shawky and Schat, 2002). DP can be diagnosed by duck embryo inoculation or cell culture (El-Samadony et al., 2013; Gao et al., 2014). Several other tests, including the passive hemagglutination assay, agar gel immunodiffusion test, commercial enzyme-linked immunosorbent assay (ELISA), and dot-ELISA, can be used to detect DPV (Hossain et al., 2005; Kataria et al., 2005; Woolcock, 2008; OIE, 2012). For genome-specific identification, such as UL30 and US4 (gD) genes, both conventional PCR and real-time PCR are the most reliable methods for DPV (Konch et al., 2009; Aravind et al., 2015). Loop-mediated isothermal amplification is the latest method adopted for DP diagnosis and is more rapid, simple, and highly sensitive compared with PCR or virus isolation techniques (Ji et al., 2009; Jiang et al., 2012; Woźniakowski and Samorek-Salamonowicz, 2014). Moreover, early diagnosis is crucial for disease prevention and control. DPV was first identified in ducks in Bangladesh in 1980 (Sarker, 1980). Since then, the disease frequently occurs every year in Bangladesh, either in an endemic or epidemic form. To date, several studies have been conducted on DPV in Bangladesh (Sarker, 1980, 1982; Khan et al., 1990; Islam, 1992; Islam et al., 1993; Akter et al., 2004; Ahamed et al., 2015; Tahmina et al., 2017; Soma et al., 2018). Although DPV is a single antigenic virus, the reasons for vaccine failure and high mortality rates in Bangladesh remain unclear (Akter et al., 2004). Therefore, it is important to compare genetic and evolutionary variations between the currently circulating field virus and the DP vaccine virus. This study aimed to molecularly characterize the circulating DPV by PCR and gene sequencing, followed by phylogenetic analysis of strains isolated from some selected Haor areas of Bangladesh. The genetic differences between field viruses and the vaccine strain were also analyzed. Materials and MethodsStudy area, sample size, and samplingFive Haor districts (Kishoreganj, Netrokona, B. Baria, Habiganj, and Sunamganj) in Bangladesh were included in the present study. The study areas were selected based on geographical location, duck population density, duck rearing practice, migratory duck movement, and disease prevalence. Kishoreganj district is located at 24.4331°North (N) and 90.7866°East (E), Netrokona district at 24.8817°N and 90.7271°E, B. Baria at 23.9608°N and 91.1115°E, Habiganj at 24.4771°N and 91.4507°E, and Sunamganj at 25.0715°N and 91.3992°E. The locations of the study areas are illustrated in a digitalized map processed in ArcGis-ArcMap software version 10.7 (ESRI, Redlands, CA). The sample size of each district is also shown in the map (Fig. 1). A complete list of duck farms in the five Haor districts was obtained from the respective District Livestock Office of Bangladesh. Considering farm clinical history and ongoing disease occurrences, 50 duck farms were included in the present study (Kishoreganj, 10; Netrokona, 11; B. Baria, 9; Habiganj, 9; and Sunamganj, 11). For this study, one duck was selected from every farm. Therefore, a total of 50 ducks were collected; of these, 35 were dead and 15 were DP-suspected sick ducks. Dead ducks with a clinical history of DP were collected immediately after death. On the contrary, sick ducks showing typical signs of DP (Baki et al., 1993; King et al., 2011), as identified by government-registered veterinary surgeons, were also collected. Sampling was carried out for 2 years from 2014 to 2016. Sample collection, transportation, and preservationSick birds were slaughtered following the rules and regulations of the ethics committee. Postmortem examination of ducks was conducted by a government-registered veterinary surgeon following the standard protocol as described in the Atlas of Avian Necropsy (Majó and Dolz, 2011). All ducks showed typical postmortem signs of DPV (Sandhu and Shawky, 2003; Konch et al., 2009). Liver, intestine, and oropharyngeal tissue samples were obtained from each bird. Therefore, a total of 150 tissue samples (liver, 50; intestine, 50; and oropharyngeal tissue, 50) were collected according to the procedure described by El-Samadony et al. (2013). All samples were collected aseptically and maintained at 4°C in an icebox during transport to the laboratory. Immediately after being transported, samples were either processed or kept at −20°C until further analysis. The study was conducted in the Virology Laboratory in the Microbiology and Hygiene Department at Bangladesh Agricultural University, Mymensingh, Bangladesh.
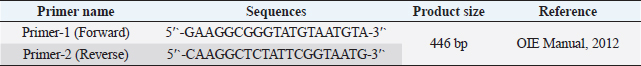
Fig. 1. Study area map of different districts of Bangladesh. Imags was extracted from DIVA-GIS (http://www.diva-gis.org/) using the Geographical Information System (GIS), The map was processed with ArcMap 10.7 software. Preparation of samplesSamples were prepared according to the procedure described in the OIE (2012). In brief, each sample was chopped into small pieces using sterile scissors and ground using a sterile mortar and pestle. In the ground sample, a sufficient amount of phosphate-buffered saline was added to obtain a 20% suspension, and the sample was filtered to remove coarse particles. The sample was centrifuged at 2,500–3,000 rpm for 10–20 minutes. The supernatant was collected and stored at −20°C until use. DNA extraction and molecular characterization of DPVDNA was extracted from the processed samples using a DNA extraction kit according to the manufacturer’s protocol (Wizard®Genomic DNA purification kit, Promega®, San Luis Obispo, CA). Previously published primers were used for the amplification of the targeted DNA segments (UL30) of DPV (OIE, 2012). Details of the primers used in the study are presented in Table 1. For amplification, a 50 μl typical reaction mixture containing 18 μl nuclease-free water, 25 μl PCR master mixture (Taq DNA polymerase, dNTPs, MgCl2, and reaction buffers) (Promega, Madison, WI), 1 μl (100 nmol/μl) forward primer, 1 μl (100 nmol/μl) reverse primer, and 5 μl DNA template was used. PCR was carried out under the following conditions: initial denaturation at 94°C for 2 minutes, followed by 35 cycles of denaturation at 94°C for 1 minutes, annealing at 55°C for 1 minutes, extension at 72°C for 2 minutes, and a final extension at 72°C for 7 minutes. PCR products (5 μl) were loaded into 2% agarose gel wells, and 1 μl of a 100-bp DNA marker was loaded as a DNA ladder and electrophoresed. Finally, DNA was stained with ethidium bromide and visualized under an ultraviolet transilluminator (Biometra, Germany). Nucleotide sequencing and phylogenetic analysisTwo PCR-positive samples, one obtained from Karimganj in the Kishoreganj district and another from Dharamapasha in the Sunamganj district, were selected and purified using the Wizard®SV Gel and PCR Clean-Up System (Promega, San Luis Obispo, CA) according to the manufacturer’s instructions and sent for sequencing. Sequencing was carried out commercially in Malaysia using the cycle sequencing method (First BASE Laboratories Sdn Bhd, Selangor, Malaysia). A total of 20 partial UL30 gene sequences from DPV were retrieved from NCBI’s GenBank online database. Pairwise alignment, followed by multiple sequence alignment of all 20 reference genome sequences, including the existing vaccine strain, BAU/KA/DPV(B1)/2014, and BAU/KA/DPV(B4)/2014, was carried out using the built-in Clustal V program of MEGA 5.2.2 software (Tamura et al., 2013). A phylogenetic tree was constructed using the neighbor-joining method (Saitou and Nei, 1987). The reliability of the branching point was evaluated using a bootstrap test (n=1,000). Table 1. Primer for PCR to detect UL30 gene of DPV.
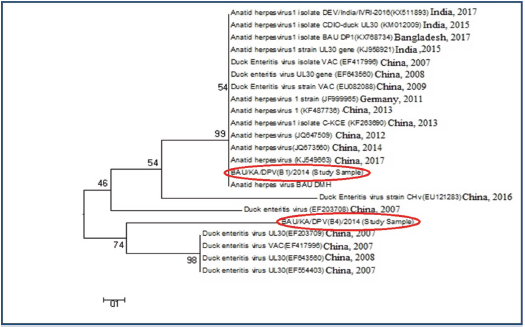
Ethical approvalThe study protocol was approved by the Animal Welfare and Ethics Committee of Bangladesh Agricultural University, Mymensingh, Bangladesh for clarity. Verbal permission from the farm owners was also obtained before bird collection. ResultsMolecular detection of DPV by PCR using a specific primerOf the 150 samples, 72% (36/50) of the liver samples, 64% (32/50) of the intestinal samples, and 44% (22/50) of the oropharyngeal tissue samples were found to be DPV-positive using PCR. Area-wise occurrence of DPV was higher in Habiganj for both liver (77.78%; 7/9) and intestinal (77.78%; 7/9) samples. Additionally, B. Baria exhibited the highest occurrence of DPV in orophayngeal tissue samples (55.56%; 5/9) compared with other study areas. On the contrary, the lowest occurrence of DPV was observed in B. Baria for both liver and intestine samples and in Habiganj for oropharyngeal tissue samples (Table 2 and Fig. 2). Among the three sample types collected, liver samples were superior to intestinal or oropharyngeal tissue samples for DPV detection by PCR. No intestinal or oropharyngeal samples were positive for DPV when the liver samples from the same animal tested negative (data not shown). Construction of the phylogenetic treeThe nucleotide sequences obtained in the present study were aligned with the sequences of other DPV strains available in GenBank, and a phylogenetic tree was generated. The obtained nucleotide sequence was submitted to GenBank (MN968777 and MN968780). A phylogenetic tree based on the UL30 gene is shown in Figure 3. The neighbor-joining tree based on a partial UL30 gene sequence from BAU/KA/DPV(B1)/2014 and BAU/KA/DPV (B4)/2014 with the sequences of DPV clustered with the DPV sequences from GenBank. This finding confirmed that the sequenced samples from Haor areas of Bangladesh had DPV. Isolate BAU/KA/DPV(B1)/2014 showed the highest similarity with the isolates of the Anatid herpesvirus (KJ549663) China/2017, Anatid herpesvirus BAU DMH/Bangladesh/2015, Anatid herpesvirus (JQ673560) China/2014, and Anatid herpesvirus (JQ647509) China/2012 and formed a sub-cluster. The sequenced strain BAU/KA/DPV(B4)/2014 showed the highest similarity with the duck enteritis virus (EF203709) China/2007, duck enteritis virus VAC (EF417996) China/2007, duck enteritis virus (EF643560) China/2008, and duck enteritis virus (EF564403) China, 2007. It was evident from the phylogenetic tree that two local circulatory serotypes of DPV were clustered with 20 NCBI sequences. Of these 20 NCBI sequences, 14 isolates originated from China, 3 from India, 2 from Bangladesh, and 1 from Germany (Fig. 2). Two isolates of DPV BAU/KA/DPV(B1)/2014 and BAU/KA/DPV(B4)/2014 showed the highest (98%) similarity with each other (data not shown). Table 2. Molecular detection of DPV from suspected field samples by PCR.
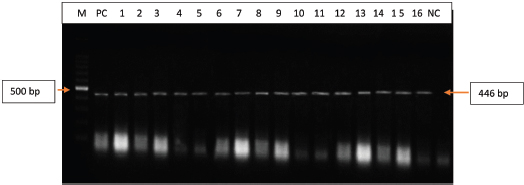
Fig. 2. Polymerase chain reaction (PCR) amplification of the UL30 gene of DPV isolated from field samples. Lane M=100 bp DNA marker; PC=Positive control, Lane 1–16: Isolates of DPV (field samples); NC=Negative control.
Fig. 3. Phylogenetic tree of the UL30 genes of DPV isolates from Bangladesh and reference sequences from GenBank. The tree was constructed by the neighbor-joining method using the Clustal V program of MEGA 5.2.2 software. The sequence of study isolates from Bangladesh is indicated in red circles and reference sequences are indicated by their accession number. Comparison between the UL30 gene of the vaccine strain and DPV isolatesWhen compared with the nucleotide sequence of the vaccine strain (accession no. EU082088), the sequence of the study sample BAU/KA/DPV(B1)/2014 showed a high variability of DPV isolate nucleotide (246 NT) sequences, which may affect the protein function and additional drug sensitivity (Fig. 4). DiscussionDP, a highly contagious viral disease, is distributed globally, specifically in duck-rearing areas. It negatively impacts the economy of the duck industry by reducing egg production and increasing mortality rates (Converse and Kidd, 2001; Sandhu and Metwally, 2008). Over time, DPV is rapidly developing a vaccine resistance in the duck population by acquiring new characteristics (Diao et al., 2006). This is one of the greatest challenges to control DPV in commercial duck farming. However, molecular characterization and genome sequencing are important to understand DPV properly and to produce an effective vaccine against the virus (Li et al., 2006; Liu et al., 2007). Immunization is considered one of the most effective tools to control DP (Lian et al., 2011; Huang et al., 2014), but vaccines are usually manufactured locally in Bangladesh without proper surveillance. Therefore, it is important to identify the virus through a genome sequencing method to develop an effective vaccine.
Fig. 4. Comparison of UL30 gene of DPV in the study sample (BAU/KA/DPV(B1)/2014) with vaccine strains (Accession no. EU082088) showing high variability of nucleotide (246NT) sequences of DPV isolates. The expected PCR amplicon appeared at 446 bp in the targeted DNA polymerase UL30 gene. The DNA polymerase gene encodes UL30 protein, which is highly conserved in DPV (Wallace et al., 2000; OIE, 2012; Wu et al., 2012; Ahamed et al., 2015). UL30 is involved in DNA replication and nucleotide metabolism in DPV (Li et al., 2006; Neher et al., 2019). It is widely used to construct phylogenetic trees and to indicate a distinct division among Alphaherpesvirinae, Betaherpesvirinae, and Gammaherpesvirinae (Thureen and Keeler, 2006). In many studies, the UL30 gene was targeted because of its highly conserved nature (Hansen et al., 2000; Xuefeng et al., 2008; Zou et al., 2010; Wu et al., 2012). In this study, by targeting the UL30 gene via PCR, 90 (60%) samples were found to be positive for DPV. The higher prevalence of DPV might be because of problems with the collection of such tissue samples, distribution of viruses in such tissues, or processing of samples for PCR. PCR results clearly indicated a higher positive rate of DPV in liver samples, followed by that in the intestine and oropharyngeal tissue samples. PCR clearly demonstrated that liver samples were superior among the three different types of samples collected for DPV detection. No intestinal or oropharyngeal samples were positive for DPV when the liver samples from the same animal tested negative. Therefore, our data suggest that liver samples are the best choice for isolation and detection of DPV in ducks. The overall occurrence recorded in this study was higher than that in a previous study conducted by Ahamed et al. (2015), who confirmed an overall 18.09% DPV occurrence in cloacal and visceral organ samples from Bangladesh. Our study revealed similar results for the overall DPV detection rates among different Haor areas, although the highest detection rates of DPV in both liver and intestine samples were recorded in Habiganj district and in oropharyngeal tissue samples were recorded in B. Baria district. However, similarities in the geographical and ecological characteristics of selected hoar areas may provide similar environmental conditions for the survival and transmission of DPV. This finding may be consistent with the area-wise findings of our study. Previously, Ahamed et al. (2015) recorded DPV detection rates of 6.62%, 28.75%, and 4.55% in Rajshahi, Netrokona, and Mymensingh districts, respectively, which are contradictory to our findings. A study conducted by Soma et al. (2018) reported a DPV prevalence of 71.42% in Netrokona, 66.67% in Sunamganj, and 50% in Mymensingh; these findings are nearly analogous to those of our study. The results indicate that DPV is highly circulated in Haor areas. In Haor areas, duck farm owners usually follow traditional management practices with improper housing systems and inappropriate boundary systems and nearly lack preventive measures. Additionally, wild and migratory birds in Haor areas can transmit DPV to domestic ducks because migratory waterfowls play a key role in DPV transmission (Dhama et al., 2017). Moreover, intermixing of recovered ducks and newly purchased ducks is a common practice of duck farm owners in the Haor areas of Bangladesh (Khan et al., 2018). After PCR amplification, two isolates from Kishoreganj and Sunamganj were sequenced. The UL30 gene of DPV was selected to determine whether any genetic variation existed between the field virus and the vaccine strain. The phylogenetic tree analysis employing the neighbor-joining method using the partial UL30 nucleotide sequences of BAU/KA/DPV(B1)/2014 and BAU/KA/DPV(B4)/2014 and another 20 nucleotide sequences from the GenBank clustered with each other, which confirmed that the study samples from the Haor areas of Bangladesh were DPVs. The phylogenetic tree showed that the sequenced strain of DPV from Kishoreganj had the highest similarity with Chinese and Bangladeshi isolates. Another identified DPV from Sunamganj showed a genetic relationship with several Chinese strains of DPV. Thus, the Bangladeshi isolates and the Chinese isolates of DPV from GenBank may have a common ancestry. This result is consistent with that of a previous study in Bangladesh (Ahamed et al., 2015). A variety of duck breeds are being imported from neighboring countries, such as China and India, every year. Farmers in Bangladesh introduce newly purchased ducks to their farms without following quarantine measures, which might play an important role in the spreading of DP. The similarity in the nucleotide sequences of DPV from the duck populations may be the cause of the same origin and distribution of the study viruses and viral strains in GenBank. Compared to the nucleotide sequence of the vaccine strain (accession no. EU082088), the BAU/KA/DPV(B1)/2014 sample showed high nucleotide variability (246 nucleotides) in the DPV isolates, which may affect protein function and antimicrobial drug sensitivity. The vaccine strain of DPV, namely “duck enteritis virus strain VAC (EU082088),” was isolated and characterized in 2009 in China. Conversely, BAU/KA/DPV (B1)/2014 was isolated and characterized in Bangladesh. This variation may be because of the difference in geographical and ecological factors between Bangladesh and China. However, owing to the 7-years gap (2009–2016) between these isolations, the field viruses could have partially mutated. Although the rate of mutation in the herpesvirus gene is very slow, the local isolates of DPV BAU/KA/DPV(B1)/2014 and BAU/KA/DPV(B4)/2014 showed 98% similarity with each other. The two isolates, one obtained from the duck farms in Kishoreganj district and another from Sunamganj district, showed 98% similarity in UL30 gene sequencing. Unfortunately, these viruses have very high variability when compared with the vaccine strain used in this study. Therefore, this area of study needs to be further investigated for better clarification. ConclusionThis study revealed that DPV is highly circulated among ducks in the Haor areas of Bangladesh. This study also highlights that circulated DPVs have a close relationship with Chinese and Indian isolates. When compared with the vaccine strain used in the study, high variability was observed using the identified isolates. Therefore, new strains are prevalent in the study areas. The present study provides evidence of molecular characterization and genome sequencing of DPV. These findings may aid in developing and selecting effective vaccines to implement effective measures to control the outbreak of DP in the Haor areas of Bangladesh. AcknowledgmentsThis research was supported by grants from the National Agricultural Technology Project, Bangladesh. Conflict of interestThe authors declare that there is no conflict of interest. Authors’ contributionsK.A.K and M.A.I carried out the experiments. K.A.K and A.A.M.S wrote the first draft of the manuscript. M.A.B helped in the sample collection and experiment. M.S.I., M.G.S, and M.T.H helped in data analysis. S.S and M.A.I conceived, designed the experiments, and finalized the manuscript. ReferencesAhamed, M.M., Hossain, M.T., Rahman, M., Nazir, K.H.M.N.H., Khan, M.F.R., Parvej, M.S., Ansari, W.K., Noor, M.N., Chiste, A.A., Amin, K.B., Hossen, M.L., Ahmed, S. and Rahman, M.B. 2015. Molecular characterization of duck plague virus isolated from Bangladesh. J. Adv. Vet. Anim. Res. 2, 296–303. Ahmed, N. and Huque, Q.M.E. 1994. Backyard poultry feeding system in Bangladesh. Asian Livest. 7, 73–79. Akter, S., Islam, M.A., Hossain, M.T., Begum, M.I.A., Amin, M.M. and Sadekuzzaman, M. 2004. Characterization and pathogenicity of duck plague virus isolated from natural outbreaks in ducks of Bangladesh. Bangladesh J. Vet. Med. 2, 107–111. Aravind, S., Kamble, N.M., Gaikwad, S.S., Shukla, S.K., Saravanan, R., Dey, S. and Mohan, C.M. 2015. Protective effects of recombinant glycoprotein D based prime boost approach against duck enteritis virus in mice model. Microb. Pathog. 88, 78–86. Baki, M.A., Dewan, M.L. and Mondal, M.M.H. 1993. An investigation on the causes of mortality of ducks in Bangladesh. Progress. Agric. 4, 27–33. Baudet, A. 1923. Mortality in ducks in the Netherlands caused by a filtrable virus; fowl plague. Tijdschr Diergeneeskd. 50, 455–459. Bos, A. 1942. Some new cases of duck pest. Tijdschr Diergeneeskd. 69, 372–381. Cai, M.S., Deng, S.X. and Li, M.L. 2013. Comparison of the immune responses in BALB/c mice following immunization with DNA-based and live attenuated vaccines delivered via different routes. Vaccine 31, 1353–1356. Calnek, B.W., Barnes, H.J., Beard, C.W., McDougald, L.R. and Saif, Y.M. 1997. Diseases of poultry, 10th ed. Ames, IA: Iowa State University Press, pp: 675–683. Campagnolo, E.R., Banerjee, M., Panigrahy, B. and Jones, R.L. 2001. An outbreak of duck viral enteritis (Duck Plague) in domestic Muscovy ducks (Cairina moschata domesticus) in Illinois. Avian Dis. 45, 522–528. Chen, L., Yu, B., Hua, J., Ye, W., Ni, Z., Yun, T., Deng, X. and Zhang, C. 2013. Construction of a full-length infectious bacterial artificial chromosome clone of duck enteritis virus vaccine strain. Virol. J. 10, 328. Converse, K.A. and Kidd, G.A. 2001. Duck plague epizootics in the United States, 1967-1995. J. Wildl. Dis. 37, 347–357. Davison, S., Converse, K.A., Hamir, A.N. and Eckorage, R.J. 1993. Duck viral enteritis in domestic Muscovy ducks in Pennsylvania. Avian Dis. 37, 1142–1146. Dhama, K., Kumar, N., Saminathan, M., Tiwari, R., Karthik, K., Kumar, M.A., Palanivelu, M., Shabbir, M.Z., Malik, Y.S. and Singh, R.K., 2017. Duck virus enteritis (duck plague)–a comprehensive update. Vet. Q. 37, 57–80. Diao, Y.X., Lu, G.X., Zheng, F.Y., Yang, J.H., Chen, Q.P. and Liu, Y.Z., 2006. Isolation and identification and characteristics of the pathogen of a new duck plague. Chin. J. Vet. Sci. 26, 136–139. El-Samadony, H.A., Tantawy, L.A., Salama, E. and Afaf, A.K. 2013. Molecular characterization of circulating duck viral enteritis in Egypt during 2012-2013. Br. J. Poult. Sci. 2, 38–44. Fadly, A.M., Glisson, J.R., McDougald, L.R., Nolan, L. and Swayne, D.E. 2008. Duck virus enteritis. Dis. Poult. 12, 384–393. Gao, X., Jia, R., Wang, M., Zhu, D., Chen, S., Lin, M., Yin, Z., Wang, Y., Chen, X. and Cheng, A. 2014. Construction and identification of cDNA library for use in the yeast two hybrid system from duck embryonic fibroblast cells post infected with duck enteritis virus. Mol. Biol. Rep. 41, 467–475. Gough, R.E. 1984. Laboratory confirmed outbreaks of duck virus enteritis in the United Kingdom from 1977 to 1982. Vet. Rec. 114, 262–265. Hanan, A., Ghada, M., Sadek, E., Afan, E.G., Nahed, I.M., Ekram, S.M. and Norhan, N.M. 2014. Preparation and evaluation of combined inactivated duck vaccine against salmonellosis, duck plague and duck hepatitis. Nat. Sci. 12, 142–147. Hansen, W.R., Nashold, S.W., Docherry, D.E., Brown, S.E. and Knudson, D.L. 2000. Diagnosis of duck plague in waterfowl by polymerase chain reaction. Avian Dis. 44, 266–274. Hossain, M.K., Ahmed, M., Kabir, H., Sarker, M.R.R., Jail, M.A. and Adhikary, G.N. 2004. Poultry diseases at Rajshahi in Bangladesh. J. Anim. Vet. Adv. 3, 656–658. Hossain, M.T., Islam, M.A., Amin, M.M. and Islam, M.A. 2005. Comparative efficacy of the conventional and experimentally developed duck plague vaccine. Int. J. Poult. Sci. 4, 369–371. Huang, J., Jia, R., Wang, M., Shu, B., Yu, X., Zhu, D., Chen, S., Yin, Z., Chen, X. and Cheng, A. 2014. An attenuated duck plague virus (DPV) vaccine induces both systemic and mucosal immune responses to protect ducks against virulent DPV infection. Clin. Vaccine Immunol. 21, 457–462. Huque, K.S., Saleque, M.A. and Khatun, R. Commercial poultry production in Bangladesh. In Proceedings of 7th International show and seminar organized by World’s Poultry Science Association (WPSA), Dhaka, Bangladesh, 2011. ICTV. 2017. International committee on taxonomy of viruses. Virus Taxonomy. Available via http://ictvonline.org/virustaxonomy.asp (Accessed 22 March 2020). Islam, M.R. 1992. Pathogenicity of two local isolates of duck plague virus. Bangladesh J. Microbiol. 9, 61–66. Islam, M.R., Nessa, J. and Halder, K.M. 1993. Detection of duck plague virus antigen in tissues by immunoperoxidase staining. Avian Pathol. 22, 389–393. Ji, J., Du, L.Q., Xie, Q.M., Cao, Y.C., Zuo, K.J., Xue, C.Y., Ma, J.Y., Chen, F. and Bee, Y.Z. 2009. Rapid diagnosis of duck plagues virus infection by loop-mediated isothermal amplification. Res. Vet. Sci. 87, 53–58. Jiang, T., Liu, J., Deng, Y.Q., Su, J.L., Xu, L.J., Liu, Z.H., Li, X.F., Yu, X.D., Zhu, S.Y., Gao, G.F., Qin, E.D. and Qin, C.F. 2012. Development of RT-LAMP and real-time RT-PCR assays for the rapid detection of the new duck Tembusu-like BYD virus. Arch. Virol. 157, 2273–2280. Kaleta, E.F., Kuczka, A., Kuhnhold, A., Bunzenthal, C., Bonner, B.M., Hanka, K., Redmann, T. and Yilmaz, A. 2007. Outbreak of duck plague (duck herpesvirus enteritis) in numerous species of captive ducks and geese in temporal conjunction with enforced biosecurity (in-house keeping) due to the threat of avian influenza A virus of the subtype Asia H5N1. Deut. Tierarztl. Wochenschr. 114, 3–11. Kataria, J.M., Mohan, C.M., Dey, S., Dash, B.B. and Dhama, K. 2005. Diagnosis and immunoprophylaxis of economically important poultry diseases: a review. Indian J. Anim. Sci. 75, 555–567. Kathryn, A., Converse, A. and Gregory, A.K. 2001. Duck plague epizootics in the United States. J. Wildl. Dis. 37, 347–357. Khan, K.A., Saha, S., Hossain, M.T., Haque, M.E., Haq, M.M. and Islam, M.A., 2018. Epidemiological investigation of recurrent outbreaks of duck plague in selected Haor (wetland) areas of Bangladesh. J. Anim. Vet. Adv. 5, 131–139. Khan, M.S.R., Sarker, A.J., Siddique, M.A.B., Hossain, W.I.M.A. and Begum, F. 1990. Passive haemaglutination and complement fixation test for the detection of duck plague virus. Bangladesh Vet. J. 24, 15–21. King, A., Lefkowita, E. and Adams, M.J. 2011. Virus taxonomy: ninth report of the international committee on taxonomy of viruses. Amsterdam, Netherlands: Elsevier, pp: 111–114. Konch, C., Upadhyaya, T.N., Goswami, S. and Dutta, B. 2009. Studies on the incidence and pathology of naturally occurring duck plague in Assam. Indian J. Vet. Pathol. 33, 213–215. Li, H., Liu, S. and Kong, X. 2006. Characterization of the genes encoding UL24, TK and gH proteins from duck enteritis virus (DEV): a proof for the classification of DEV. Virus Genes. 33, 221–227. Lian, B., Cheng. A., Wang, M., Zhu, D., Luo, Q., Jia, R., Liu, F., Han, X. and Chen, X. 2011. Induction of immune responses in ducks with a DNA vaccine encoding duck plague virus glycoprotein C. Virol. J. 8, 214. Liu, S., Chen, S., Li, H. and Kong, X. 2007. Molecular characterization of the harpers simplex virus 1 (HSV-1), homologues, UL25 to UL30, in duck enteritis virus (DEV). Gene. 401, 88–96. Majó, N. and Dolz, R. 2011. Atlas of avian necropsy: microscopic diagnosis and sampling. Pontevedra, Spain: Servet Veterinary Publishing Company, pp: 10–11. Neher, S., Barman, N.N., Bora, D.P., Deka, D., Tamuly, S., Deka, P., Bharali, A. and Das, S.K. 2019. Detection and isolation of Duck Plague virus from field outbreaks in Assam, India. Indian J. Anim. Res. 53, 790–798. OIE. 2012. Manual of diagnostic tests and vaccines for terrestrial animals. Duck virus enteritis. Chapter 2.3.7. Paris, France: OIE. Richter, J.H.M. and Horzinek, M.C. 1993. Duck plague. In: Virus infections of birds. Eds., McFerran, J.B. and McNulty, M.S. Amsterdam, The Netherlands: Elsevier Science Publishers B.V., pp: 77–90. Saitou, N. and Nei, M. 1987. The neighbor-joining method: a new method for reconstructing phylogenetics tress. Mol. Biol. Evol. 4, 406–425. Sandhu, T.S. and Metwally SA 2008. Duck virus enteritis (duck plague). In: Diseases of poultry. 12th ed. Saif, Y.M., Fadly, A.M., Glisson, J.R., McDougald, L.R., Nolan, L.K. and Swayne, D.E. Oxford, UK: American Wiley-Blackwell, pp: 384–393. Sandhu, T.S. and Shawky, S.A. 2003. Duck virus enteritis (duck plague). In: Diseases of poultry. 11th ed. Saif, Y.M., Barnes, H.J., Glission, J.R., Fadly, A.M., McDougald, L.R. and Swayne, D.E. Ames, IA: Iowa State University Press, pp: 354–363. Sarker, A.J. 1980. Duck plague in Bangladesh. Indian Vet. J. 57, 1–5. Sarker, A.J. 1982. Duck plague in Bangladesh. Isolation and identification of the etiological agent. Indian Vet. J. 59, 669–674. Shawky, S. and Schat, K.A. 2002. Latency sites and reactivation of duck enteritis virus. Avian Dis. 46, 308–313. Shawky, S., Sandhu, T. and Shivaprasad, H.L. 2000. Pathogenicity of a low-virulence duck virus enteritis bisolate with apparent immunosuppressive ability. Avian Dis. 44, 590–599. Soma, S.S., Nazir, K.H.M.N.H., Rahman, M.T., Rahman, M.M. Ara, M.S., Sultana, R., Haydar, M. R., Siddique, M.P. and Rahman, M.B. 2018. Isolation and molecular detection of duck plague virus for the development of vaccine seed. Asian Australas. J. Biosci. Biotechnol. 3, 78–85. Tahmina, S., Hossain, K.M., Rahman, H., Hossain, A., Saha, A.K., Anjuman, S. and Giasuddin. 2017. Adaptation of duck plague virus (DPV) in baby hamster kidney cell line (BHK-21) towards vaccine development. Ind. J. Curr. Res. 9, 49733–49737. Tamura, K., Stecher, G., Peterson, D., Filipski, A. and Kumar, S. 2013. MEGA6: Molecular evolutionary genetic analysis version 6.0. Mol. Biol. Evol. 30, 2725–2729. Thureen, D.R. and Keeler, C.L. 2006. Psittacid herpesvirus 1 and infectious laryngotracheitis virus: comparative genome sequence analysis of two avian alphaherpesviruses. J. Virol. 80, 7863–7872. Wallace, R., Hansen, A., Sean, W., Nashold, A., Douglas, E., Docherty, A., Susan, E., Brown, B., Dennis, L. and Knudson, B. 2000. Diagnosis of duck plague in waterfowl by polymerase chain reaction. Avian Dis. 44, 266–274. Wang, G., Qu, Y., Wang, F., Hu, D., Liu, L., Li, N., Yue, R., Li, C. and Liu, S. 2013. The comprehensive diagnosis and prevention of duck plague in northwest Shandong province of China. Poult. Sci. 92, 2892–2898. Wang, J., Hoper, D., Beer, M. and Osterrieder, N. 2011. Complete genome sequence of virulent duck enteritis virus (DEV) strain 2085 and comparison with genome sequences of virulent and attenuated DEV strains. Virus Res. 160, 316–325. Woolcock, P.R. 2008. Duck virus enteritis. Manual of diagnostic tests and vaccines for terrestrial animals. Chapter 2.3.7. Paris, France: OIE. Woźniakowski, G. and Samorek-Salamonowicz, E. 2014. First survey of the occurrence of duck enteritis virus (DEV) in free-ranging Polish water birds. Arch. Virol. 159, 1439–1444. Wu, Y., Cheng, A., Wang, M., Yang, Q., Zhu, D., Jia, R., Chen, S., Zhou, Y., Wang, X. and Chen, X. 2012. Complete genomic sequence of Chinese virulent duck enteritis virus. J. Virol. 86, 5965. Xuefeng, Q., Xiaoyan, Y., Anchun, C., Mingshu, W., Dekang, Z. and Renyong, J. 2008. The pathogenesis of duck virus enteritis in experimentally infected ducks: a quantitative time-course study using TaqMan polymerase chain reaction. Avian Pathol. 37, 307–310. Yuan, G.P., Cheng, A.C., Wang, M.S., Liu, F., Han, X.Y., Liao, Y.H. and Xu, C. 2005. Electron microscopic studies of the morphogenesis of duck enteritis virus. Avian Dis. 49, 50–55. Zou, Q., Sun, K., Cheng, A., Wang, M., Xu, C., Zhu, D., Jia, R., Luo, Q., Zhou, Y., Chen, Z. and Chen, X. 2010. Detection of anatid herpesvirus 1 gC gene by TaqMan™ fluorescent quantitative real-time PCR with specific primers and probe. Virol. J. 7, 37. | ||
| How to Cite this Article |
| Pubmed Style MKAK, Islam MA, Sabuj AAM, Basher MA, Islam MS, Hossain MG, Hossain MT, Saha S, . Molecular Characterization of Duck Plague Virus from selected Haor (Wetland) areas of Bangladesh. Open Vet J. 2021; 11(1): 42-51. doi:10.4314/ovj.v11i1.8 Web Style MKAK, Islam MA, Sabuj AAM, Basher MA, Islam MS, Hossain MG, Hossain MT, Saha S, . Molecular Characterization of Duck Plague Virus from selected Haor (Wetland) areas of Bangladesh. https://www.openveterinaryjournal.com/?mno=129053 [Access: July 27, 2024]. doi:10.4314/ovj.v11i1.8 AMA (American Medical Association) Style MKAK, Islam MA, Sabuj AAM, Basher MA, Islam MS, Hossain MG, Hossain MT, Saha S, . Molecular Characterization of Duck Plague Virus from selected Haor (Wetland) areas of Bangladesh. Open Vet J. 2021; 11(1): 42-51. doi:10.4314/ovj.v11i1.8 Vancouver/ICMJE Style MKAK, Islam MA, Sabuj AAM, Basher MA, Islam MS, Hossain MG, Hossain MT, Saha S, . Molecular Characterization of Duck Plague Virus from selected Haor (Wetland) areas of Bangladesh. Open Vet J. (2021), [cited July 27, 2024]; 11(1): 42-51. doi:10.4314/ovj.v11i1.8 Harvard Style , M. K. A. K., Islam, M. A., Sabuj, A. A. M., Basher, M. A., Islam, M. S., Hossain, M. G., Hossain, M. T., Saha, S. & (2021) Molecular Characterization of Duck Plague Virus from selected Haor (Wetland) areas of Bangladesh. Open Vet J, 11 (1), 42-51. doi:10.4314/ovj.v11i1.8 Turabian Style , Md. Kamrul Ahmed Khan, Md. Alimul Islam, Abdullah Al Momen Sabuj, Md. Abul Basher, Md. Saiful Islam, Md. Golzar Hossain, Muhammad Toffazzal Hossain, Sukumar Saha, and . 2021. Molecular Characterization of Duck Plague Virus from selected Haor (Wetland) areas of Bangladesh. Open Veterinary Journal, 11 (1), 42-51. doi:10.4314/ovj.v11i1.8 Chicago Style , Md. Kamrul Ahmed Khan, Md. Alimul Islam, Abdullah Al Momen Sabuj, Md. Abul Basher, Md. Saiful Islam, Md. Golzar Hossain, Muhammad Toffazzal Hossain, Sukumar Saha, and . "Molecular Characterization of Duck Plague Virus from selected Haor (Wetland) areas of Bangladesh." Open Veterinary Journal 11 (2021), 42-51. doi:10.4314/ovj.v11i1.8 MLA (The Modern Language Association) Style , Md. Kamrul Ahmed Khan, Md. Alimul Islam, Abdullah Al Momen Sabuj, Md. Abul Basher, Md. Saiful Islam, Md. Golzar Hossain, Muhammad Toffazzal Hossain, Sukumar Saha, and . "Molecular Characterization of Duck Plague Virus from selected Haor (Wetland) areas of Bangladesh." Open Veterinary Journal 11.1 (2021), 42-51. Print. doi:10.4314/ovj.v11i1.8 APA (American Psychological Association) Style , M. K. A. K., Islam, M. A., Sabuj, A. A. M., Basher, M. A., Islam, M. S., Hossain, M. G., Hossain, M. T., Saha, S. & (2021) Molecular Characterization of Duck Plague Virus from selected Haor (Wetland) areas of Bangladesh. Open Veterinary Journal, 11 (1), 42-51. doi:10.4314/ovj.v11i1.8 |