| Research Article | ||
Open Vet J. 2023; 13(10): 1290-1298 Open Veterinary Journal, (2023), Vol. 13(10): 1290–1298 Original Research Monitoring of recessive defects associated with low reproductive performance in dairy cattle in UruguayAndrea Branda-Sica1*, Rody Artigas2, Elena de Torres3, Evangelina Kinley3, Paula Nicolini4, María Teresa Federici1 and Silvia Llambí21Instituto Nacional de Investigación Agropecuaria, Sistema Ganadero Extensivo—Salud Animal, Canelones, Uruguay 2Facultad de Veterinaria, Unidad Académica de Genética y Mejora Animal, Universidad de la República, Montevideo, Uruguay 3Facultad de Veterinaria, Campo Experimental N°2, Universidad de la República, San José, Uruguay 4Centro Universitario de Tacuarembó, Instituto Superior de la Carne, Universidad de la República, Tacuarembó, Uruguay *Corresponding Author: Andrea Branda-Sica. Instituto Nacional de Investigación Agropecuaria, Sistema Ganadero Extensivo—Salud Animal, Canelones, Uruguay. Email: abranda [at] inia.org.uy Submitted: 02/07/2023 Accepted: 12/09/2023 Published: 31/10/2023 © 2023 Open Veterinary Journal
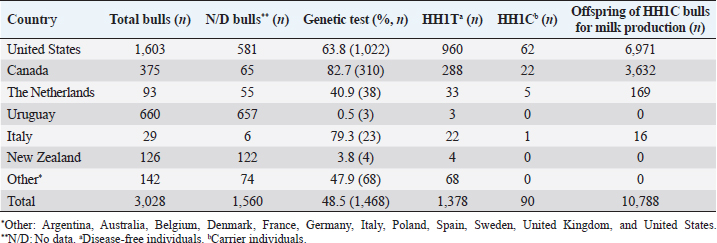
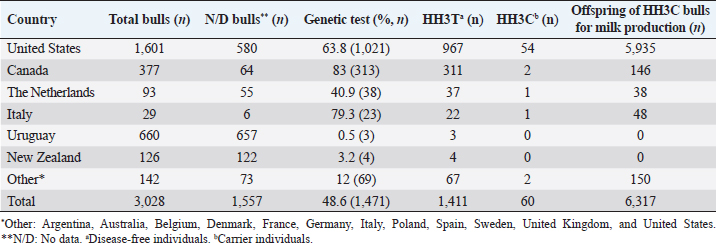
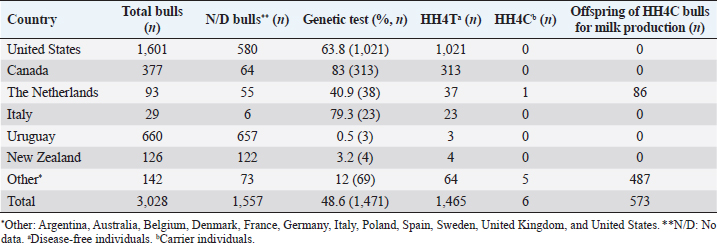
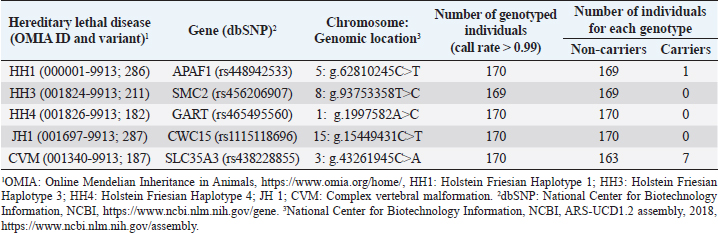
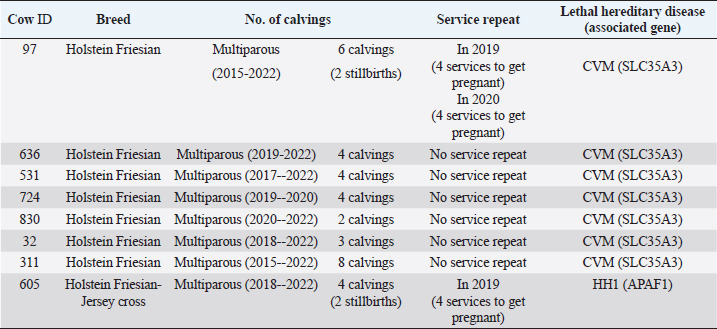
AbstractBackground: Most dairy cattle breeds originate show an average generational inbreeding rate of 1%, which favors the occurrence of recessive defects associated with low reproductive performance. Aim: The objective of this study was to monitor recessive defects associated with low reproductive performance in dairy cattle. Methods: To monitor bulls carrying the Holstein Friesian haplotype (HH) 1, HH3, and HH4 haplotypes, we analyzed the records of 3,028 national and imported Holstein Friesian bulls from the 2021 updated sires’ catalog published by “Evaluaciones Genéticas Lecheras”; and to determine the presence of these mentioned haplotypes, as well as Jersey haplotype (JH) 1 and complex vertebral malformation (CVM), were genotype with the GeneTitan® 2,500 single nucleotide polymorphism (SNP) bovine chip, estimate their frequencies and evaluate their impact on the fertility of 100 Holstein Friesian cows and 70 Holstein Friesian-Jersey crosses belonging to an experimental dairy. Results: From a total of 1,468 (48.5%) bulls with genetic information from the sires’ catalog for HH1 and 1,471 (48.6%) for HH3 and HH4, we found 90 (6.1%) carriers for HH1, 60 (4.1%) for HH3, and 6 (0.4%) for HH4, respectively. By genotyping with the chip, we calculated the herd frequency of the mutant alleles and herd prevalence of carriers for HH1 and CVM as q=0.003 and 0.022; 0.59% and 4.3% (call rate >0.99), respectively. No mutant alleles were found for HH3, HH4, and JH1 in the analyzed population. We examined reproductive data by observing the presence of CVM and HH1 mutant alleles in repeat cows with an average of four services to achieve pregnancy. Conclusion: This study demonstrated the presence of recessive defects associated with low reproductive performance in the analyzed population, which can affect the health and productivity of dairy cattle. Therefore, cows and bulls should be closely monitored through genetic testing to lower the incidence of recessive defects in dairy cattle. Keywords: Artificial insemination, Bos taurus, Fertility, Hereditary diseases. IntroductionMost dairy cattle breeds originate from a small number of founder animals through artificial insemination with a strong selection for a few productive traits (Fritz et al., 2013; Liebig et al., 2022). Consequently, these breeds show an average generational inbreeding rate of 1% that favors the appearance of recessive defects responsible for embryonic, fetal, and perinatal mortality because it increases homozygosity levels (Van Raden et al., 2011b; Fritz et al., 2013). The implementation of routine genotyping of hundreds and thousands of animals within popular US dairy breeds using single nucleotide polymorphism (SNP) chips has provided comprehensive genomic data for all current breeding populations (Van Raden et al., 2011b; Howard et al., 2017). Van Raden et al. (2011b) were the first to propose examining this type of data to identify genomic regions in homozygosity-deficient animals. In their work, they identified and listed five haplotypes out of 11 candidates with detrimental effects on fetal development, which could lead to abortion and fetal death in three dairy cattle breeds: Holstein Friesian (HH1, HH3, and HH4), Jersey (JH1), and Brown Swiss (BH1). After two years, other new lethal haplotypes were reported in Holstein Friesian cattle, and in the meantime, some were solved at the molecular level, such as HH1 ( APAF1, Adams et al., 2016), HH3 ( SMC2, Daetwyler et al., 2014; McClure et al., 2014), HH4 ( GART, Fritz et al., 2013) and HH5 ( TFB1M, Schütz et al., 2016). In the Danish Holstein population, a haplotype associated with a missense variant in the SLC35A3 gene that causes complex vertebral malformation (CVM) was identified (Thomsen et al., 2006). It was demonstrated that, in the homozygous state, the mutation typically results in intrauterine death. Sonstegard et al. (2013) identified the mutation in the CWC15 gene that is associated with the JH1 haplotype and negatively impacts reproduction in Jersey cattle. Recently, Häfliger et al. (2022) updated the SNP positions from the new Bos taurus genome version ARS-UCD1.2 (NCBI, ARS-UCD1.2 assembly, 2018; Rosen et al., 2020) in the genomic regions of the haplotypes previously identified by other authors (Thomsen et al., 2006; Sonstegard et al., 2013; Daetwyler et al., 2014; McClure et al., 2014; Adams et al., 2016). For all bovine breeds, 54 haplotypes with homozygous deficits have been reported; in 25 of them, the mutations associated with the responsible genes have been identified (OMIA, 2011). In the Holstein Friesian breed, 11 haplotypes have been identified; these mutations reduce the pregnancy rate and produce an increase in stillbirths and spontaneous abortion that aggravate the already critical situation of fertility loss (Van Raden et al., 2011b; Fritz et al., 2013; McClure et al., 2014; Adams et al., 2016; Schütz et al., 2016; Fritz et al., 2018; Hozé et al., 2020; Häfliger et al., 2022). Given the economic importance of producing offspring for the selection of breeders and generating genetic influence of the resulting cows, the main objectives of this study were: (a) to monitor bulls with genetic information for the haplotypes HH1, HH3, and HH4, which are associated with low reproductive efficiency by analyzing the 2021 Holstein Friesian sires’ catalog; and (b) to determine the presence of the mutant alleles of these haplotypes, as well as JH1 and CVM, with the GeneTitan® 2,500 SNP bovine chip of Affymetrix platform. We calculated their frequencies and evaluated their impact on fertility in Holstein Friesian cows and Holstein Friesian-Jersey crosses from a dairy farm of the experimental field of the Veterinary Faculty of the “Universidad de la República” (Libertad, San José, Uruguay). Materials and MethodsAnalysis of the 2021 Holstein Friesian sires’ catalogWe analyzed the records of 3,028 national and imported Holstein Friesian bulls, born between 1964 and 2016, from the 2021 updated sire catalog published by “Evaluaciones Genéticas Lecheras,” available at www.geneticalechera.com.uy. Each record was evaluated using the following national and international databases: (a) ABS Global: https://absbullsearch.absglobal.com/; (b) DairyNZ: https://www.dairynz.co.nz/animal/animal-evaluation/bull-team/; and (c) “Evaluaciones Genéticas Lecheras (Uruguay).” Sampling and genetic materialDNA from 100 Holstein Friesian cows and 70 Holstein Friesian-Jersey crosses was extracted from blood using the Wizard® Genomic DNA Purification Kit (Promega) following the manufacturer’s specifications. DNA concentration, quality, and purity were assessed with Quant-iT™ PicoGreen at 260 nm (Thermo Scientific, USA), considering an OD260/OD280 ratio between 1.8 and 2.0 and an OD260/OD230 ratio greater than 1.5. Genetic analysis of genomic data with the GeneTitan® bovine chip.These DNA samples were genotyped at “Genexa ADN Evolutivo” (Montevideo, Uruguay) with the GeneTitan® 2,500 SNP bovine chip, with Axiom Microarray Genotyping Technology platform from Affymetrix. With these DNA samples, Axiom technology was fine-tuned using the B. taurus reference genome sequence version, ARS-UCD1.2 (NCBI, UCSC bosTau9 version, ARS-UCD1.2 assembly, 2018; Rosen et al., 2020; Häfliger et al., 2022), which is more accurate and has been unified for more effortless transfer and interpretation of results. The Affymetrix platform consists of small DNA molecules attached to fixed locations on the chip, which recognize the specific alleles of an SNP. The different alleles or nucleotides are detected by differential hybridization of the DNA sample. In order to identify those Holstein Friesian cows and Holstein Friesian-Jersey crosses carriers of the hereditary diseases: HH1 (rs448942533), HH3 (rs456206907), HH4 (rs465495560), JH1 (rs1115118696), and CVM (rs438228855), we classified as carriers those genotypes with a call rate greater than 99%, thus ensuring the high quality of the genomic data of the analyzed population. Allelic and genotypic frequencies were calculated by direct counting according to the method of Nei (1987). Evaluation of reproductive dataWe evaluated reproductive data for Holstein Friesian and Holstein Friesian-Jersey crossbred cows carrying the allelic variants previously identified by genotyping with the GeneTitan® bovine chip. These cows have had conventional reproductive management with predominant artificial insemination, gestation diagnosis no later than 70 days post-insemination, and no hormonal treatments during the experimental period. Data were provided through the diary of the experimental field of the Veterinary Faculty. Ethical approvalAll procedures were carried out following the rules and standards of the Ethics Committee on the Use of Animals and approved by the “Comisión Honoraria de Experimentación Animal de la Universidad de la República,” Uruguay (protocol from CEUA n° 1526). ResultsAnalysis of the 2021 Holstein Friesian sires’ catalogWe analyzed the Holstein Friesian sires’ catalog of the genetic evaluations updated to 2021, observing that 87.12% (n=2,638) of the genetic contribution came mainly from the US, Canada, and Uruguay for haplotypes HH1 (Table 1), HH3 (Table 2), and HH4 (Table 3). Of the 3,028 bulls analyzed, only 48.5% (n=1,468) had a recorded genetic test for the HH1 haplotype (Table 1), and another 48.6% (n=1,471) for the HH3 (Table 2) and HH4 (Table 3) haplotypes. Of these, 6.1% (n=90) were carriers for haplotype HH1 (HH1C) from the US, Canada, Netherlands, and Italy; 4.1% (n=60) for HH3 (HH3C) from the US, Canada, the Netherlands, Italy, and other countries; and 0.4% (n=6) for HH4 (HH4C) from the Netherlands and other countries. When evaluating the progeny of HH1C bulls for dairy production in Uruguay, we observed there were 10,788 daughters included in the national genetic evaluation system (Table 1). We also observed them for HH3C and HH4C bulls, with 6,317 (Table 2) and 573 daughters (Table 3), respectively. Genotyping with the GeneTitan® bovine chipGenotyping detected the following variants: HH1 in a single Holstein Friesian-Jersey cross cow, and CVM in seven Holstein Friesian cows in a total of 170 genotyped cows (call rate >0.99) (Table 4). The frequency of the mutant allele for HH1 in the population analyzed was low, q =0.003; and the prevalence of carriers was 0.59% (call rate >0.99). For CVM, it was q =0.022 and 4.3% (call rate >0.99). No mutant alleles were found for HH3, HH4, and JH1 in the analyzed population. Analysis of reproductive data of cows carrying the associated mutationsWe analyzed the reproductive data using the number of inseminations in cows carrying the mutations that had been genotyped with the GeneTitan® bovine chip. Two cows, CVM and HH1 carriers, gave birth to dead calves; these cows had an average of four service repeats to achieve pregnancy (Table 5). Table 1. Genetic status for the HH1 haplotype of bulls used as sires in Uruguayan dairy genetic evaluation systems until 2021 according to country of origin.
Table 2. Genetic status for the HH3 haplotype of bulls used as sires in Uruguayan dairy genetic evaluation systems until 2021 according to country of origin.
Table 3. Genetic status for the HH4 haplotype of bulls used as sires in Uruguayan dairy genetic evaluation systems until 2021 according to country of origin.
Table 4. Number of individuals genotyped free and carriers of lethal hereditary diseases using the GeneTitan® bovine chip; the genes associated with their identification in NCBI, and their location in the genome.
Table 5. Number of service repeats and calving in cows carrying the associated mutations previously identified with the GeneTitan® bovine chip.
DiscussionThe sires’ catalog of the dairy genetic evaluations includes bulls with at least one daughter included in the genetic evaluation system for the Holstein Friesian breed in Uruguay. After analysis of the catalog, it appears a strong component of genetic material mainly from the US and Canada; these had been already found in other investigations (Artigas et al., 2020) that showed a predominance of these countries, where presumably the founding mutations of the HH1 and HH3 haplotypes originated (Van Raden et al., 2011b; Van Raden et al., 2012; Fritz et al., 2013; Fritz et al., 2018; Häfliger et al., 2022). It has been noted that the HH4 mutation came from Europe (Fritz et al., 2013; Cole et al., 2016), as it was not found in the bulls from the United States and Canada from the dairy genetic evaluation. After analyzing all the bulls with genetic information in the Holstein Friesian breed gene pool, we detected 90 animals carrying the HH1 haplotype (HH1C=6.1%), 60 carrying HH3 (HH3C=4.1%), and six carrying HH4 (HH4C=0.4%). The number of bulls carrying these haplotypes could be higher, given the low number of animals with genetic information available in public databases regarding their genetic status for the disease, including 99.5% of Uruguayan Holstein Friesian bulls. A similar pattern has been found for brachyspina inherited disease (Artigas et al., 2020). The high proportion of bulls without molecular diagnostic tests in the sire catalog can be explained by the fact that this analysis includes bulls before diagnostic tests for mutations in the APAF1 (HH1), SCM2 (HH3), and GART (HH4) genes were published (Fritz et al., 2013; McClure et al., 2014; Adams et al., 2016; Cole et al., 2016). Bulls’ carriers of the HH1C mutation from the 2021 sires´ catalog contributed genetically to Holstein Friesian breed improvement programs, having many daughters (n=10,788) in national dairy genetic evaluation systems. The same happened with the daughters of HH3C (n=6,317) and HH4C (n=573) bulls. Since the inheritance mechanism is autosomal recessive, we assume that, on average, 50% of the daughters (n=5,394) were HH1C carriers, just as for HH3C (n=3,158) and HH4C (n=286), thus indicating the presence of the mutant alleles in the Uruguayan selection nucleus. Since all daughters (n=10,788 for HH1C; n=6,317 for HH3C; n=573 for HH4C) were evaluated for milk production traits, they left offspring at the national level. Among the 26 HH1C bulls used in Uruguay, stand out O-Bee Manfred Justice (HOLUSAM000122358313), Tesk Holm Valiant Rockie (HOLUSAM000001841366), Lemax Pawnee Memorial (HOLUSAM000001765326), End-Road PVF Boliver (HOLUSAM000123586443) and MJR Blackstar Emory (HOLUSAM000002114601), direct descendants of Walkway Chief Mark (HOLUSAM000001773417), Milu Betty Ivanhoe Chief (HOLUSAM000001578139), SWD Valiant (HOLUSAM000001650414), and Pawnee Farm Arlinda Chief (HOLUSAM000001427381), which are the main disseminators of the mutation in the APAF1 (apoptotic peptidase activating factor) gene that blocks approximately one-third of the encoded protein (Larkin et al., 2012; Van Raden et al., 2012; Adams et al., 2016; Cole et al., 2016). A substitution of a thymine for a cytosine (c.1741C>T) characterizes this mutation (Adams et al., 2016). This mutation causes fetal and embryonic loss between 60 and 200 days of gestation and a reduced conception rate in cows; consequently, fertility is reduced in carrier bulls (Adams et al., 2016). Of the HH3C bulls used in Uruguay, stands out Arlinda Melwood (HOLUSAM000001879149), a direct descendant of Glendell Arlinda Chief (HOLUSAM000001556373), another major disseminator of the SMC2 mutation (McClure et al., 2014; Cole et al., 2016). This mutation in the SMC2 (structural maintenance of chromosomes) gene is characterized by a thymine-to-cytosine substitution (c.3404T>C) in exon 24 (Daetwyler et al., 2014; McClure et al., 2014). Häfliger et al. (2022) updated the position of the SNP responsible for this mutation (rs45206907) on chromosome 8 (93,753,358) according to the new version of the reference genomic sequence assembly (NCBI, ARS-UCD1.2 assembly, 2018). The SMC2 gene plays an essential role in DNA repair and chromosome condensation and segregation during cell division. This point mutation changes amino acid 1135 from phenylalanine to serine (F1135S) and alters the function of the NTPase domain of the encoded protein. Daetwyler et al. (2014) analyzed 1,000 bull genomes to identify the SMC2 mutation, to which embryonic loss was ascribed as a negative effect. Of the HH4C used in Uruguay, Jocko Besne (HOLFRAM005694028588) is a son of Besne Buck (HOLFRAM004486041658) from Europe, from which he inherited the GART mutation (Fritz et al., 2013; Cole et al., 2016). The GART enzyme phosphoribosylglycinamide synthetase is required for purine nucleotide synthesis. The GART mutation is associated with decreased fertility and embryonic mortality in the first month of gestation (Fritz et al., 2013). Bulls carrying haplotypes HH1 (APAF1), HH3 (SMC2), and HH4 (GART) have been used to produce offspring for the selection of sires to be used in artificial insemination, thus having a substantial genetic influence on the resulting cow population in the different dairy farms in Uruguay. Although the mutations associated with haplotypes HH1 (APAF1), HH3 (SMC2), HH4 (GART), JH1 (CWC15), and CVM (SLC35A3) were identified 10 years ago, this study was the first report in Uruguay that evaluates reproductive data genomic data from genotyping with the GeneTitan® bovine chip, which allowed a much more accurate imputation compared to that of other commercial chips. This technology allowed the detection of two mutations associated with the APAF1 (HH1) and SLC35A3 (CVM) genes, which affect fetal development and may cause abortion and fetal death in Holstein Friesian cows and Holstein Friesian-Jersey crosses. In the case of the APAF1 (HH1) mutation, we obtained a herd prevalence of 0.59% with a call rate close to 100%. This value was low compared to that reported by Briano-Rodríguez et al. (2021) (4.4%), who performed genotyping with the commercial GeneSeek Genomic Profiler -GGP- Bovine 50K chip, with which they detected 16 Holstein Friesian calves carrying the APAF1 (HH1) mutation in eight dairy farms in Uruguay. Therefore, the APAF1 (HH1) mutation was still dispersed in Uruguay’s Holstein Friesian dairy herd. The prevalence of APAF1 (HH1) carrier cows in Germany was similar to that of this study (1.8%, Schütz et al., 2016). In the USA, Holstein Friesian cows carrying the APAF1 (HH1) mutation were found at a higher frequency of 4.5% and 2.25% (Van Raden et al., 2011b; Adams et al., 2016, respectively). However, it had been observed that between 1980 and 1990, the frequency of carrier cows was 0.08, with a subsequent decrease to approximately 0.03 in 2010 (Van Raden et al. 2011b). VanRaden et al. (2011a) determined the negative effect of this haplotype on reproduction, estimating an impact on conception rate of -0.35%, and evaluated the 60-day non-return rate (NR60), which was -1.1. It was estimated that this mutation caused more than 5,00,000 abortions and a loss of $ 420 million to the dairy industry (Adams et al., 2016). The SLC35A3 (CVM) mutation was reported in the OMIA catalog (001340-9913) and has been studied in cattle of the Holstein Friesian breed since 2006 (Thomsen et al., 2006; Ruść and Kamiński, 2007; Meydan et al., 2010) and traced back to the elite North American sire Carlin-M Ivanhoe Bell (HOUSA000001667366), who received the lethal recessive mutation from his sire Penstate Ivanhoe Star (HOLUSAM000001441440) (Chu et al., 2008). CVM disease was primarily caused by a defining mutation in the membrane transporter protein UDP-N-acetyl glucosamine of the SLC35A3 gene (Thomsen et al., 2006) and results in the substitution of valine for phenylalanine (V180F) at position 180 (Ruść and Kamiński, 2007). The genotyping of the SNP responsible for this disease (rs438228855, located at position 43,261,945 of bovine chromosome 3) was performed with the GeneTitan® bovine chip. The genotyping was highly accurate and efficient, with a herd prevalence of carrier cows of 4.3% (with a call rate of 100%). This prevalence was intermediate compared to other reports made in Uruguay using other genotyping PCR-HRM/melting methodologies (6.45%, Branda-Sica et al., 2019) and GeneSeek Genomic Profiler -GGP- 50K bovine chip (2.09%, Briano-Rodriguez et al., 2021). This study found that the herd prevalence of CVM carriers was similar to that of cows in Turkey (3.4%, Meydan et al., 2010). Nevertheless, the prevalence was low when compared with other countries, such as Denmark (31.0%, Thomsen et al., 2006), Poland (24.8%, Rusc, and Kaminski, 2007), and Japan (13.0%, Ghanem et al., 2008). The reason for these differences was differences in the regions of origin of the sampled population, although it might also be influenced using semen from bulls’ carriers of the mutation over time. Most countries have developed improvement programs to decrease the frequency of CVM carriers in the cattle population (Ruść and Kamiński, 2007). Hence, in some Holstein Friesian populations in Uruguay, the frequency of CVM carriers still seems to be high regarding the other autosomal recessive mutations. Acquiring repeat breeder cows that are carriers of CVM and HH1 may be attributed to ovarian dysfunction (Ghanem et al., 2017). In a previous study, Ghanem et al. (2010) found that progesterone concentration tended to decrease during the estrous cycle in CVM carrier cows due to luteal dysfunctions, which could reduce the conception rate and increase the frequency of repeat cow syndrome. Due to the above reasons, Ghanem et al. (2017) studied fertility in APAF1 mutation (HH1) carrier cows by measuring progesterone concentration in skim milk as a tool to indicate an ovarian function in cows, finding that progesterone concentrations did not differ significantly (p =0.8) between HH1 carrier and non-carrier cows at the first 45 days after artificial insemination. All cows validated in this study had a normal homozygous genotype for the diseases associated with haplotypes HH3 (SMC2), HH4 (GART), and JH1 (CWC15), with no mutant alleles identified in the population analyzed. However, some studies associated the lethal effects of HH3 (SMC2), HH4 (GART), and JH1 (CWC15) haplotypes with embryonic losses immediately after conception, which are not phenotypically observable; nevertheless, the reduction of conception rate associated with these mutations occurs before 60 days, while embryonic losses associated to HH1 (APAF1) and CVM occur throughout the entire gestation (Van Raden et al., 2011a; Van Raden et al., 2012; Norman et al., 2012; Fritz et al., 2013). Van Raden et al. (2012) reported an HH3 heterozygous cows (SMC2) frequency of 4.7% and a 60-day non-return rate (NR60) of -3.1% for this haplotype. Häfliger et al. (2022) detected a purely recessive embryonic lethal effect of SMC2 mutation that allows indirect detection in the non-return rate in Canadian Holstein Friesian heifers. In Germany, Schütz et al. (2016) detected HH3 and HH4 carriers with frequencies of 5.1% and 4%, respectively. Fritz et al. (2013) reported frequencies of 3.6% for HH4 haplotype, which was associated with a reduction in births in French Holstein Friesian cows, where they observed a reduction in calving rate of -5.8% in heifers and -1.74% in carrier cows when mated to carrier males and daughters of HH4 carrier males. In Uruguay, Briano-Rodriguez et al. (2021) found mutant alleles of HH3 (3.13%) and HH4 (1.04%) haplotypes in a population of Holstein Friesian calves born in 2016. Today, these calves would have become breeders with genetic information and should have been registered in the catalog of parents of the dairy genetic evaluation of the Holstein Friesian breed. In any case, the female calves could have remained as replacements for the farm. For the Jersey breed, there are no reports on the presence of JH1 (CWC15) disease carriers in Uruguay. This disease was reported by US ARS-USDA scientists in 2013 (Sonstegard et al., 2013), and its origin was traced to the elite North American bull Observer Chocolate Soldier (JERUSAM000000596832), born in 1962 (Van Raden et al., 2011a). This bull sired 1,454 daughters for milk production, and consequently, its impact was further magnified with 107 sons and 715 grandsires used in artificial insemination to generate >50,000 granddaughters and >2,00,000 great-granddaughters. As a result, the frequency of JH1 heterozygotes experienced an initial rapid increase, remaining stable at 20%–25% since 1980. Van Raden et al. (2004) examined 52,449 fertility records of JH1 heterozygous bulls mated to daughters of JH1 heterozygous bulls and found a conception success rate of 33.3% versus 37.0% for 2,90,373 recent mating of normal bulls to daughters of normal bulls. The reduction in conception rate of 3.7% confirmed JH1 as a recessive lethal disease, in rough agreement with the expected reduction of 4.6%. The conception success rate was 36.3% in 57,523 mating of heterozygous bulls to daughters of normal bulls, with a slight reduction of 0.7% potentially caused by heterozygous dams with JH1 maternal inheritance (Sonstegard et al., 2013). ConclusionThis study demonstrated the presence of recessive defects associated with low reproductive performance in the analyzed population, which can have a negative impact on fertility, calf viability, and overall herd productivity. Therefore, cows and bulls should be closely monitored through genetic testing to lower the incidence of recessive defects in dairy cattle. Furthermore, it is essential to implement preventive measures such as genetic counseling for producers, education on the consequences of these defects, and promoting responsible use of assisted reproduction to minimize the risk of transmitting these undesirable traits. It is recommended to conduct studies aimed at quantifying the economic losses in dairy farms with the presence of Holstein Friesian haplotypes. In addition, pedigree and genotyping studies of animals should be carried out as a tool to prevent mating between carriers of certain haplotypes. To prevent the spread of JH1 haplotype, it is advisable to analyze the limited set of 90 records from domestic and imported Jersey bulls born between 1978 and 2012. This data can be sourced from the dairy genetic evaluation available at www.geneticalechera.com.uy. The analysis should aim to identify the presence of the mutant allele within the Jersey 2022 sires’ catalog. AcknowledgmentsThe authors are thankful to the National Agency for Research and Innovation (ANII), the National Institute for Agricultural Research (INIA), and the Sectorial Commission for Scientific Research of the University of the Republic (CSIC-UdelaR) for the support and funding of this research study. The authors appreciate the technical support received from our collaborators, Federico Giannitti and Mariana Ramírez. To the translation agency DNA Translations for the translation into English. Conflict of interestThe authors declare that there is no conflict of interest regarding the publication of this article. Authors contributionsA.B.S. and S.L. designed the experimental procedures. A.B.S., R.A., E.T., E.K., and S.L. performed the experimental work. A.B.S., R.A., P.N., and S.L. conceptualized and designed the study. All authors read and approved the final manuscript. FundingThe research leading to the results presented in this publication received funding from ANII (POS_NAC_2017_1_141239), INIA (projects PL_027 N-23398 and BT_21 N-23330), and CSIC-UdelaR. Data availabilityThe data supporting the findings of this study are available within the manuscript. Any other data are available from the corresponding author upon reasonable request. ReferencesAdams, H.A., Sonstegard, T.S., VanRaden, P.M., Null, D.J., Van Tassell, C.P., Larkin, D.M., and Lewin, H.A. 2016. Identification of a nonsense mutation in APAF1 that is likely causal for a decrease in reproductive efficiency in Holstein dairy cattle. J. Dairy Sci. 99(8), 6693–6701. Artigas, R., Federici, M.T., Vázquez, N., Alcántara, M., Ramírez, M., Guerra, S., Dutra, F., and Llambí, S. 2020. Identificación por catálogo y detección molecular de bovinos Holstein portadores de braquiespina en Uruguay. FAVE Cs. Vet. 19(2), 50–54. Branda-Sica, A., Nicolini, P., Federici, M.T., and Llambí, S. 2019. Identification of Holstein cow carriers of complex vertebral malformation by high resolution melting curves (HRM). Arch. Vet. Sci. 24(4), 62–70. Briano-Rodríguez, C., Romero, A., Llambí, S., Branda-Sica, A., Federici, M., Giannitti, F., Caffarena, D., Schild, C., Casaux, M.L., and Dutra, F. 2021. Lethal and semi-lethal mutations in Holstein calves in Uruguay. [Mutações letais e semi-letais em bezerros da raça Holandesa no Uruguai]. Animal Production, Cienc. Rural. 51(7), e20200734. Chu, Q., Sun, D., and Yu, Y. 2008. Identification of complex vertebral malformation carriers in Chinese Holstein. J. Vet. Diagn. Invest. 20(2), 228–230. Cole, J.B., Null, D.J., and VanRaden, P.M. 2016. Phenotypic and genetic effects of recessive haplotypes on yield, longevity, and fertility. J. Dairy Sci. 99(9), 7274–7288. Daetwyler, H.D., Capitan, A., Pausch, H., Stothard, P., van Binsbergen, R., Brondum, R., Liao, X., Djari, A., Rodriguez, S.C., Grohs, C., Esquerré, D., Bouchez, O., Rossignol, M.N., Klopp, C., Rocha, D., Fritz, S., Eggen, A., Bowman, A., Coote, D., Chamberlain, A.J., Anderson, C., Van Tassell, C.P., Hulsegge, I., Goddard, M.E., Guldbrandtsen, B., Lund, M.S., Veerkamp, R.F., Boichard, D.A., Fries, R., and Hayes, B.J. 2014. Whole genome sequencing of 234 bulls facilitates maping of monogeneic and complex traits in cattle. Nat. Genet. 46(8), 858–865. Fritz, S., Hoze, C., Rebours, E., Barbat, A., Bizard, M., Chamberlain, A., Escouflaire, C., Vander Jagt, C., Boussaha, M., Grohs, C., Allais-Bonnet, A., Philippe, M., Vallée, A., Amigues, Y., Hayes, B.J., Boichard, D., and Capitan, A. 2018. An initiator codon mutation in SDE2 causes recessive embryonic lethality in Holstein cattle. J. Dairy Sci. 101(7), 6220–6231. Fritz, S., Capitan, A., Djari, A., Rodriguez, S.C., Barbat, A., Baur, A., Grohs, C., Weiss, B., Boussaha, M., Esquerré, D., Klopp, C., Rocha, D., and Boichard, D. 2013. Detection of Haplotypes associated with prenatal death in dairy cattle and identification of deleterious mutations in GART, SHBG and SLC37A2. PLoS One. 8(6), e65550. Ghanem, M.E., Akita, M., Suzuki, T., Kasuga, A., and Nishibori, M. 2008. Complex vertebral malformation in Holstein cows in Japan and its inheritance to crossbred F1 generation. Anim. Reprod. Sci. 103(3–4), 348–354. Ghanem, M.E., Nishibori, M., Isobe, N., and Hisaeda, K. 2017. Detection of the APAF1 mutation in Holstein cows and mummified fetuses in Japanese dairy herds. Reprod. Domest. Anim. 53(1), 137–142. Ghanem, M.E., Suzuki, T., Kasuga, A., and Nishibori, M. 2010. Effect of complex vertebral malformation on luteal function in Holstein cows during oestrous cycle and early pregnancy. Reprod. Domest. Anim. 45(4), 729–733. Häfliger, I.M., Spengeler, M., Seefried, F.R., and Drögemüller, C. 2022. Four novel candidate causal variants for deficient homozygous haplotypes in Holstein cattle. Sci. Rep. 12(1), 5435. Howard, J.T., Pryce, J.E., Baes, C., and Maltecca, C. 2017. Invited review: inbreeding in the genomics era: inbreeding, inbreeding depression, and management of genomic variability. J. Dairy Sci. 100(8), 6009–6024. Hozé, C., Escouflaire, C., Mesbah-Uddin, M., Barbat, A., Boussaha, M., Deloche, M.C., Boichard, D., Fritz, S., and Capitan, A. 2020. Short communication: a splice site mutation in CENPU is associated with recessive embryonic lethality in Holstein cattle. J. Dairy Sci. 100(1), 607–612. Larkin, D.M., Daetwyler, H.D., Hernandez, A.G., Wright, C., Hetrick, L.A., Boucek, L., Bachman, S.L., Band, M.R., Akraiko, T.V., Cohen-Zinder, M., Thimmapuram, J., Macleod, I.M., Harkins, T.T., McCague, J.E., Goddard, M.E., Hayes, B.J., and Lewin, H.A. 2012. Wholegenome resequencing of two elite sires for the detection of haplotypes under selection in dairy cattle. PNAS. 109(20), 7693–7698. Liebig, B.E., Bishop, J.V., McSweeney, K.D., Van Campen, H., González-Berrios, C.L., Hansen, T.R. and Thomas, M.G. 2022. Direct genomic value daughter pregnancy rate and services per conception are associated with characteristics of day-16 conceptuses and hormone signaling for maternal recognition of pregnancy in lactating Holstein cows. Appl. Anim. Sci. 38(2), 157–169. McClure, M., Bickhart, D., Null, D., VanRaden, P., Xu, L., Wiggans, G., Liu, G., Schroeder, S., Glasscock, J., Armstrong, J., Cole, J.B., Van Tassell, C.P., and Sonstegard, T.S. 2014. Bovine exome sequence analysis and targeted SNP genotyping of recessive defects BH1, HH2 and HH3 reveal a putative causative mutation in SMC2 for HH3. PLoS One. 9(3), e92769. Meydan, H., Yildiz, M.A., and Agerholm, J.S. 2010. Screening for bovine leukocyte adhesion deficiency, deficiency of uridine monophosphate synthase, complex vertebral malformation, bovine citrullinaemia, and factor XI deficiency in Holstein cows reared in Turkey. Acta Vet. Scand. 52, 56. NCBI, ARS-UCD1.2 assembly. 2018. National Center for Biotechnology Information, ARS-UCD1.2 Assembly. https://www.ncbi. nlm.nih.gov/assembly/GCF_002263795.1/ Nei, M. 1987. Molecular evolutionary genetics. Nueva York: Columbia University Press, p: 512. Norman, H.D., Miller, R.H., Wright, J.R., Hutchison, J.L. and Olson, K.M. 2012. Factors associated with frequency of abortions recorded through dairy herd improvement test plan. J. Dairy Sci. 95(7), 4074–4084. OMIA. 2011. Online Mendelian Inheritance in Animals, Faculty of Veterinary Science, University of Sydney, Australia. https://www.omia.org/home/ Rosen, B.D., Bickhart, D.M., Schnabel, R.D., Koren, S., Elsik, C.G., Tseng, E., Rowan, T.N., Low, W.Y., Zimin, A., Couldrey, C., Hall, R., Li, W., Rhie, A., Ghurye, J., McKay, S.D., Thibaud-Nissen, F., Hoffman, J., Murdoch, B.M., Snelling, W.M., McDaneld, T.G., Hammond, J.A., Schwartz, J.C., Nandolo, W., Hagen, D.E., Dreischer, C., Schultheiss, S.J., Schroeder, S.G., Phillippy, A.M., Cole, J.B., Van Tassell, C.P., Liu, G., Smith, T.P.L., and Medrano, J.F. 2020. De novo assembly of the cattle reference genome with single-molecule sequencing. GigaScience. 9(3), giaa021. Ruść, A., and Kamiński, S. 2007. Prevalence of complex vertebral malformation carriers among polish Holstein Friesian bulls. J. Appl. Genet. 48(3), 247–252. Schütz, E., Wehrhahn, C., Wanjek, M., Bortfeld, R., Wemheuer, W.E., Beck, J., and Brenig, B. 2016. The Holstein Friesian Lethal Haplotype 5 (HH5) results from a complete deletion of TFB1M and Cholesterol Deficiency (CDH) from an ERV-(LTR) insertion into the coding region of APOB. PLoS One. 11(6), e0154602. Sonstegard, T.S., Cole, J.B., VanRaden, P.M., Van Tassell, C.P., Null, D.J., Schroeder, S.G., Bickhart, D., and McClure, M.C. 2013. Identification of a nonsense mutation in CWC15 associated with decreased reproductive efficiency in Jersey cattle. PLoS One. 8(1), e54872. Thomsen, B., Horn, P., Panitz, F., Bendixen, E., Petersen, A.H., Holm, L.E., Nielsen, V.H., Agerholm, J.S., Arnbjerg, J., and Bendixen, C. 2006. A missense mutation in the bovine SLC35A3 gene, encoding a UDP-N-acetylglucosamine transporter, causes complex vertebral malformation. Genome Res. 16(1), 97–105. Van Raden, P.M., Null, D.J., Sonstegard, T.S., Adams, H.A., Van Tassell, C.P., and Olson, K.M. 2012. Fine mapping and discovery of recessive mutations that cause abortions in dairy cattle. J. Dairy Sci. 95(Suppl. 2), ii–iii(abstract LB6). Van Raden, P.M., Null, D.J., Olson, K.M., and Hutchison, J.L. 2011a. Reporting of haplotypes with recessive effects on fertility. Interbull Bulletin, 44, 117–121. Van Raden, P.M., Null D.J., Olson, K.M., and Hutchison, J.L. 2011b. Harmful recessive effects on fertility detected by absence of homozygous haplotypes. J. Dairy Sci. 94(12), 6153–6161. | ||
| How to Cite this Article |
| Pubmed Style Branda-Sica A, Artigas R, Torres Ed, Kinley E, Nicolini P, Federici MT, Llambí S. Monitoring of recessive defects associated with low reproductive performance in dairy cattle in Uruguay. Open Vet J. 2023; 13(10): 1290-1298. doi:10.5455/OVJ.2023.v13.i10.8 Web Style Branda-Sica A, Artigas R, Torres Ed, Kinley E, Nicolini P, Federici MT, Llambí S. Monitoring of recessive defects associated with low reproductive performance in dairy cattle in Uruguay. https://www.openveterinaryjournal.com/?mno=159408 [Access: July 27, 2024]. doi:10.5455/OVJ.2023.v13.i10.8 AMA (American Medical Association) Style Branda-Sica A, Artigas R, Torres Ed, Kinley E, Nicolini P, Federici MT, Llambí S. Monitoring of recessive defects associated with low reproductive performance in dairy cattle in Uruguay. Open Vet J. 2023; 13(10): 1290-1298. doi:10.5455/OVJ.2023.v13.i10.8 Vancouver/ICMJE Style Branda-Sica A, Artigas R, Torres Ed, Kinley E, Nicolini P, Federici MT, Llambí S. Monitoring of recessive defects associated with low reproductive performance in dairy cattle in Uruguay. Open Vet J. (2023), [cited July 27, 2024]; 13(10): 1290-1298. doi:10.5455/OVJ.2023.v13.i10.8 Harvard Style Branda-Sica, A., Artigas, . R., Torres, . E. d., Kinley, . E., Nicolini, . P., Federici, . M. T. & Llambí, . S. (2023) Monitoring of recessive defects associated with low reproductive performance in dairy cattle in Uruguay. Open Vet J, 13 (10), 1290-1298. doi:10.5455/OVJ.2023.v13.i10.8 Turabian Style Branda-Sica, Andrea, Rody Artigas, Elena de Torres, Evangelina Kinley, Paula Nicolini, María Teresa Federici, and Silvia Llambí. 2023. Monitoring of recessive defects associated with low reproductive performance in dairy cattle in Uruguay. Open Veterinary Journal, 13 (10), 1290-1298. doi:10.5455/OVJ.2023.v13.i10.8 Chicago Style Branda-Sica, Andrea, Rody Artigas, Elena de Torres, Evangelina Kinley, Paula Nicolini, María Teresa Federici, and Silvia Llambí. "Monitoring of recessive defects associated with low reproductive performance in dairy cattle in Uruguay." Open Veterinary Journal 13 (2023), 1290-1298. doi:10.5455/OVJ.2023.v13.i10.8 MLA (The Modern Language Association) Style Branda-Sica, Andrea, Rody Artigas, Elena de Torres, Evangelina Kinley, Paula Nicolini, María Teresa Federici, and Silvia Llambí. "Monitoring of recessive defects associated with low reproductive performance in dairy cattle in Uruguay." Open Veterinary Journal 13.10 (2023), 1290-1298. Print. doi:10.5455/OVJ.2023.v13.i10.8 APA (American Psychological Association) Style Branda-Sica, A., Artigas, . R., Torres, . E. d., Kinley, . E., Nicolini, . P., Federici, . M. T. & Llambí, . S. (2023) Monitoring of recessive defects associated with low reproductive performance in dairy cattle in Uruguay. Open Veterinary Journal, 13 (10), 1290-1298. doi:10.5455/OVJ.2023.v13.i10.8 |