| Research Article | ||
Open Vet J. 2024; 14(4): 1019-1028 Open Veterinary Journal, (2024), Vol. 14(4): 1019–1028 Original Research Epitope mapping and a candidate vaccine design from canine distemper virusCamila Pereira dos Santos1, Jerlane Tarcilia Gomes Telles1, Georgia de Freitas Guimarães1, Laura Helena Vega Gonzales Gil2, Amanda Mota Vieira1, Jose Wilton Pinheiro Junior1, Carlos Eduardo Calzavara-Silva3 and Rita de Cássia Carvalho Maia1*1Department of Veterinary Medicine, Federal Rural University of Pernambuco, Recife, Brazil 2Department of Virology and Experimental Therapy, Aggeu Magalhães Research Center, Recife, Brazil 3Laboratory of Cellular and Molecular Immunology, Rene Rachou Institute – Fiocruz Minas, Belo Horizonte, Brazil *Corresponding Author: Rita de Cássia Carvalho Maia. Department of Veterinary Medicine, Federal Rural University of Pernambuco, Recife, Brazil. Email: rita.carvalho [at] ufrpe.br Submitted: 25/12/2023 Accepted: 11/03/2024 Published: 30/04/2024 © 2024 Open Veterinary Journal
AbstractBackground: Canine distemper (CD) is a worldwide spread disease that has been described in 12 families of mammals, especially in the Carnivora order, being better studied in domestic canines where vaccination represents the best means of control. CD is controlled by vaccination, but many cases of the disease still occur in vaccinated animals. Aim: The aim of this work was to study antigen-specific epitopes that can subsidize the development of a new vaccine approach. Methods: Mapping of T cell reactive epitopes for CD virus (CDV) was carried out through enzyme-linked immunospot assays using 119 overlapped synthetic peptides from the viral hemagglutinin protein, grouped in 22 pools forming a matrix to test the immune response of 32 animals. Results: Evaluations using the criteria established to identify reactive pools, demonstrated that 26 animals presented at least one reactive pool, that one pool was not reactive to any animal, and six pools were the most frequent among the reactive peptides. The crisscrossing of the most reactive pools in the matrix revealed nine peptides considered potential candidate epitopes for T cell stimulation against the CDV and those were used to design an in-silico protein, containing also predicted epitopes for B cell stimulation, and further analyzed using immune epitope databases to ensure protein quality and stability. Conclusion: The final in silico optimized protein presents characteristics that qualify it to be used to develop a new prototype epitope-based anti-CDV vaccine. Keywords: CDV, ELISPOT, Hemagglutinin, Peptides, Vaccine. IntroductionCanine distemper (CD) is considered an important threat among the infectious diseases of domestic canines and some other carnivores, due to its high mortality and morbidity rates (Elia et al., 2015; Costa et al., 2019; Sheikh et al., 2021). CD virus (CDV) is enveloped, single-stranded, negative sense and nonsegmented-RNA, belonging to the Morbillivirus genus of the Paramyxoviridae family of viruses (Von Messling et al., 2003; Costa et al., 2019). The virus genome comprises six proteins: “N” (nucleoprotein), “O” (phosphoprotein), “M” (matrix protein), and “L” (large protein); and two integral membrane proteins: “H” (hemagglutinin) and “F” (fusion protein), and the last two proteins are responsible for the major variabilities and immune system evasions (Curran et al., 1991, Haas et al., 1997). Although the vaccine strains from CDV have not changed in the past 60 years, there is a residual potential for new antigenic field strains of the virus to emerge around the world (Kapil and Yeary, 2011). Therefore, due to the global distribution of the infection and the enlarged susceptibility of different species, including humans (Sakai et al., 2013), the necessity to develop more current, efficient, and safer vaccines to both protect against and eradicate the virus has increased (Patel et al., 2012; Ramirez et al., 2021). In Brazil, CD is an endemic disease involved in both economic and emotional losses. Although vaccination is stimulated and performed in domiciled dogs, the stray dog population in both urban and rural areas is a means of keeping the virus circulating and mutating (Duque-Valencia et al., 2019; Yoak et al., 2014). Whereas vaccination continues to be the main prophylactic measure against the disease (Rikula et al., 2007), its failures are common and attributed to both the vaccine and the host, since it has been observed that they cannot readily protect against field strains even when properly applied (Haas et al., 1997). This line of reason also relies on the fact that variation in the H protein between vaccine and field strains can accentuate the loss of protection (Nielsen et al., 2012; Findlow et al., 2020). On the other hand, H protein is considered a key protein for CDV since it is responsible for virus adsorption to the host cells through its main receptor, signaling lymphocyte activation molecule (SLAM or CD150). Following hemagglutinin interaction with CD150, leading to virus fusion, pores are formed, and the RNA complex is injected into the cytosol of the host cell. This cellular membrane fusion is also important to cell-to-cell dissemination of the virus, determining the intensification of the symptoms, suggesting that an adequate immune response against that protein can readily prevent an infection from this virus (Martella et al., 2011; Plattet et al., 2016; Jiang et al., 2019). Moreover, prevention of CDV epidemics has become increasingly important since it has been demonstrated that lethal outbreaks in nonhuman primates could readily adapt to the use of human SLAM receptors (Lundegaard et al., 2012). Peptide-based vaccines are the new-generation target to deliver vaccines more effectively using identified peptides from a known epitope to induce a more efficient immune response against infectious agents (Lundegaard et al., 2012). Targeting T cell epitopes in this approach is highly desirable since enhanced T cell response is crucial for virus clearance, even in the presence of antibodies, in convalescent dogs (Kiecker et al., 2004; Miller et al., 2009). Therefore, in the present study, the epitope mapping of protein H reactive epitopes recognized by T CD4 and T CD8 from CDV was carried out using canine interferon (IFN) gamma enzyme-linked immunospot (ELISPOT) to identify target epitopes and propose a putative in silico vaccine design with enhanced immunogenicity. Materials and MethodsPopulation of studyThe population of the study comprised 32 dogs of all ages and breeds from the endemic metropolitan region of Recife, Pernambuco, Brazil. The exclusion criteria included animals belonging to any of the following situations: indication to use anti-CDV serum, presence of cancer or autoimmune diseases, or undergoing corticoid or immunossupressive treatment. Blood was collected from animals and tutors signed the informed consent form, with accessible language and in duplicate. SerologySerology to detect the antibody levels produced by animals, indicating exposure to CDV antigens, in this study was performed by immuno-chromatography using the commercial Anigen Rapid CDV Ab Test®, Anigen, Animal Genetics, Inc., which detects qualitative and quantitative anti-CDV IgG antibodies in the blood, serum, or plasma samples. For each animal, a total blood sample was used following the instructions from the manufacturer. The results were interpreted accordingly, and the titles were classified as Undetectable or No measurable, Low (1:16), Medium (1:64), or High (1:128) titer. Separation of peripheral mononuclear blood cells (PMBCs)PMBCs separation was performed using a ficoll-hypaque gradient (Ficoll Paque PLUS—GE Healthcare Life Sciences). The blood samples were diluted in phosphate saline buffer, pH 7.2, sterile, 1:1 ratio. Afterward, the diluted blood was transferred carefully to a tube with Ficoll in the same proportion of the initial blood volume, without mixing them, and was centrifuged for 30 seconds, at 800 × g. The ring of PMBCs was transferred to another tube completing the volume to 15ml, centrifuging again for 5 minutes at 800 × g. After discarding the supernatant, 1 ml of ammonium-chloride-potassium (ACK) lysing buffer was added to lyse the red blood cells for 3 minutes, homogenizing every minute. Afterward, the volume was completed to 10 ml using Roswell Park Memorial Institute (RPMI) medium and centrifuged again for 5 minutes at 800 × g. The resulting supernatant was discarded and 1mL of serum-free culture medium (SFM) was added to re-suspend the pellet. A 100 μl aliquot was added to 900 μl of SFM to quantify and analyze cell viability using ViCell® (Cellular Viability Analyzer, BiosystemsTM, BD). Another 4 ml of SFM was added to the pellet and the cellular concentration adjusted to 2 × 106 cells/well. Synthetic peptidesOne hundred and nineteen peptides were synthesized (GenScript© USA Inc.) based on the protein H sequence from the onderstepoort strain of the CDV (Fig. 1), the most used strain in Brazilian vaccines. Those were arranged in 15 mers with overlapping of 10 amino acid residues, representing a proper stimulus for CD4 and TCD8 cells, considering that the amino acid core of each peptide is sufficient to fill major histocompatibility complex (MHC) classes I (8-9-mers) and II (11-15-mers) clefts (Kiecker et al., 2004). The peptides were diluted in 10% DMSO to a final concentration of 10 μl/ml. Subsequently, those peptides were grouped in pools and combined into a matrix (11 × 11) to ensure that each peptide must be present in two different pools (Chart 1) (Roederer and Koup, 2003), thus allowing the deduction of the likely immunogenic peptides. IFN-γ ELISPOTThe IFN-γ ELISPOT assays were performed using the Canine IFN-gamma ELISpot kit, from R&D Systems, Inc, as described in the protocol below. The wells were filled with 200 μl of sterile culture medium and incubated for 20 minutes at room temperature. After removing the medium, 100 μl of RPMI with the peptides and 100 μl of either cells or controls were added to each well. Phorbol 12-myristate 13-acetate (PMA) with ionomycin and cells was used as a positive control, and cells alone were used as negative controls. The plates were incubated in a CO2 humidifier incubator at 37°C, overnight. Each well was drained and washed with a washing buffer (250–300 μl) four times by blot drying.
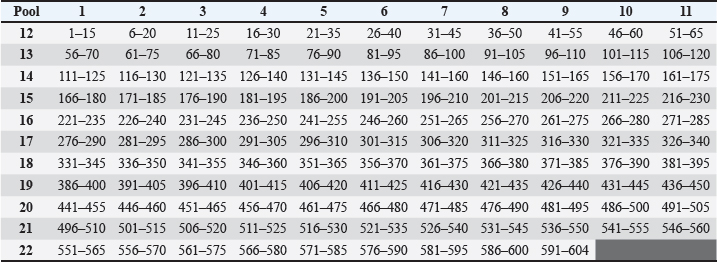
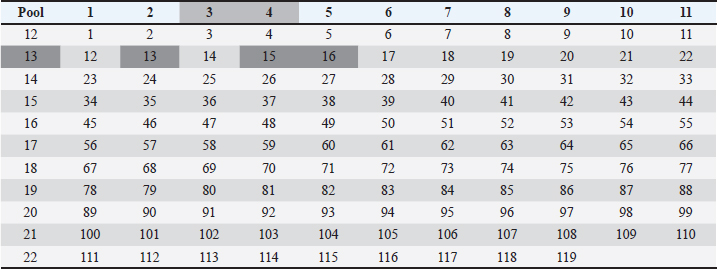
Fig. 1. Sequence, one letter coded, of canine distemper virus protein H. Fonte: http://www.ncbi.nlm.nih.gov/protein/BAF03641.1. Chart 1. Matrix of individual peptides in pools from canine distemper virus protein H.
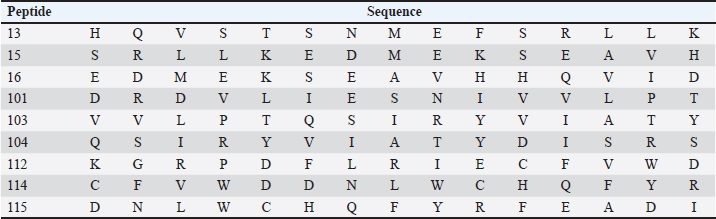
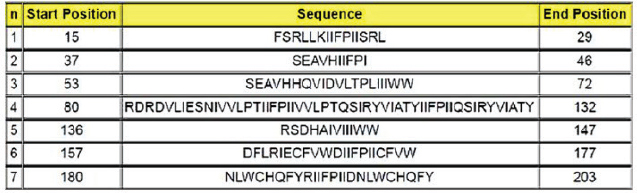
Subsequently, 100μL of detection antibody was added to each well and the plate was incubated at 2°C to 8°C, overnight. The following washes were performed as described above and followed by the addition of 100 μl of diluted streptavidin alkaline phosphatase (AP) to each well and incubated for 2 hours at room temperature. The washes were performed as before. The BCIP/NBT chromogen was added to each well (100 μl) and incubated for one hour at room temperature, protected from light. The chromogen solution was discarded from the microplates and those were washed with deionized water and blot dried. After removing the flexible plastic drain from the bottom of the plate, and cleaning the plate completely using a paper towel, the plates were dried at 37°C for 15–30 minutes. Finally, the plate was read using the immunospot series 3B analyzer ELISPOT (Cellular Technologies Ltd., Shaker Heights, OH) with the software immunospot version 3.0 (Cellular Technologies Ltd). Screening of immunogenic peptidesThe immunogenicity of the CDV H protein peptides was determined by IFN-γ ELISPOT performed ex vivo using peripheral blood mononuclear cells (PBMCs) from volunteers. The statistical criteria for identifying the reactive pools of peptides was based on the combination of the following equations (de Melo et al., 2013): Mean of the number of spots (peptides) minus 2 times the standard deviation > mean of the number of spots from the negative control; Mean of the number of spots > mean of the number of spots from negative control plus 2 times the standard deviation; Mean of the number of spots minus the mean of number of spots from negative control > 10. In silico protein designThe in silico protein design was based on the sequences of the nine epitopes indicated as most reactive for T cells by the ELISPOT assay from the CDV envelope protein H (Table 1). To complete the design, predicted epitopes for B cells obtained from the original H protein sequence were added, in addition, to insert sequences to ensure protein stability. The protein analysis was initially performed in the Immune Epitope and Database and Analysis Resources (https://www.iedb.org/), and furthermore, the sequences were studied using the Database from http://imed.med.ucm.es/Tools/index.html. Table 1. Sequence of protein H peptides from canine distemper virus considered potential candidates of T cell epitopes using IFN-γ ELISPOT assay.
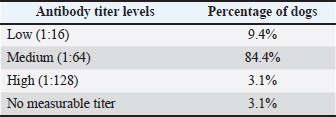
Ethical approvalThe present work has been approved by the Animal Ethics Committee from the Federal Rural University of Pernambuco (UFRPE number 44/2014) and was performed in line with the principles of the Declaration of Helsinki. ResultsSerologyThe production of antibodies in vaccinated dogs was accessed to demonstrate that the response to the CDV antigen was detectable and to demonstrate that the presence of antibodies alone may not be a guarantee of T cell activation and protection. The results showed that in the serological profile from the studied population, 84.4% (27/32) of the dogs showed medium antibody titer levels (1:64–1:16), 9.36% (3/32) showed a low antibody titer (1:16), 3.12% (1/32) showed high titer (1:64), and 3.12% (1/32) did not show any measurable titer (Table 2). IFN-γ ELISPOTOur results showed that eight animals presented reactivity to only one pool of peptides and eighteen animals showed reactivity for more than two pools, amongst those, one animal showed reactivity of 17 from the 22 pools analyzed. Six animals excluded from this study presented reactivity below those criteria in all pools. The identification of reactive pools following the aforementioned criteria allowed the verification of the frequency in which those pools appear in the studied population, and that one pool (11) showed no reactivity in any assay (Table 3). Table 2. Antibody levels of dogs vaccinated against canine distemper.
Table 3. Frequency of pools showing reactivity for the studied population.
Using the cutoff at 25% to identify the most frequent pools in the present study, six pools of peptides were identified: 2, 4, 5, 13, 21, and 22. The intersection of those pools revealed nine peptides (13, 15, 16, 101, 103, 104, 112, 114, and 115) considered the strongest candidates for T cell epitope activation (Chart 2). The sequences from those peptides are shown in Table 1. Chart 2. Most reactive pools after statistics (crosshatched) and peptides considered potential candidates to T cell epitopes (gray).
Protein designThe selected epitopes were then used to construct an artificial chimerical protein. The chimerical protein has as core the selected epitopes surrounded by amino acid residues (C and/or N terminals) found at the original protein sequence and spaced by nonantigenic/immunogenic amino acids whose sequences were exclusively defined for the chimerical protein presented here. The protein in silico optimization analyzes were performed using the specific programs for each evaluated parameter (Fig. 2). The candidate protein design generated a product under the Patent number BR102022013555-0. Analyzing the putative protein for its immunogenicity, such as antigen binding sites (Fig. 3) and sequences of antigenic determinants in the protein with their initial and final positions (Fig. 4). Moreover, as a relevant part of the immune defense process for the eventual construction of an effective vaccine and for detection of antibodies when developing a diagnostic test, the protein has binding and recognition sites for B cells, replicated from the original protein, which produces antibodies, as demonstrated in the Figure 5, which shows both the regions, and their sequences present in the putative protein. DiscussionThe present work aimed to determine the epitope mapping of T cell immune response against CDV elicited in dogs from the city of Recife, Brazil, and study a in silico vaccine candidate. Previous studies for the identification of relevant epitopes of this agent were performed for the nucleocapsid, phosphoprotein, or fusion proteins (Dean et al., 2004). The virus binding factor, the H protein from the Onderstepoort strain, was chosen in this article to be mapped based on previous studies which demonstrated that this polymer has an important role in the cellular immune stimulus, since it controls host specificity and cellular tropism, in addition to induce the majority of CDV neutralizing antibodies (Von Messling et al., 2003; Kapil and Yeary, 2011). Among the tools commonly used in the search for epitopes the ELISPOT offers many advantages related to direct ex vivo immune diagnostic monitoring, allowing automation and high throughput screening of data, presenting faster results (Anthony and Lehmann, 2003; Lehmann and Zhang, 2012). It also has the advantage over traditional assays (such as flow cytometry) with the possibility of faster, cheaper, and more sensible results for direct quantification of effector cells (Letsch and Scheibenbogen, 2003; Schwarz et al., 2022). The animals enrolled in this study were immunized at least once in their lives and at least one month before the blood draw. The vaccines were live modified (Nobivac®Canine, MSD Saúde Animal; Vencomax®, Laboratórios Vencofarma do Brasil, LTDA; Vanguard Plus®, Laboratórios Pfizer Ltda; Bio Max®, Lema Injex Biologic) or recombinant vaccines (Recombiteck®C6/CV, Merial Inn) mostly raised against the onderstepoort strain of CDV. The interval between the vaccination and the blood draw to obtain the PMBCs to be used in the ex vivo proliferation assays is crucial to detect the T cell activity (Ghosh et al., 2001), ergo, it is understandable that a previous immune stimulation with this virus is an interesting parameter to perform the IFN-γ ELISPOT assay. Since the IFN-γ is not expressed in peripheral blood leukocytes in dogs with CD (Beineke et al., 2009), when using this technique, our results conveyed faithfully the quantification of T cell activation that recognizes the immunogenic peptides. In the present work, IgG titers were determined against CDV to confirm the stimulation of B cells against the virus in vaccinated animals. Monti (2004) describes that most of the authors determine 1:100 as a satisfactory titer to protect against infection. In our study, only one animal showed titers above 1:100 (1:128). Interestingly, 25 animals presented antibody titers below 1:100, and only one animal, although immunized one month before the test, did not show any measurable levels of antibody, but all of them were positive to at least one pool of peptides. These findings demonstrate the importance of the cellular immune response in CD even in the absence of humoral immunity (Diallo, 1990).
Fig. 2. Chimerical protein in silico designed. Green–amino acids contained in the sequence of the original protein that is not part of the selected peptides; Underlined–identified epitopes that favor T cell response; Red–nonantigenic/immunogenic spacer amino acid residues; Bold–selected epitopes that favor B cells.
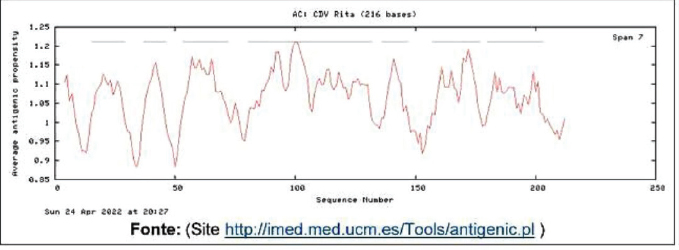
Fig. 3. Demonstrate the average antigenic propensity binding sites of the putative protein.
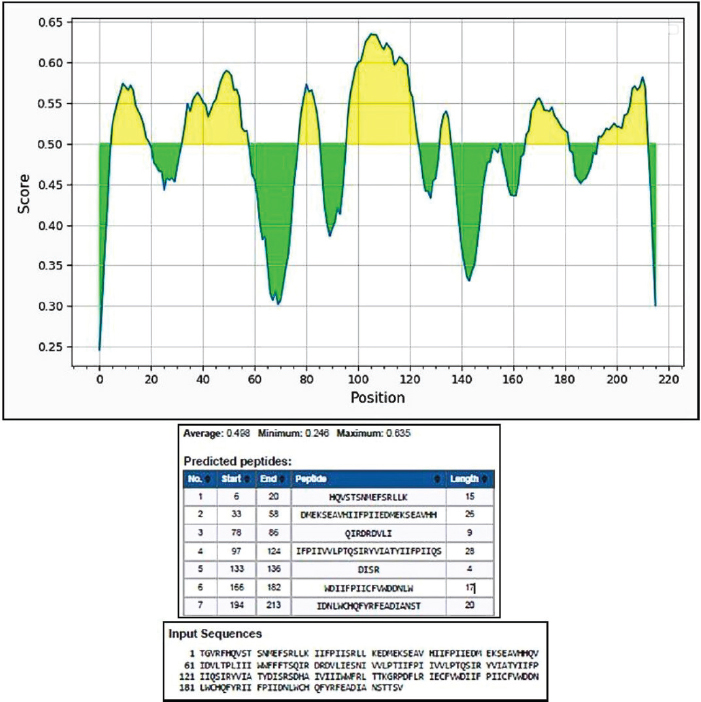
Fig. 4. Sequences of antigenic determinants in the protein with their initial and final positions. On the other hand, animals (number 6) that did not present reactivity to any pool and were excluded from this study, exhibited a medium titer of antibodies (between 1:16 and 1:64). Moreover, analysis of the high background seen on their 96-well plate indicated that they probably had elevated nonspecific immune response, low cellular viability or no specific response (de Souza, 2011). This result shows that the vaccine’s ability to elicit antibody productions does not guarantee T cell stimulation, essential for virus immune defense. It has also been observed in this study that six peptide pools were recognized by CD4 and CD8 containing-cell cultures, which could suggest that those epitopes can be recognized by either T cell subset, just as Dean et al. (2004) demonstrated when performing the epitope mapping for the feline immunodeficiency virus. In the present work, we present nine peptides originating from six different pools identified as potential T cell epitope candidates (Chart 2) for a new protein with enhanced immunogenicity.
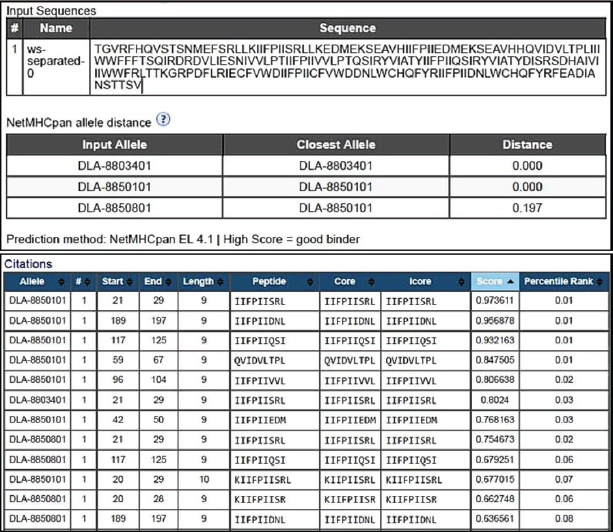
Fig. 5. B cell binding and recognition sites maintained from the original H protein and present in the putative protein. The strategy of using peptide-based in silico-designed vaccines is highly desired due to some features which can be cited as safety, lacking both infectious and allergenic agents, low cost for production, storage, and transportation, and in many cases single-peptide immune stimulation, and in addition, considering the extensive amount of epitope databases existing nowadays, it has become easier to predict immunogenicity, processing and recognition, even protease degradation sites (Soria-Guerra et al., 2015; García-Machorro et al., 2022). As the proposal is based on the design of a structure that favors the binding site with T cells, Figure 6 shows the best binding sites for this protein with MHC class I of host cells, and although the figure shows only the first twelve possible binding sites for recognition of the protein, the program’s analysis showed some dozens of possible sites. This in silico analysis demonstrates the putative protein’s potential to elicit specific responses from T and B lymphocytes against distemper virus H protein-specific sequence. Therefore, our findings describe new data presented here by the epitope mapping of protein H from the CDV virus recognized by T CD4 and CD8 lymphocytes in a cohort of individuals using the IFNγ ELISPOT technique, culminating with a protein design candidate to promote the development of a safer, long-lasting and with minimal side effects vaccine against the CDV virus.
Fig. 6. Best binding sites for the putative protein with the MHC class I of host cells. ConclusionThe identification of important CDV epitopes for T cell stimulation brought by this article can guide the development of new antigenic targets based on selected T cell epitopes using the ELISPOT assay for IFN-γ. The identification of nine candidate peptides from the H protein of CDV was further studied in silico to elicit stronger and more specific immune responses by overcoming subdominant epitopes within conserved sequences, avoiding the presence of inhibitor epitopes and thus allowing even the combination of multiple epitopes from different proteins to generate protection from the pathogen and proposing the design of a protein as a vaccine candidate. AcknowledgmentsThe authors acknowledge the scientific and infrastructure support of the Oswaldo Cruz foundation–FIOCRUZ to this work. The authors are extremely grateful to Eduardo José Moura do Nascimento, for performing ELISPOT readings for the experiments at the Center for Vaccine Research, University of Pittsburgh, Pittsburgh, PA. Conflict of interestThe authors declare to have no conflict of interest and have no relevant financial or nonfinancial interests to disclose. FundingThe authors acknowledge the Foundation for the Support of Science and Technology of the State of Pernambuco (FACEPE) for their financial support [grant numbers APQ-0713-5.05/10]. Authors’ contributionsAll authors contributed to the study’s conception and design. Sample preparation, data collection, and analysis were performed by Camila Pereira dos Santos, Jerlane Tarcilia Gomes Teles, Laura Helena Vega Gonzales Gil, Carlos Eduardo Calzavara-Silva, and Rita de Cássia Carvalho Maia. Data analysis and the first draft of the manuscript were written by Camila Pereira dos Santos, Georgia de Freitas Guimarães, José Wilton Pinheiro Junior, and Amanda Mota Vieira and all authors commented on previous versions of the manuscript. All authors read and approved the final manuscript. Data availabilityThe data that support the findings of this study are not openly available due to reasons of sensitivity of intellectual property and are available from the corresponding author upon reasonable request. ReferencesAnthony, D.D. and Lehmann, P.V. 2003. T-cell epitope mapping using the ELISPOT approach. Methods 29, 260–269. Beineke, A., Puff, C., Seehusen, F. and Baumgärtner, W. 2009. Pathogenesis and immunopathology of systemic and nervous CD. Vet. Immunol. Immunopathol. 127, 1–18. Costa, V.G., Saivish, M.V., Rodrigues, R.L., Lima Silva, R.F., Moreli, M.L. and Krüger, R.H. 2019, Molecular and serological surveys of canine distemper virus: a meta-analysis of cross-sectional studies. PLoS One 14, e0217594. Curran, M., Clarke, D. and Rima, B. 1991. The nucleotide sequence of the gene encoding the attachment protein H of canine distemper virus. J. Gen. Virol. 72, 443–447. de Melo, A.B., Nascimento, E.J., Braga-Neto, U., Dhalia, R., Silva, A.M., Oelke, M., Schneck, J.P., Sidney, J., Sette, A. and Montenegro, S.M. 2013. T-cell memory responses elicited by yellow fever vaccine are targeted to overlapping epitopes containing multiple HLA-I and-II binding motifs. PLoS Negl. Trop. Dis. 7, e1938. de Souza, J.R. 2011. Mapeamento de epítopos reativos para as células T das proteínas não estruturais NS1 e NS3 do vírus dengue 3. Ph.D. Thesis, FIOCRUZ, Recife, PE, Brazil. Dean, G.A., LaVoy, A. and Burkhard, M.J. 2004. Peptide mapping of feline immunodeficiency virus by IFN-γ ELISPOT. Vet. Immunol. Immunopathol. 100, 49–59. Diallo, A. 1990. Morbillivirus group: genome organisation and proteins. Vet. Microb. 23, 155–163. Duque-Valencia, J., Forero-Muñoz, N.R., Díaz, F.J., Martins, E., Barato, P. and Ruiz-Saenz, J. 2019. Phylogenetic evidence of the intercontinental circulation of a Canine distemper virus lineage in the Americas. Sci. Rep. 9, 15747. Elia, G., Camero, M., Losurdo, M., Lucente, M.S., Larocca, V., Martella, V., Decaro, N. and Buonavoglia, C. 2015. Virological and serological findings in dogs with naturally occurring distemper. J. Virol. Methods 213, 127–130. Findlow, J., Bayliss, C.D., Beernink, P.T., Borrow, R., Liberator, P. and Balmer, P. 2020. Broad vaccine protection against Neisseria meningitidis using factor H binding protein. Vaccine 38, 7716–7727. García-Machorro, J., Ramírez-Salinas, G.L., Martinez-Archundia, M. and Correa-Basurto, J. 2022. The advantage of using immunoinformatic tools on vaccine design and development for coronavirus. Vaccines 10, 1844. Ghosh, S., Walker, J. and Jackson, D.C. 2001. Identification of canine helper T-cell epitopes from the fusion protein of canine distemper virus. Immunology 104, 58–66. Ramirez, G.S., Marin, R.S. and Saenz, R.J. 2021. A concise review on certain important veterinary viruses in the Americas. Rev. MVZ Cordoba 26(2), e1965. Haas, L., Martens, W., Greiser-Wilke, I., Mamaev, L., Butina, T., Maack, D. and Barrett, T. 1997. Analysis of the haemagglutinin gene of current wild-type canine distemper virus isolates from Germany. Virus Res. 48, 165–171. Jiang, Y., Jia, S., Zheng, D., Li, F., Wang, S., Wang, L., Qiao, X., Cui, W., Tang, L. and Xu, Y. 2019. Protective immunity against canine distemper virus in dogs induced by intranasal immunization with a recombinant probiotic expressing the viral H protein. Vaccines 7, 213. Kapil, S. and Yeary, T.J. 2011. Canine distemper spillover in domestic dogs from urban wildlife. Vet. Clin. Small Anim. Pract. 41, 1069–1086. Kiecker, F., Streitz, M., Ay, B., Cherepnev, G., Volk, H.-D., Volkmer-Engert, R. and Kern, F. 2004. Analysis of antigen-specific T-cell responses with synthetic peptides—What kind of peptide for which purpose? Hum. Imm. 65, 523–536. Lehmann, P.V. and Zhang, W. 2012. Unique strengths of ELISPOT for T cell diagnostics. Handbook of ELISPOT: methods and protocols. Berlin, Germany: Springer, pp: 3–23. Letsch, A. and Scheibenbogen, C. 2003. Quantification and characterization of specific T-cells by antigen-specific cytokine production using ELISPOT assay or intracellular cytokine staining. Methods 31, 143–149. Lundegaard, C., Lund, O. and Nielsen, M. 2012. Predictions versus high-throughput experiments in T-cell epitope discovery: competition or synergy? Expert Rev. Vaccines 11, 43–54. Martella, V., Blixenkrone-Møller, M., Elia, G., Lucente, M., Cirone, F., Decaro, N., Nielsen, L., Banyai, K., Carmichael, L. and Buonavoglia, C. 2011. Lights and shades on an historical vaccine canine distemper virus, the Rockborn strain. Vaccine 29, 1222–1227. Miller, C.H., Maher, S.G. and Young, H.A. 2009. Clinical use of interferon-γ. Ann. N. Y. Acad. Sci. 1182, 69–79. Monti, F.S. 2004. Anticorpos contra o vírus da cinomose em cães vacinados em diferentes estabelecimentos da área urbana do município de Viçosa/MG. MS Thesis. UFV, Viçosa, Brazil. Nielsen, L., Jensen, T.H., Kristensen, B., Jensen, T.D., Karlskov-Mortensen, P., Lund, M., Aasted, B. and Blixenkrone-Møller, M. 2012. DNA vaccines encoding proteins from wild-type and attenuated canine distemper virus protect equally well against wild-type virus challenge. Arch. Virol. 157, 1887–1896. Patel, J., Heldens, J., Bakonyi, T. and Rusvai, M. 2012. Important mammalian veterinary viral immunodiseases and their control. Vaccine 30, 1767–1781. Plattet, P., Alves, L., Herren, M. and Aguilar, H.C. 2016. Measles virus fusion protein: structure, function and inhibition. Viruses 8, 112. Rikula, U., Nuotio, L. and Sihvonen, L. 2007. Vaccine coverage, herd immunity and occurrence of canine distemper from 1990–1996 in Finland. Vaccine 25, 7994–7998. Roederer, M. and Koup, R.A. 2003. Optimized determination of T cell epitope responses. J. Imm. Methods 274, 221–228. Sakai, K., Yoshikawa, T., Seki, F., Fukushi, S., Tahara, M., Nagata, N., Ami, Y., Mizutani, T., Kurane, I. and Yamaguchi, R. 2013. Canine distemper virus associated with a lethal outbreak in monkeys can readily adapt to use human receptors. J. Virol. 87, 7170–7175. Schwarz, M., Mzoughi, S., Lozano-Ojalvo, D., Tan, A.T., Bertoletti, A. and Guccione, E. 2022. Current research in immunology. Curr. Res. Immunol. 3, 215–221. Sheikh, T., Ranjan, R., Jha, A.K. and Kumar, S. 2021. Canine distemper: a fatal disease seeking special intervention. J. Entomol. Zool. Stud. 9, 1411–1418. Soria-Guerra, R.E., Nieto-Gomez, R., Govea-Alonso, D.O. and Rosales-Mendoza, S. 2015. An overview of bioinformatics tools for epitope prediction: implications on vaccine development. J. Biom. Inf. 53, 405–414. Von Messling, V., Springfeld, C., Devaux, P. and Cattaneo, R. 2003. A ferret model of canine distemper virus virulence and immunosuppression. J. Virol. 77, 12579–12591. Yoak, A.J., Reece, J.F., Gehrt, S.D. and Hamilton, I.M. 2014. Disease control through fertility control: secondary benefits of animal birth control in Indian street dogs. Prev. Vet. Med. 113, 152–156. | ||
| How to Cite this Article |
| Pubmed Style Santos CP, Teles JTG, Guimarães GF, Gil LHVG, Vieira AM, Junior JWP, Calzavara-Silva CE, Maia RdCC. Epitope mapping and a candidate vaccine design from canine distemper virus. Open Vet J. 2024; 14(4): 1019-1028. doi:10.5455/OVJ.2024.v14.i4.9 Web Style Santos CP, Teles JTG, Guimarães GF, Gil LHVG, Vieira AM, Junior JWP, Calzavara-Silva CE, Maia RdCC. Epitope mapping and a candidate vaccine design from canine distemper virus. https://www.openveterinaryjournal.com/?mno=177059 [Access: October 06, 2024]. doi:10.5455/OVJ.2024.v14.i4.9 AMA (American Medical Association) Style Santos CP, Teles JTG, Guimarães GF, Gil LHVG, Vieira AM, Junior JWP, Calzavara-Silva CE, Maia RdCC. Epitope mapping and a candidate vaccine design from canine distemper virus. Open Vet J. 2024; 14(4): 1019-1028. doi:10.5455/OVJ.2024.v14.i4.9 Vancouver/ICMJE Style Santos CP, Teles JTG, Guimarães GF, Gil LHVG, Vieira AM, Junior JWP, Calzavara-Silva CE, Maia RdCC. Epitope mapping and a candidate vaccine design from canine distemper virus. Open Vet J. (2024), [cited October 06, 2024]; 14(4): 1019-1028. doi:10.5455/OVJ.2024.v14.i4.9 Harvard Style Santos, C. P., Teles, . J. T. G., Guimarães, . G. F., Gil, . L. H. V. G., Vieira, . A. M., Junior, . J. W. P., Calzavara-Silva, . C. E. & Maia, . R. d. C. C. (2024) Epitope mapping and a candidate vaccine design from canine distemper virus. Open Vet J, 14 (4), 1019-1028. doi:10.5455/OVJ.2024.v14.i4.9 Turabian Style Santos, Camila Pereira, Jerlane Tarcila Gomes Teles, Georgia Freitas Guimarães, Laura Helena Vega Gonzales Gil, Amanda Mota Vieira, Jose Wilton Pinheiro Junior, Carlos Eduardo Calzavara-Silva, and Rita de Cassia Carvalho Maia. 2024. Epitope mapping and a candidate vaccine design from canine distemper virus. Open Veterinary Journal, 14 (4), 1019-1028. doi:10.5455/OVJ.2024.v14.i4.9 Chicago Style Santos, Camila Pereira, Jerlane Tarcila Gomes Teles, Georgia Freitas Guimarães, Laura Helena Vega Gonzales Gil, Amanda Mota Vieira, Jose Wilton Pinheiro Junior, Carlos Eduardo Calzavara-Silva, and Rita de Cassia Carvalho Maia. "Epitope mapping and a candidate vaccine design from canine distemper virus." Open Veterinary Journal 14 (2024), 1019-1028. doi:10.5455/OVJ.2024.v14.i4.9 MLA (The Modern Language Association) Style Santos, Camila Pereira, Jerlane Tarcila Gomes Teles, Georgia Freitas Guimarães, Laura Helena Vega Gonzales Gil, Amanda Mota Vieira, Jose Wilton Pinheiro Junior, Carlos Eduardo Calzavara-Silva, and Rita de Cassia Carvalho Maia. "Epitope mapping and a candidate vaccine design from canine distemper virus." Open Veterinary Journal 14.4 (2024), 1019-1028. Print. doi:10.5455/OVJ.2024.v14.i4.9 APA (American Psychological Association) Style Santos, C. P., Teles, . J. T. G., Guimarães, . G. F., Gil, . L. H. V. G., Vieira, . A. M., Junior, . J. W. P., Calzavara-Silva, . C. E. & Maia, . R. d. C. C. (2024) Epitope mapping and a candidate vaccine design from canine distemper virus. Open Veterinary Journal, 14 (4), 1019-1028. doi:10.5455/OVJ.2024.v14.i4.9 |