| Original Article | ||
Open Vet J. 2023; 13(1): 74-89 Open Veterinary Journal, (2023), Vol. 13(1): 74–89 Original Research Molecular characterization of norovirus and sapovirus detected in animals and humans in Costa Rica: Zoo-anthropozoonotic potential of human norovirus GII.4 Derling José Pichardo Matamoros1,2*, Cristina Solís Worsfold3, Rocío Cortés Campos2,4, Hilda María Bolaños Acuña5, Elena Campos Chacón5 and Carlos Francisco Jiménez Sánchez2,4 1Postgraduate Program in Agricultural Sciences and Natural Resources (PPCARN), University of Costa Rica (UCR), San Pedro, Costa Rica 2Veterinary Virology Diagnostic and Research Unit (UNDIVE), School of Veterinary Medicine, National University, Barreal de Heredia, Costa Rica 3Elanco Animal Health, 800 5th St NW Fort Dodge, IA 50501, USA 4Tropical Diseases Research Program (PIET), School of Veterinary Medicine, National University, Barreal de Heredia, Costa Rica 5National Bacteriological Reference Center of the Costa Rican Institute for Teaching and Research in Health and Nutrition (CNRB-INCIENSA), San Diego, Costa Rica Submitted: 20/10/2022 Accepted: 04/01/2023 Published: 14/01/2023 *Corresponding Author: Derling José Pichardo Matamoros. Postgraduate Program in Agricultural Sciences and Natural Resources (PPCARN), University of Costa Rica (UCR), San Pedro, Costa Rica. Email: derling.pichardo [at] ucr.ac.cr © 2023 Open Veterinary Journal
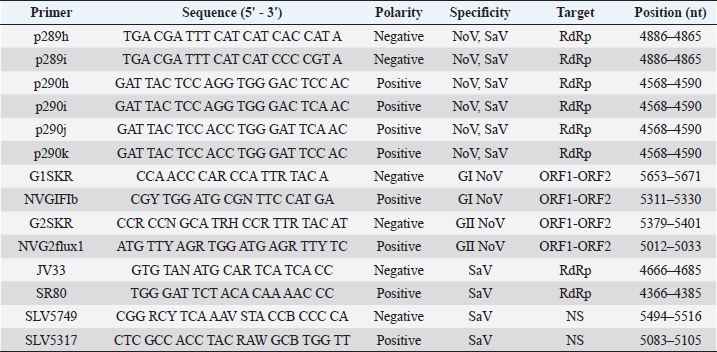
AbstractBackground: Noroviruses (NoV) and sapoviruses (SaV) are major causes of acute viral gastroenteritis in humans worldwide, as well as gastrointestinal infections in animals. However, it has not been determined whether these viruses are zoonotic pathogens. Aim: In this study, we investigated the presence of NoV and SaV in stool samples from dogs, pigs, cows, and humans to determine some aspects of the molecular epidemiology and the genetic relationship of several strains present in these species. Methods: Polymerase chain reaction and sequencing of NoV and SaV strains present in stool samples from humans and dogs with diarrhea, pigs, and cattle with and without diarrhea were carried out during fragmented periods from 2002 to 2012. Results: Of all samples analyzed, 11.6% (123/1,061) of the samples were positive for NoV and 0.88% (9/1,023) were positive for SaV. The phylogenetic analysis confirmed 16 human strains of NoV (HuNoV) belonging to HuNoV G?/GII.P2 (1), GII.4/GII.P4 (5), G?/GII.P4 (9), and GII.6/GII.P6 (1) and allowed us to verify and assign three strains of human SaV to genotypes GI.2 (1) and GII.5 (2). In dogs, eight strains of NoV [HuNoV G?/GII.P4 (4) and canine G?/GVI.P1 (4)] and two strains of canine SaV were determined. In pigs, six strains were assigned to HuNoV G?/GII.P4 and four strains to porcine SaV were assigned to genogroup GIII (2), GVIII (1), and GXI (1). In bovines, five strains were characterized as HuNoV G?/GII.P4. Conclusions: This study showed that NoV and SaV prototype strains have been present in humans and dogs in Costa Rica. Additionally, it revealed that the zoonotic potential of SaV is very limited, while the zoonotic implications for HuNoV GII.4 are stronger due to the simultaneous circulation of strains related to HuNoV GII.4 in four species, which suggests a zoo-anthropozoonosis. Keywords: Costa Rica, Norovirus, Phylogeny, Sapovirus, Zoonosis. IntroductionNorovirus (NoV) and Sapovirus (SaV) belong to the Caliciviridae family and are emerging infectious agents of global distribution. NoV are the leading epidemiological cause of foodborne nonbacterial diarrhea outbreaks and acute viral gastroenteritis in humans (Hall et al., 2014; Mans, 2019). On a smaller scale, SaV are related to outbreaks and sporadic gastroenteritis cases in humans (Magwalivha et al., 2018; Varela et al., 2019). NoV also causes diarrhea in cattle (Di Felice et al., 2016), pigs (Wang et al., 2005), and dogs (Martella et al., 2008c), while SaV causes diarrhea in pigs (Wang et al., 2007) and have been identified in dogs with diarrhea (Li et al., 2011). Given that caliciviruses are +ssRNA viruses, mutations and genetic recombination to occurs very frequently (White, 2014; Ludwig-Begall et al., 2018). As a result, serious difficulties arise when assigning the same genotype to a strain when the phylogenetic information is obtained from the gene encoding the polymerase and capsid protein VP1. However, this classification system based on both genes (polymerase/capsid) is very appropriate to characterize strains and assign them to a known genotype and P-type or identify recombinant strains and new genotypes (Farkas et al., 2004; Katayama et al., 2004; Bull et al., 2007; Dey et al., 2018; Lun et al., 2018; Cavicchio et al., 2022). Basically, NoV and SaV are classified based on the genetic similarity of these genes in three levels: genogroups, genotypes within each genogroup, and viral strains or variants within each genotype (Katayama et al., 2002; Zheng et al., 2006; Oka et al., 2015; Cavicchio et al., 2022). A more updated classification has been recently proposed suggesting expanding the number of NoV genogroups to 10 and the number of genotypes to 49 (Chhabra et al., 2019). Therefore, human NoV strains (HuNoV) are included within genogroups GI, GII, GIV, GVIII, and GIX (Chhabra et al., 2019; Parra, 2019; Ludwig-Begall et al., 2021). The sequence analysis based on VP1 recognizes nine genotypes GI (GI.1-GI.9) and 27 genotypes GII (GII.1-GII.10, and GII.12-GII.17) in humans (Katayama et al., 2002; Zheng et al., 2006; Mathijs et al., 2011; Chhabra et al., 2019), while genotypes GII.11, GII.18, and GII.19 are prototype strains in pigs (Wang et al., 2005; Nakamura et al., 2010). Genogroup GIII includes genotypes GIII.1 (Günther and Otto, 1987), GIII.2, and GIII.3 in cattle (Woode and Bridger, 1978; Dastjerdi et al., 1999; Chhabra et al., 2019). Genogroup GIV includes human genotype GIV.1 (Vinjé and Koopmans, 2000) and canine genotype GIV.2 (Martella et al., 2008c). In addition, genogroup GVI includes genotypes GVI.1 and GVI.2 in canine NoV (Chhabra et al., 2019). Human SaV (HuSaV) strains are included within genogroups GI (genotypes GI.1-GI.8), GII (genotypes GII.1-GII.8), GIV (genotype GIV.1), and GV (genotypes GV.1 and GV.2) (Farkas et al., 2004; Katayama et al., 2004; Reuter et al., 2010; Dufkova et al., 2011; Shibata et al., 2015; Diez-Valcarce et al., 2018; Xue et al., 2019). Pigs are infected by genogroups GIII, GV, GVI, GVII, GVIII, GIX, GX, and GXI (Wang et al., 2005; Nakamura et al., 2010; Reuter et al., 2010; Dufkova et al., 2011; Kuroda et al., 2017; Diez-Valcarce et al., 2018) and dogs by genogroup GXIII (Kuroda et al., 2017; Diez-Valcarce et al., 2018). The zoonotic potential of NoV and SaV has been described in many studies. Porcine GII NoV are genetically and antigenically related to HuNoV (Wang et al., 2005, 2007), and the presence of HuNoV GII.3, GII.13 (Nakamura et al., 2010), and GII.4 has been demonstrated in naturally infected pigs (Mattison et al., 2007; Nakamura et al., 2010). The close genetic similarity between GII.4 strains identified in dogs that have been in direct contact with humans with HuNoV genetic variants GII.4-2006b (98.6% similarity) and GII.4-2008 (100% similarity) suggests that HuNoV GII.4 caused an infection in the gastrointestinal tract (Summa et al., 2012). That observation has also been supported by serological evidence found in dogs exposed to virus-like particles similar to HuNoV GI and GII attached to gastrointestinal tissue and by HuNoV, the latter indicated a productive infection in dogs (Caddy et al., 2014, 2015). HuNoV GII.4 has also been reported in cattle (Mattison et al., 2007), and HuNoV GII.4-HS66 has been shown to infect and induce an immune response in pigs (Cheetham et al., 2006) and gnotobiotic calves (Souza et al., 2008). Strains of HuNoV GII.4 are a more dominant cause of outbreaks and epidemics of gastroenteritis in humans due to their ability to infect and circulate in domestic animals (dogs, pigs, and cattle) and their ability to evolve faster than other NoV genotypes given the frequent mutations and recombinations in the polymerase region and capsid protein VP1 (Bull et al., 2007; Bull and White, 2011; White, 2014; Parra, 2019). However, more studies are needed to elucidate the zoonotic potential of NoV. The strain of porcine SaV (PoSaV) GVIII/MI-QW19 is related to the strains of HuSaV GII/Mc10, GII/C12, and GII/Cruise Ship, with which it shares a 66.3% identity in a sequence fragment of 95 amino acids in the polymerase (Wang et al., 2007). PoSaV GVIII is also genetically related to the HuSaV GIV/Hou7-1181 strain, with which it shares a 66% identity in the polymerase (Reuter et al., 2010). In addition, PoSaV GVIII strain 43/06-18p3 is 50% genetically similar to HuSaV GI and GV in the polymerase gene (Martella et al., 2008b), and HuSaV GV has been detected in pig feces (Nakamura et al., 2010). Consequently, the possibility of co-infection with PoSaV GVIII and HuSaV (GI, GII, GIV, or GV) suggests the occurrence of zoonotic transmission or pig/human recombinant strains (Bank-Wolf et al., 2010). As of today, the presence of strains associated with the NoV or SaV that cause diarrhea in humans in Costa Rica has not been well documented. There is no evidence of common strains between humans and animals. Available studies do not include an analysis of the four species, and it has not been confirmed that animals in direct contact with humans are reservoirs of viral strains with public health impact. Therefore, knowing about the reservoirs and molecular epidemiology of these viruses is necessary to reduce the impact of gastroenteritis outbreaks and to clarify whether HuNoV GII.4 strains have zoonotic potential. The objective of this study was to identify NoV and SaV strains in human, canine, porcine, and cattle stool samples to establish the genetic similarity between circulating strains in the metropolitan area of Costa Rica. Materials and MethodsStool samplesThis research included an ambispective descriptive analysis. A total of 1,061 fecal samples were collected in Costa Rica during the 2002–2012 period. Residual human samples (240 stool samples) were obtained from adults and children under 5 years of age during acute gastroenteritis outbreaks. The samples were previously evaluated for rotavirus and other pathogens by hospitals in the Costa Rican metropolitan area and the National Bacteriological Reference Center of the Costa Rican Institute for Teaching and Research in Health and Nutrition as described in the literature (Bourdett-Stanziola et al., 2008). As far as animal samples, canine diarrheal fecal samples belonged to dogs (278 samples) that had been in direct contact with humans from the greater metropolitan area of Costa Rica and were sent to the Veterinary Virology Diagnostics and Research Unit (UNDIVE) through a commercial microbiological laboratory (ACOPSA, Heredia, Costa Rica). Fecal samples from pigs (194 samples) of all ages with and without diarrhea were obtained from intensive breeding farms. Bovine stool samples (349 samples) were collected from specialized dairy farms from calves and cows with and without diarrhea. Samples were stored at −70°C until processing. RNA extraction and retro-transcriptionStool samples were suspended in 20% (w/v) PBS at pH 7.2, and once cleared, 250 and 50 µl aliquots were collected. Five 50 µl aliquots were combined to extract viral RNA based on a final volume of 250 µl. Nucleic acid was extracted from each group and each individual sample (when necessary) using Trizol (Ambion, USA) following the manufacturer’s instructions. Subsequently, RNA was diluted with 25 μl RNase free-H2O (Fermentas, Lithuania) and was immediately subjected to a reverse transcription reaction using the RevertAid H Minus First Strand cDNA Synthesis Kit (Fermentas, Lithuania) following the manufacturer’s instructions and using the 2720 Thermal Cycler equipment (Applied Biosystems, USA). The cDNA obtained was stored at −70°C until use. Detection of NoV and SaV by RT-PCRThe presence of NoV and SaV was initially evaluated in a mix of five samples and then in each individual sample if found positive. The screening was performed to amplify the RdRp region by PCR using p290hijk(+)/p289hi(−)(Invitrogen, USA) primer pairs to detect NoV and SaV in humans and animals, while the SR80(+)/JV33(−) (Invitrogen, USA) primer pairs were used to detect SaV in the four species (Table 1). The PCR reaction mix was prepared based on a final volume of 50 µl using 25 µl PCR Master Mix (2X), #K0171 (Fermen tas, Lithuania), 1 µl of each sense/antisense primer, 2–5 µl cDNA, and RNAase-free water (Fermentas, Lithuania) to complete the volume. PCR conditions were performed as described in the literature for NoV (Jiang et al., 1999; Reuter et al., 2010) and SaV (Vinjé et al., 2000; Reuter et al., 2010). The expected product was 319 bp and 320 bp for NoV and SaV, respectively. The cDNA polymerase-based RT-PCR of positive samples was analyzed using a second RT-PCR to amplify the ORF1-ORF2 junction in NoV and the NS segment of the VP1 protein in SaV. The PCR reaction mix was prepared similarly to that described above. HuNoV genotyping was performed using specific genogroup degenerate primers NVG1F1b(+)/G1SKR(−) for HuNoV GI and NVG2flux1(+)/G2SKR(−) for HuNoV GII (Invitrogen, USA) in separate tubes. Reaction conditions were determined as described in the literature (Kojima et al., 2002; Nordgren et al., 2008; Nakamura et al., 2010). Expected products for GI NoV and GII NoV were 381 bp and 390 bp, respectively. Regarding SaV, SLV-5317(+) and SLV-5749(−) primer pairs (Invitrogen, USA) (Table 1) were used, and the PCR cycle was partially modified from Hansman et al. (2004) as follows: 94°C × 3 minutes, 40 cycles of 94°C× 1 minute, 48°C× 1.5 minutes, and 72°C × 1 minute and a final extension of 72°C × 10 minutes. The expected amplicon size was 434 bp. Table 1. Primers used to detect NoV and SaV by RT-PCR.
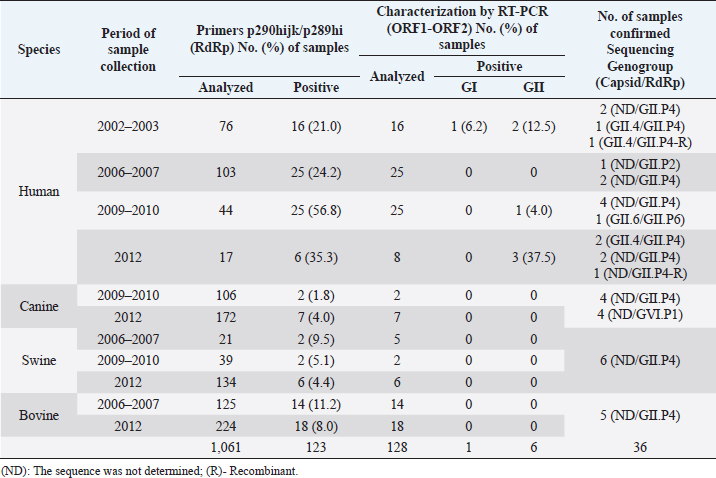
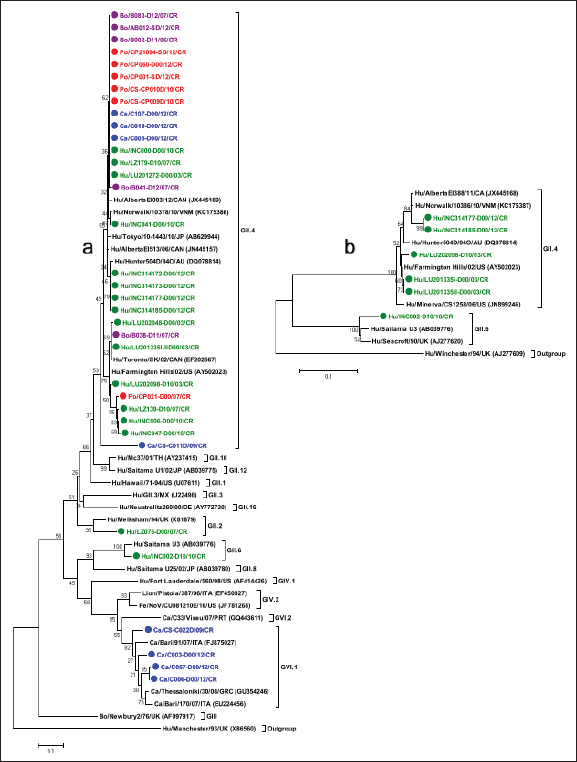
The strain of HuNoV LU201335I-IID00/2003/CR (GenBank accession number: KM057715) was used as a positive control in each round of PCR to detect NoV, while the strain of HuSaV SJ164-D11/2007/CR (GenBank accession number: KM001680) was used as a positive control in each round of PCR to detect SaV. In addition, RNAase-free water (Fermentas, Lithuania) was used as a negative control in each round of PCR. Amplified products by PCR were separated in 2% agarose gel containing 2 µl GelRed Nucleic Acid Stain 10,000X Water (Biotium, USA) and displayed using image capture and UV illuminator (UVP BioDoc-ItTM, USA) equipment. The DNA products obtained were stored at −70°C until sequencing. Sequencing and sequence analysisA total of 110 positive samples directly from the 1% agarose gel were purified using the QIAquick gel extraction kit (QIAGEN, Inc) following the manufacturer’s instructions, and 46 samples positive to NoV and 10 positives to SaV were selected as a representation for sequencing according to period and species, band quality on the agarose gel, and DNA concentration (ng/μl). The partial sequences of DNA nucleotides corresponding to the polymerase, NS segment, and ORF1-ORF2 junction were determined by direct cycles in both directions using the strains described above for each genus and the BigDye Terminator v3.1 Cycle Sequencing Kit (Applied Biosystems, USA), following the manufacturer’s instructions. Amplified fragments were purified using the QIAquick PCR Purification Kit (QIAGEN, Inc), and the nucleotide sequence was obtained using the 3130 Genetic Analyzer automated sequencer (Applied Biosystems, USA). DNA sequences were analyzed and edited with the MEGA v6.06 software and aligned using ClustalW 1.6 (Tamura et al., 2013). Phylogenetic analysisThe strain’s genotype was assigned taking into consideration the genetic classification based on the range of nucleotide similarity determined for the partial sequence of the gene encoding polymerase or the capsid region, the international classification and nomenclature, and reference strains for each genotype reported in different studies for humans, dogs, pigs, and cattle (Katayama et al., 2002; Zheng et al., 2006; Wang et al., 2007; Martella et al., 2008a, 2008c; Nakamura et al., 2010; Reuter et al., 2010; Kuroda et al., 2017; Chhabra et al., 2019). Phylogenetic trees were built using MEGA X (Kumar et al., 2018), including for each genus the reference genotypes found with the Basic Local Alignment Search Tool (Altschul et al., 1990). The genetic distance between different genotypes was analyzed using the Kimura 2-parameter model as a method for nucleotide substitution (Kimura, 1980). Phylogenetic trees were created using the Neighbor-Joining method (Saitou and Nei, 1987), and the statistical significance was obtained by 2,000 bootstrap repetitions (Efron and Gong, 1983). Accession numbersThe NoV and SaV sequences obtained in this study were added to the GenBank with the following accession numbers: canine GVI.P1 NoV from KM057711 to KM057714, human GII NoV from KM057715 to KM057730, canine GII.P4 NoV from KM057731 to KM057734, porcine GII.P4 NoV from KM057735 to KM057740, bovine GII.P4 NoV from KM057741 to KM057745, recombinant strain of human GII NoV KP067788, human GI.2 SaV KM001680, human GII.5 SaV KJ418887 and KM001679, canine SaV KM001673 and KM001674, and PoSaV from KM001675 to KM001678. Ethical approvalThe residual human diarrheal stool samples were sent from hospitals and public health reference centers located in the Costa Rican metropolitan area to the UNDIVE, School of Veterinary Medicine of National University, as part of the cooperation plan to monitor outbreaks of diarrhea in the country to detect the presence of Calicivirus by ensuring the privacy of patients and in accordance with Article 42 of the current Health Surveillance Regulation (Decree No. 40556-S, La Gaceta, Scope No. 2016, August 23rd. 2017). Meanwhile, the feces samples of animals were obtained from local diagnostic centers, cattle or pig farms with the prior consent of the producer and considering the Canadian Council on Animal Care guidelines on: the care and use of farm animals in research, teaching, and testing (Canadian Council on Animal Care, 2009). ResultsDetection of NoV and SaV in humans and animals by RT-PCRThe use of RT-PCR with the degenerate primer pair p290hijk(+)/p289hi(−) targeting the NoV and SaV RdRp region allowed the identification of 123 positive samples for NoV from 1,061 stool samples analyzed, and 2 HuSaV strains and 2 PoSaV strains were characterized by sequencing. In addition, the use of RT-PCR with a specific primer pair SR80(+)/JV33(−) targeting the SaV polymerase allowed the identification of 5 positive samples from the 1,023 stool samples analyzed. The 123 samples positive for NoV were genotyped with a second RT-PCR using degenerate primers NVG1f1b(+)/G1SKR(−) and NVG2flux1(+)/G2SKR(−) targeting the ORF1-ORF2 junction of genogroups GI and GII, respectively. This additional assay identified one human sample as GI NoV and six human samples as GII NoV; however, no animal samples were classified as GI or GII. The presence of SaV by RT-PCR targeting the NS segment of capsid protein VP1 with primers SLV-5317(+)/SLV-5749(−) was confirmed only in one human sample. A complete summary of these results is shown in Tables 2 and 3. NoV and SaV genotypes characterized in humans and animalsTo determine the nucleotide sequence similarity (ntS) between the NoV and SaV identified in humans and animals, 56 positive samples were selected from the products obtained with the primers used in this study to confirm their presence and assign the genotype. High-quality nucleotide (nt) sequences were obtained from 50 samples, while 36 samples were associated with NoV (17 in humans, 8 in dogs, 6 in pigs, and 5 in cattle) and 9 with SaV (3 in humans, 2 in dogs, and 4 in pigs). No sequence was determined in five of the samples. The sequence analysis was based on a 274 nt fragment of the polymerase for the sequences obtained with primers p290hijk(+)/p289hi(−) and a 332 nt fragment of the ORF1-ORF2 junction of the VP1 protein for sequences obtained with primers NVG1f1b(+)/G1SKR(−) and NVG2flux1(+)/G2SKR(−). On the other hand, in the case of SaV, the comparative sequence analysis was based on a 280 nt fragment of the polymerase for sequences obtained with primers SR80(+)/JV33(−), a 286 nt fragment of the polymerase for sequences obtained with primers p290hijk(+)/p289hi(−), and 388 nt of the NS subregion for sequences obtained with primers SLV-5317(+)/SLV-5749(−). A total of 17 human samples were verified as HuNoV using sequence analysis. Using the phylogenetic analysis, strain LZ075-D/07/CR detected in the study was classified within P-type GII.P2 because of its 86.7% ntS with the GII.2/Melksham/94 UK strain in the polymerase region; meanwhile, strain INC002-D10/10/CR showed a 94.3% ntS with the GII.6/SaitamaU3 Japanese strain in the polymerase region and was confirmed as a HuNoV GII.6 strain according to the phylogenetic information obtained from the ORF1-ORF2 junction, as it showed a greater association with reference strains GII.6/Seacroft/90/UK (95.2% ntS) and GII.6/SaitamaU3/JP (94.7% ntS), so it is a GII.6/GII.P6 strain. A total of 14 sequences were grouped within P-type GII.P4 based on polymerase phylogenetic information. This subgroup showed 95.9% ntS among groups when compared to the representative strains of GII.P4 reported in different countries (Farmington Hills/02/USA, Toronto/SK/02/CAN, AlbertaEI513/06/CAN, AlbertaEI003/12/CAN, Tokyo/10-1443/10/JP, Norwalk/10378/10/VNM, and Hunter504D/04O/AU). In addition, the presence of five strains was confirmed in genotype GII.4 according to the phylogenetic information obtained from the ORF1-ORF2 junction, all of which were highly associated with reference strains GII.4 AlbertaEI388/CAN (97.2% ntS), Norwalk10386/10/VNM (96.2%–97.2% ntS), and Farmington Hills/02/USA (95.7%–99.5% ntS) (Table 4, Fig. 1a and b). On the other hand, a recombinant strain of HuNoV GII.4 (INC313846-D/2012/CR) was determined to be present in the gene encoding the polymerase (GenBank accession number: KP067788). Table 2. Presence of NoV in human, canine, swine, and bovine fecal samples in Costa Rica during the 2002–2012 period.
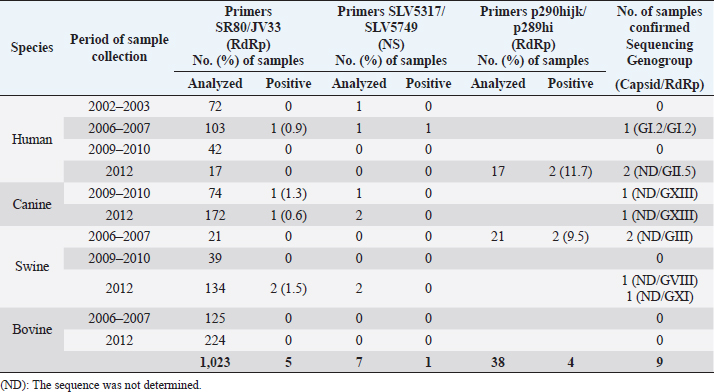
Table 3. Presence of SaV in human, canine, swine, and bovine fecal samples in Costa Rica during the 2002–2012 period.
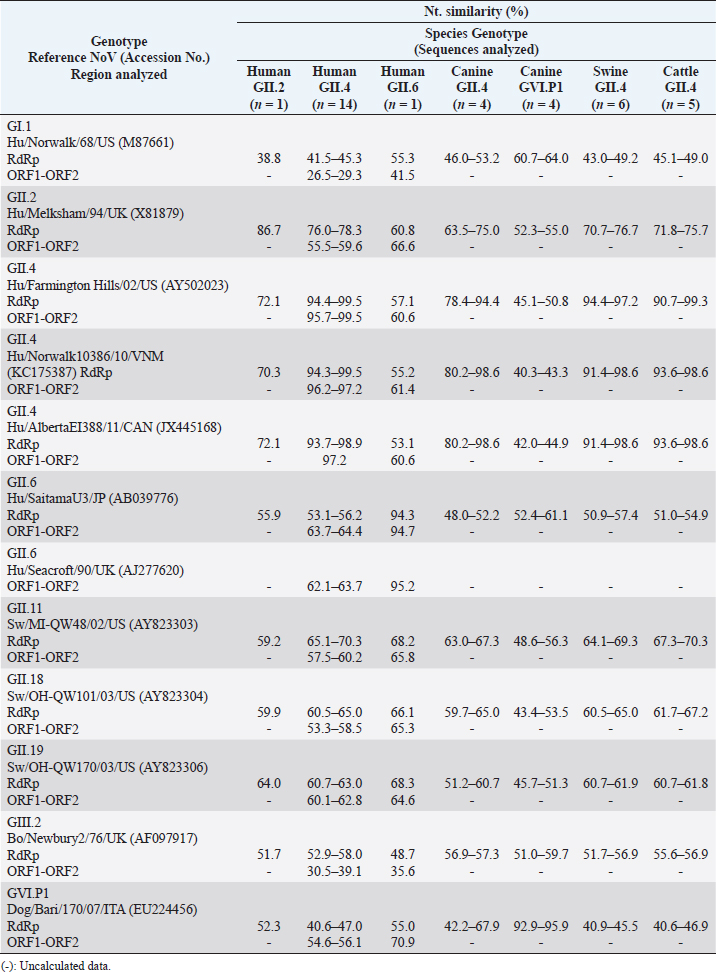
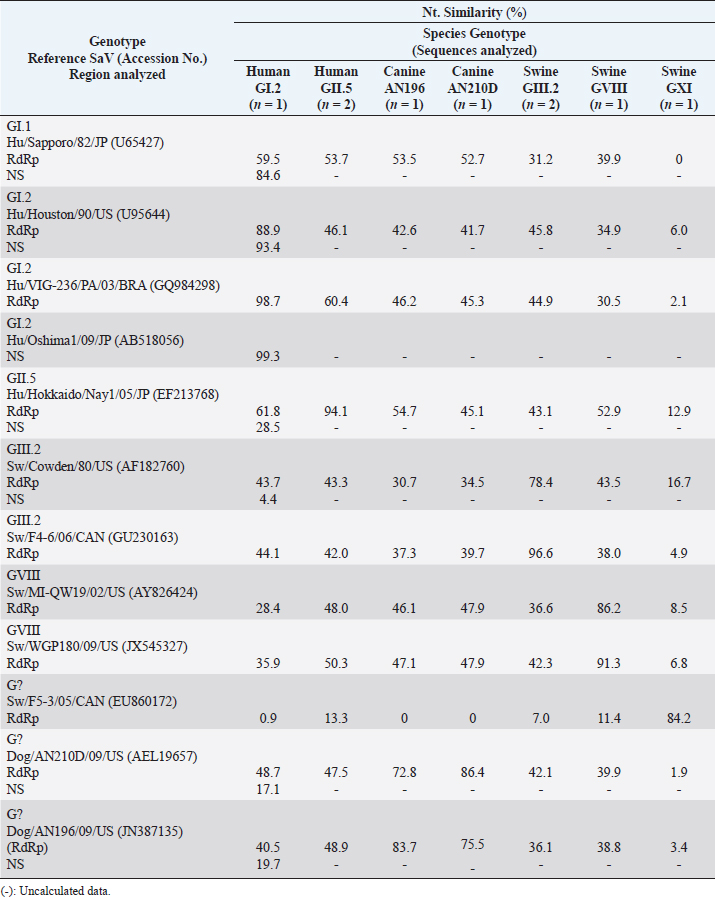
In animals, the sequence analysis was based solely on phylogenetic information obtained from the gene encoding the polymerase. Eight nt sequences obtained from canine samples were associated with NoV, of which strains CS-C011D/09/CR, C005-D00/12/CR, C010-D00/12/CR, and C107-D00/12/CR were grouped with reference strains of HuNoV GII.P4 (92.4% ntS between groups); meanwhile, the other four strains (CS-C022D/09/CR, C003-D00/12/CR, C006-D00/12/CR, and C057-D00/12/CR) showed a closer association with the representative strains of the canine P-types GVI.P1 (86.8% ntS between groups). In addition, six nt sequences obtained from pig fecal samples and five from cattle samples showed to be very similar to HuNoV GII.P4, with 96.4% and 95.5% ntS among groups with HuNoV GII.P4 reference strains, respectively (Table 4, Fig. 1a). Strain HuSaV SJ164-D11/2007/CR identified in this study was very similar to the GI.2/VIG-236/PA strain from Brazil (98.7% ntS) (Aragão et al., 2010) and the GI.2/Houston/90 strain from the USA (88.9% ntS). The canine strains identified here (C014-D10/2009/CR and C104-D00/2012/CR) were genetically similar to the AN196 (83.7% ntS) and AN210D (86.4% ntS) SaV strains recorded in the USA, respectively (Li et al., 2011). In addition, porcine strain CP12-SD21/2012/CR was closely associated with USA PoSaV strains GVIII/WGP180 (91.3% ntS) and GVIII/MI-QW19 (86.2% ntS) (Wang et al., 2006). Porcine strain CP20-SD21/2012/CR showed greater genetic similarity to unknown genogroup (G?) of Canada’s porcine strain F5-3 (84.2% ntS) (L’Homme et al., 2009) and, together with Brazil’s BRA21-RS strain (Barry et al., 2008), formed a new genogroup (GXI) (Fig. 2a and detailed information in Table 5). The nt sequence obtained with the SLV5317(+)/SLV5749(-) primers allowed us to verify that the HuSaV SJ164-D11/07/CR strain was human GI.2 SaV according to the phylogenetic information of the NS segment of the VP1 protein. However, it showed greater genetic similarity to Japan’s GI.2/Oshima1/09 reference strain (99.3% ntS) (Miyoshi et al., 2010) (Table 5, Fig. 2b). From the four sequences obtained with primers p290hijk(+)/p289hi(−) (Table 3), two were classified as PoSaV GIII (strains BOP007-D10/2007/CR and BOP017-D10/2007/CR) because they exhibited a high degree of genetic similarity with reference strains from Canada GIII/F4-6/06 (96.6% ntS) and from the USA GIII/Cowden/80 (78.4% ntS) (Wang et al., 2005); meanwhile, the other two are associated mostly with HuSaV (strains INC313871-D00/12/CR and INC313934-D00/12/CR), grouping with genotype GII.5 strains and showing greater genetic relationship with the GII.5/Hokkaido/Nay1/05 strain from Japan (94.1% ntS) associated with gastroenteritis outbreaks (Hansman et al., 2007) (Table 5, Fig. 2a). Table 4. Genetic similarity between NoV strains detected in humans, canines, swine, and cattle with prototype NoV strains of the four species.
Fig. 1. Phylogenetic analysis of NoV. (a): The tree was constructed using the 274 nt partial sequence of the polymerase. (b): The tree was constructed using the 332 nt partial sequence of the ORF1-ORF2 junction. Statistical significance was determined using the bootstrap method from 2,000 resamples; values are displayed above the node. The GenBank accession number of the reference strains is shown in parentheses. The Hu/SaV/Manchester/93/UK (X86560) and Hu/Winchester/94/UK (AJ277609) strains were used as an outgroup for trees (a) and (b), respectively. The strains detected in the study are color-coded (human strains are green, canine blue, swine red, and bovine purple).
Fig. 2. Phylogenetic analysis of SaV. (a): The tree was constructed using the 280 nt partial sequence of the polymerase. (b): The tree was constructed using the 388 nt partial sequence of the NS subregion. Statistical significance was determined using the bootstrap method from 2,000 resamples; values are displayed above the node. The GenBank accession number of the reference strains is shown in parentheses. The Hu/NoV/Norwalk/68-93/US (M87661) and Mc10/00/TH (AY237420) strains are used as an outgroup for trees (a) and (b), respectively. The strains detected in the study are color-coded (human strains are green, swine red, and canine blue). Table 5. Genetic similarity between SaV strains detected in humans, canines, and swine with prototype SaV strains of the three species.
Phylogenetic comparison of NoV and SaV in humans and animalsAll strains identified in humans were compared with those detected in animals, and the high prevalence of the HuNoV GII.P4 in Costa Rican species was noticeable. The strains detected in dogs, pigs, and cattle showed a high degree of genetic association with the HuNoV GII.P4 strains characterized in humans, showing 91.9%, 96.0%, and 95.1% ntS between groups, respectively. On the other hand, the ntS identified in the study in human HuNoV GII.P4 strains was 89.8%–100%, 79.4%–100% in canine HuNoV GII.P4 strains, 91.4%–100% in porcine HuNoV GII.P4 strains, 87.6%–100% in cattle HuNoV GII.P4 strains, and 83.3%–100% in CaNoV GVI.P1 strains. Regarding the similarity between human, canine, and PoSaV, the HuSaV GII.5 strains detected here showed 51% ntS in the RdRp region with the porcine GVIII/CP12-SD21/12/CR strain. In comparison, swine GIII strains showed greater similarity to human strain GI.2/Houston/90 from the USA (45.8% ntS), and canine strain C014-D10/2009/CR showed 53.5% ntS with the HuSaV GI.1/Sapporo/82 strain from Japan. DiscussionFew studies have sought to explore the molecular epidemiology of NoV and SaV in humans and animals simultaneously. This article describes a comprehensive and integrated analysis of the detection, molecular characterization, and phylogeny between NoV and SaV identified in human, canine, porcine, and cattle fecal samples from 2002 to 2012 fragmented periods. Different studies have documented the HuNoV genetic diversity in the pediatric population as a cause of acute diarrhea and vomiting, being genogroup GII more prevalent than GI and genotype GII.4 is the most prevalent within genogroup GII (Bull and White, 2011; Hasing et al., 2019; Mans, 2019). In this study, the sequence analysis confirmed the presence of HuNoV G?/GII.P2, G?/GII.P4, GII.4/GII.P4, and GII.6/GII.P6 in human diarrheal samples, dominated by HuNoV G?/GII.P4 and GII.4/GII.P4 during the periods evaluated, which being consistent with other reports in which HuNoV GII.4 is the main cause of human gastroenteritis (Bucardo et al., 2008; Gómez-Santiago et al., 2012). Canine P-types GVI of NoV strains were found in less than seven-month-old dogs with diarrhea. Our findings were similar to those reported in puppies with diarrhea for the Bari/91/07 strain in Italy (Martella et al., 2009), the C33/Viseu/07 strain in Portugal (Mesquita et al., 2010), and the Thessaloniki/30/08 strain in Greece (Ntafis et al., 2010). Canine strains CS-C011D/09/CR, C005-D00/12/CR, C010-D00/12/CR, and C107-D00/12/CR were mostly associated with reference strain HuNoV GII.P4/Farmington Hills/02/USA. The circulation of HuNoV GII in dogs in Brazil (Sokel and Kale, 2019), canine strains IC-09, 261-10, and 3-09 in Finland were related to genetic variants GII.4-2006b and GII.4-2008, which were associated with the human GII.4/Bristol/93/UK reference strain (Summa et al., 2012). Like so, the transmission of HuNoV GII.e-GII.4 Sydney from human to canine have also been reported (Charoenkul et al., 2020). On the other hand, knowledge about the ability of HuNoV to attach to in vitro canine tissues and the presence of anti-HuNoV antibodies in dogs suggest the possibility of productive infection (Caddy et al., 2014, 2015). Therefore, given that this research study has shown that HuNoV GII.4 strains have circulated in domestic dogs, HuNoV zoo-anthropozoonotic implications are very strong, and the risk to generate recombinant strains between HuNoV and CaNoV present in dogs is high, while the zoonotic potential for GIV and GVI NoV has been reported to be limited (Ford-Siltz et al., 2019). Regarding pigs, direct contact with HuNoV has been demonstrated by serological findings in the pig population of Nicaragua (Bucardo et al., 2016), and the circulation of strains related to HuNoV GII.4, GII.3, and GII.13 in Japan (Nakamura et al., 2010) has been described. In this study, strains associated with the HuNoV GII.4 genotype were found in diarrheal samples. Strains CP021-D00/07, CS-CP009D/10, CS-CP010D/10, CP001-SD/12, CP060-D00/12, and CP21094-SD/12 showed great ntS with human strain GII.P4/Farmington Hills/02/USA. In addition, porcine strains very similar to the reference strain GII.P4/Farmington Hills/02/USA were reported in Canada (Mattison et al., 2007). Although it has been shown that the HuNoV GII.4/HS66/01/USA and variant GII.4-2006b can be replicated and cause acute disease in gnotobiotic pigs (Cheetham et al., 2006; Bui et al., 2013), in this study, the evidence found in the field confirms that HuNoV GII.P4 infects pigs and that the absence of PoNoV prototypes could be associated with the circulation of an unknown strain common between humans and animals in Costa Rica, unlike the study described by Cavicchio et al. (2020). This study also demonstrates the presence of HuNoV GII.P4 in cow feces. Previously, the circulation of the GII.4/CE-M-06-0509/07 strain has been reported in Canada, which is closely associated with the reference strain Farmington Hills/02/USA (Mattison et al., 2007). In addition, it has been proven that the HuNoV GII.4/HS66 can cause infections and disease in cattle (Souza et al., 2008), although the evidence found in this study suggests that cattle could spread the virus, Oliver et al. (2003) did not obtain evidence to support that HuNoV circulates between these two species. Genetically variable NoV strains related to HuNoV have been reported present in animals, which may indicate that animals are NoV reservoirs or are being infected by humans directly or indirectly (Wang et al., 2007; Scipioni et al., 2008; Martella et al., 2009; Bank-Wolf et al., 2010; Caddy et al., 2015). In this sense, this study shows the circulation of strains associated with HuNoV GII.P4 in the four species evaluated, which suggests that humans facilitate animal infection by indirect contact through the consumption of water and food contaminated with human waste, resulting in zoo-anthropozoonosis caused by recombinant strains G?/GII.P4, which are capable of infecting dogs, pigs, and cattle due to the occurrence of essential and necessary changes in the VP1 region of genotype GII.4 that allowed it to stabilize as described by Sato et al. (2017) and with an impressive capacity to infect other species. The foregoing could also be explained by a co-infection between human and animal strains along with the ability of RNA polymerase to switch templates and promote the development of recombinant RNA viruses (White, 2014). Given that recombination is an evolutionary mechanism used by NoV (Ludwing-Begall et al., 2018) that causes major changes in the viral genome and that intergenogroup recombinations have been recorded between GII.9/GI.7 (Reuter et al., 2010) and GI.3/GII.4 (Nayak et al., 2008), it is likely that the undetermined phylogenetic information from the capsid region in most strains characterized in this study as HuNoV GII.P4 in humans and animals is associated with recombinant virus polymerase of prototype genotypes of each animal, or it is a common strain with G?/GII.P4. Consequently, we would agree with the hypothesis described by Villabruna et al. (2019), which states that NoV may not be host restricted and might be able to jump the species barrier, especially HuNoV GII.4 due to their ample capacity to generate high replication rates (Ludwing-Begall et al., 2021), resulting in the zoo-anthropozoonotic events that have long occurred in Costa Rica. Therefore, further research is essential to determine the phylogenetic information of the VP1 region and clarify the significance of these findings. Regarding SaVs, the low genetic diversity of HuSaV observed in humans in this study is comparable to what was reported in Mexico (Gómez-Santiago et al., 2012), Argentina (Gomes et al., 2008), Brazil (Aragão et al., 2010), and Nicaragua (Bucardo et al., 2012) where genotypes GI.1, GI.2, and GII.1 HuSaV were reported in under 5-year-old children with diarrhea. Unlike those records, this study describes the presence of genotype GII.5. In pigs, the identified genetic diversity of PoSaV (genogroups GIII, GVIII, and GXI) was broader than the one reported in Brazil (genogroups GIII and GXI) (Barry et al., 2008), while in dogs, Li et al. (2011) proposed that the AN210D and AN196 CaSaV strains found in the USA should be classified into a new genogroup (GIX). In addition, this study identified two CaSaV strains (C014-D10/09 and C104-D00/12), which were very similar to the strains described above and, like those strains, were grouped within the same genogroup (Fig. 2a). Soma et al. (2015) reported the presence of CaSaV that were very similar to the USA strains (72.5%–86.5% ntS), and Bodnar et al. (2016) also described two CaSaV strains that were closely related to strain AN210D (90.4% ntS) in the polymerase. Therefore, the evidence found in this and other studies demonstrates the clear segregation of CaSaV as a new genogroup GXIII (Kuroda et al., 2017). Due to the phylogenetic information obtained from the polymerase in other studies, considering GIX as a CaSaV genogroup is very controversial because pigs are infected by genogroup GIX (Wang et al., 2005; Martella et al., 2008a; Nakamura et al., 2010; Reuter et al., 2010; Dufkova et al., 2011; Diez-Valcarce et al., 2018). No SaV were detected in cattle in this investigation. Similarly, Smiley et al. (2003) and Mijovski et al. (2010) reported the absence of SaV in cattle. On the other hand, there was not a strong genetic similarity of polymerase among the human, canine, and PoSaV identified in this research or among the host species, unlike Oka et al. (2015), who suggested not using the polymerase region for genotyping because it is less diverse. So, unlike NoV, the zoonotic potential of SaV is limited because the SaV in animals were distant from human strains, as indicated by Eu Lim et al. (2020) for PoSaV. Overall, the results of this study suggest that NoV and SaV should be included in epidemiological surveillance programs. Since the prevalence of genotypes varies geographically, active monitoring of NoV and SaV circulating strains in humans and animals will provide evidence to optimize prevention measures for gastroenteritis cases and promote vaccine formulation. AcknowledgmentsWe would like to express our sincere thanks to Dr. Mario Baldi and MSc. Martha Piche for their invaluable collaboration in collecting stool samples from pigs and cattle for 2012; Dra. Magaly Caballero (ACOPSA laboratory, Heredia, Costa Rica) for providing residual canine fecal samples for the 2009–2012 period, and Dra. Sabine Huter (Epidemiological Surveillance Section-Senasa) for facilitating the agricultural census of the San Carlos region, Alajuela, Costa Rica. We would also like to thank Ing. Reynaldo Pereira for his assistance in sequencing nucleic acids and Dr. Stephen Baker (London School of Hygiene and Tropical Medicine, London, United Kingdom) for his observations. Our special thanks to the German Academic Exchange Service (DAAD) for their regional scholarship program for graduate studies, Vice-rectory for Research of National University for the translation from Spanish to English of this article, and Postgraduate Program in Agricultural Sciences and Natural Resources (PPCARN), University of Costa Rica (UCR) for their national scholarship program. Conflict of interestThe authors declare that there is no conflict of interest. Author’s contributionsDesigned the project and wrote the article, D.P., and C.J.; Ran the stool sample analysis, D.P., C.S. and R.C.; Performed sequencing and phylogenetic analysis of the data, D.P.; critically reviewed the scientific content of the manuscript, C.S., H.B., and E.C. All authors read and approved the final manuscript. FundingThis research was funded by the Tropical Disease Research Program (PIET); Virology Laboratory, School of Veterinary Medicine of National University of Costa Rica; Network for Research and Training in Tropical Diseases in Central America (NeTropica) [Proyecto N° 05-N-2010], and the German Academic Exchange Service (DAAD). ReferencesAltschul, S.F., Gish, W., Miller, W., Myers, E.W. and Lipman, D.J. 1990. Basic local alignment search tool. J. Mol. Biol. 215, 403–410. Aragão, G.C., Oliveira, D.D., dos Santos, M.C., Mascarenhas, J.D., Oliveira, C.D., Linhares A.D. and Gabbay, Y.B. 2010. Molecular characterization of norovirus, sapovirus and astrovirus in children with acute gastroenteritis from Belém, Pará, Brazil. Rev. Pan-Amaz. Saude. 1, 149–158. Bank-Wolf, B.R., König, M. and Thiel, H-J. 2010. Zoonotic aspects of infections with noroviruses and sapoviruses. Vet. Microbiol. 140, 204–212. Barry, A.F., Alfieri, A.F. and Alfieri, A.A. 2008. High genetic diversity in RdRp gene of Brazilian porcine sapovirus strains. Vet. Microbiol. 131, 185–191. Bodnar, L., Di Martino, B., Di Profio, F., Melegari, I., Lanave, G., Lorusso, E., Cavalli, A., Elia, G., Bányai, K., Marsilio, F., Buonavoglia, C. and Martella, V. 2016. Detection and molecular characterization of sapoviruses in dogs. Infect. Genet. Evol. 38, 8–12. Bourdett-Stanziola, L., Jiménez, C. and Ortega-Barria, E. 2008. Diversity of human rotavirus G and P genotypes in Panama, Costa Rica, and the Dominican Republic. Am. J. Trop. Med. Hyg. 79, 921–924. Bucardo, F., Nordgren, J., Carlsson, B., Paniagua, M., Lindgren, P-E., Espinoza, F. and Svensson, L. 2008. Pediatric norovirus diarrhea in Nicaragua. J. Clin. Microbiol. 46, 2573–2580. Bucardo, F., Carlsson, B., Nordgren, J., Larson, G., Blandon, P., Vilchez, S. and Svensson, L. 2012. Susceptibility of children to sapovirus infections Nicaragua, 2005-2006. Emerg. Infect. Dis. 18, 1875–1878. Bucardo, F., González, F., Reyes, Y., Blandón, P., Saif, L.J. and Nordgren, J. 2016. Seroprevalence in household raised pigs indicate high exposure to GII noroviruses in rural Nicaragua. Zoonoses Public Health 63, 600–607. Bui, T., Kocher, J., Li, Y., Wen, K., Li, G., Liu, F., Yang, X., LeRoith, T., Tan, M., Xia, M., Zhong, W., Jiang, X. and Yuan, L. 2013. Median infectious dose of human norovirus GII.4 in gnotobiotic pigs is decreased b simvastatin treatment and increased by age. J. Gen. Virol. 94, 2005–2016. Bull, R.A., Tanaka, M.M. and White, P.A. 2007. Norovirus recombination. J. Gen. Virol. 88, 3347–3359. Bull, R.A. and White, P.A. 2011. Mechanisms of GII.4 norovirus evolution. Trends Microbiol. 19, 233–240. Caddy, S.L., Breiman, A., Le Pendu, J. and Goodfellow, I. 2014. Genogroup IV and VI canine noroviruses interact with histoblood group antigens. J. Virol. 88, 10377–10391. Caddy, S.L., de Rougemont, A., Emmott, E., El-Attar, L., Mitchell, J.A., Hollinshead, M., Belliot, G., Le Pendu, J. and Goodfellow, I. 2015. Evidence for human norovirus infection of dogs in the United Kingdom. J. Clin. Microbiol. 53, 1873–1883. Cavicchio, L., Laconi, A., Piccirillo, A. and Beato, M.S. 2022. Swine norovirus: past, present, and future. Viruses 14, 537. Cavicchio, L., Tassoni, L., Laconi, A., Cunial, G., Gagliazzo, L., Milani, A., Campalto, M., Di Martino, G., Forzan, M., Monne, I. and Beato, M.S. 2020. Unrevealed genetic diversity of GII norovirus in the swine population of North East Italy. Sci. Rep. 10, 9217. Canadian Council on Animal Care. 2009. CCAC guidelines on: the care and use of farm animals in research, teaching and testing. Available via https://www.ccac.ca/Documents/Standards/Guidelines/Farm_Animals.pdf Charoenkul, K., Nasamran, C., Janetanakit, T., Tangwangvivat, R., Bunpapong, N., Boonyapisitsopa, S. and Amonsin, A. 2020. Human norovirus infection in dogs, Thailand. Emerg. Infect. Dis. 26, 350–353. Cheetham, S., Souza, M., Meulia, T., Grimes, S., Han, M.G. and Saif, L.J. 2006. Pathogenesis of a genogroup II human norovirus in gnotobiotic pigs. J. Virol. 80, 10372–10381. Chhabra, P., de Graaf, M., Parra, G.I., Chan, M.C-W., Green, K., Martella, V.,Wang, Q., White, P.A., Katayama, K., Vennema, H., Koopmans, M.P.G. and Vinjé, J. 2019. Updated classification of norovirus genogroups and genotypes. J. Gen. Virol. 100, 1393–1406. Dastjerdi, A.M., Green, J., Gallimore, C.I., Brown, D.W. and Bridger, J.C. 1999. The bovine newbury agent-2 is genetically more closely related to human SRSVs than to animal caliciviruses. Virology 254, 1–5. Dey, S.K., Sumiya, M.K., Shaha, M., Haque, R., Okitsu, S. and Ushijima, H. 2018. Intragenogroup recombination in the complete genome sequence of human sapovirus circulating in Bangladesh. Genome. Announc. 6, e00388–18. Diez-Valcarce, M., Castro, C.J., Marine, R.L., Halasa, N., Mayta, H., Saito, M., Tsaknaridis, L., Pan, CY., Bucardo, F., Becker-Dreps, S., Lopez, MR., Magaña, LC., Ng T.F.F. and Vinjé, J. 2018. Genetic diversity of human sapovirus across the Americas. J. Clin. Virol. 104, 65–72. Di Felice, E., Mauroy, A., Pozzo, F.D., Thiry, D., Ceci, C., Di Martino, B., Marsilio, F. and Thiry, E. 2016. Bovine noroviruses: a missing component of calf diarrhoea diagnosis. Vet. J. 207, 53–62. Dufkova, L., Kulich, P. and Prodelalova, J. 2011. Molecular characterization of a porcine sapovirus strain isolated from a piglet with diarrhoea. Vet. Med. 56, 409–415. Efron, B. and Gong, G. 1983. A Leisurely look at the bootstrap, the jackknife, and cross-validation.Am. Stat. 37, 36–48. Eu Lim, L., Byung-Joo, P., Hee-Seop, A., Sang-Hoon, H., Hyeon-Jeong, G., Dong-Hwi, K. and In-Soo, C. 2020. Detection and genetic analysis of zoonotic hepatitis E virus, rotavirus, and sapovirus in pigs. Korean J. Vet. Res. 60, 61–68. Ford-Siltz, L.A., Mullis, L., Sanad, Y.M., Tohma, K., Lepore, C.J., Azevedo, M. and Parra, G.I. 2019. Genomics analyses of GIV and GVI noroviruses reveal the distinct clustering of human and animal viruses. Viruses 11, E204. Farkas, T., Zhong, W.M., Jing, Y., Huang, P.W., Espinosa, S.M., Martinez, N., Morrow, A.L., Ruiz-Palacios, G.M., Pickering, L.K. and Jiang, X. 2004. Genetic diversity among sapoviruses. Arch. Virol. 149, 1309–1323. Gomes, K.A., Stupka, J.A., Diana, A. and Parra, G.I. 2008. Caracterización molecular de calicivirus aislados de brotes de gastroenteritis ocurridos en la Argentina durante los años 2005 y 2006. Rev. Argent. Microbiol. 40, 222–228. Gómez-Santiago, F., Ribas-Aparicio, R.M. and García-Lozano, H. 2012. Molecular characterization of human calicivirus associated with acute diarrheal disease in mexican children. Virol. J. 9, 54. Günther, H. and Otto, P. 1987. Diarrhea in young calves. 7. "Zackenvirus" (Jena agent 117/80)-a new diarrhea pathogen in calves. Arch. Exp. Vet. 41, 934–938. Hall, A.J., Wikswo, M.E., Pringle, K., Gould, L.H. and Parashar, U.D. 2014. Vital signs: Foodborne norovirus outbreaks-United States, 2009-2012. Morb. Mortal Wkly. Rep. 63, 491–495. Hansman, G.S., Katayama, K., Maneekarn, N., Peerakome, S., Khamrin, P., Tonusin, P.K.S. and Ushijima, H. 2004. Genetic diversity of norovirus and sapovirus in hospitalized infants with sporadic cases of acute gastroenteritis in Chiang Mai, Thailand. J. Clin. Microbiol. 42, 1305–1307. Hansman, G.S., Ishida, S., Yoshizumi, S., Miyoshi, M., Ikeda, T., Oka, T. and Takeda, N. 2007. Recombinant sapovirus gastroenteritis, Japan. Emerg. Infect. Dis. 13, 786–788. Hasing, M.E., Lee, B.E., Qiu, Y., Xia, M., Pabbaraju, K., Wong, A., Tipples, G., Jiang, X. and Pang, X.L. 2019. Changes in norovirus genotype diversity in gastroenteritis outbreaks in Alberta, Canada: 2012-2018. BMC Infect. Dis. 19, 177. Jiang, X., Huang, P.W., Zhong, W.M., Farkas, T., Cubitt, D.W. and Matson, D.O. 1999. Design and evaluation of a primer pair that detects both norwalk-like and sapporo-like caliciviruses by RT-PCR. J. Virol. Methods 83, 145–154. Katayama, K., Miyoshi, T., Uchino, K., Oka, T., Tanaka, T., Takeda, N. and Hansman, G.S. 2004. Novel recombinant sapovirus. Emerg. Infect. Dis. 10, 1874–1876. Katayama, K., Shirato-Horikoshi, H., Kojima, S., Kageyama, T., Oka, T., Hoshino, F., Fukushi, S., Shinohara, M., Uchida, K., Suzuki, Y., Gojobori, T. and Takeda, N. 2002. Phylogenetic analysis of the complete genome of 18 Norwalk-like viruses. Virology 299, 225–239. Kimura, M. 1980. A simple method for estimating evolutionary rates of base substitutions through comparative studies of nucleotide sequences. J. Mol. Evol. 16, 111–120. Kojima, S., Kageyama, T., Fukushi, S., Hoshino, F.B., Shinohara, M., Uchida, K., Natori, K., Takeda, N. and Katayama, K. 2002. Genogroup-specific PCR primers for detection of norwalk-like viruses. J. Virol. Methods. 100, 107–114. Kumar, S., Stecher, G., Li, M., Knyaz, C. and Tamura, K. 2018. MEGA X: Molecular evolutionary genetics analysis across computing platforms. Mol. Biol. Evol. 35, 1547–1549. Kuroda, M., Masuda, T., Ito, M., Naoi, Y., Doan, Y.H., Haga, K., Tsuchiaka, S., Kishimoto, M., Sano, K., Omatsu, T., Katayama, Y., Oba, M., Aoki, H., Ichimaru, T., Sunaga, F., Mukono, I., Yamasato, H., Shirai, J., Katayama, K., Mizutani, T., Oka, T. and Nagai, M. 2017. Genetic diversity and intergenogroup recombination events of sapoviruses detected from feces of pigs in Japan. Infect. Genet. Evol. 55, 209–217. L’Homme, Y., Sansregret, R., Plante-Fortier, É., Lamontagne, A-M., Lacroix, G., Ouardani, M., Deschamps, J., Simard, G. and Simard, C. 2009. Genetic diversity of porcine norovirus and sapovirus: Canada, 2005-2007. Arch. Virol. 154, 581–593. Li, L., Pesavento, P.A., Shan, T., Leutenegger, C.M., Wang, C. and Delwart, E. 2011. Viruses in diarrhoeic dogs include novel kobuviruses and sapoviruses. J. Gen. Virol. 92, 2534–2541. Ludwig-Begall, L.F., Mauroy, A. and Thiry, E. 2018. Norovirus recombinants: recurrent in the field, recalcitrant in the lab—a scoping review of recombination and recombinant types of noroviruses. J. Gen. Virol. 99, 970–988. Ludwig-Begall, L.F., Mauroy, A. and Thiry, E. 2021. Noroviruses-the state of the art, nearly fifty years after their initial discovery. Viruses 13, 1541. Lun, J.H., Hewitt, J., Sitabkhan, A., Eden, J.S., Enosi Tuipulotu, D., Netzler, N.E., Morrell, L., Merif, J., Jones, R., Huang, B., Warrilow, D., Ressler, K.A., Ferson, M.J., Dwyer, D.E., Kok, J., Rawlinson, W.D., Deere, D., Crosbie, N.D. and White, P.A. 2018. Emerging recombinant noroviruses identified by clinical and waste water screening. Emerg. Microbes Infect. 7, 50. Lyoo, EL., Park, B.J., Ahn, H.S., Han, S.H., Go, H.J., Kim, D.H., Lee, J.B., Park, S.Y., Song, C.S., Lee, S.W. and Choi, I.S. 2020. Detection and genetic analysis of zoonotic hepatitis E virus, rotavirus, and sapovirus in pigs. Korean J. Vet. Res. 60, 61–68. Magwalivha, M., Kabue, J.P., Traore, A.N. and Potgieter, N. 2018. Prevalence of human sapovirus in low and middle income countries. Adv. Virol. 2018, 5986549. Mans, J. 2019. Norovirus infections and disease in lower-middle and low-income countries, 1997-2018. Viruses 11, E341. Martella, V., Bányai, K., Lorusso, E., Bellacicco, A.L., Decaro, N., Mari, V. and Buonavoglia, C. 2008a. Genetic heterogeneity of porcine enteric caliciviruses identified from diarrhoeic piglets. Virus Genes 36, 365–373. Martella, V., Lorusso, E., Banyai, K., Decaro, N., Corrente, M., Elia, G., Cavalli, A., Radogna, A., Costantini, V., Saif, L.J., Lavazza, A., Di Trani, L. and Buonavoglia, C. 2008b. Identification of a porcine calicivirus related genetically to human sapoviruses. J. Clin. Microbiol. 46, 1907–1913. Martella, V., Decaro, N., Lorusso, E., Radogna, A., Moschidou, P., Amorisco, F.Lucente, M.S, Desario C, Mari V, Elia G, Banyai K, Carmichael LE and Buonavoglia, C. 2009. Genetic heterogeneity and recombination in canine noroviruses. J. Virol. 83, 11391–11396. Martella, V., Lorusso, E., Decaro, N., Elia, G., Radogna, A., D’Abramo, M., Desario, C., Cavalli, A., Corrente, M., Camero, M., Germinario, C.A., Bányai, K., Di Martino, B., Marsilio, F., Carmichael, L.E. and Buonavoglia, C. 2008c. Detection and molecular characterization of a canine norovirus. Emerg. Infect. Dis. 14, 1306–1308. Mathijs, E., Denayer, S., Palmeira, L., Botteldoorn, N., Scipioni, A., Vanderplasschen, A., Thiry, E. and Dierick, K. 2011. Novel norovirus recombinants and of GII.4 sub-lineages associated with outbreaks between 2006 and 2010 in Belgium. Virol. J. 8, 310–322. Mattison, K., Shukla, A., Cook, A., Pollari, F., Friendship, R., Kelton, D., Bidawid, S. and Farber, J.M. 2007. Human noroviruses in swine and cattle. Emerg. Infect. Dis. 13, 1184–1188. Mesquita, J.R., Barclay, L., Nascimento, M.S. and Vinjé, J. 2010. Novel norovirus in dogs with diarrhea. Emerg. Infect. Dis. 16, 980–982. Mijovski, J.Z., Poljsak-Prijatelj, M., Steyer, A., Barlic-Maganja, D. and Koren, S. 2010. Detection and molecular characterisation of noroviruses and sapoviruses in asymptomatic swine and cattle in Slovenian farms. Infect. Genet. Evol. 10, 413–420. Miyoshi, M., Yoshizumi, S., Kanda, N., Karino, T., Nagano, H., Kudo, S., Okano, M. and Ishida, S. 2010. Different genotypic sapoviruses detected in two simultaneous outbreaks of gastroenteritis among schoolchildren in the same school district in Hokkaido, Japan. Jpn. J. Infect. Dis. 63, 75–78. Nakamura, K., Saga, Y., Iwai, M., Obara, M., Horimoto, E., Hasegawa, S., Kurata, T., Okumura, H., Nagoshi, M. and Takizawa, T. 2010. Frequent detection of noroviruses and sapoviruses in swine and high genetic diversity of porcine sapovirus in Japan during fiscal year 2008. J. Clin. Microbiol. 48, 1215–1222. Nayak, M.K., Balasubramanian, G., Sahoo, G.C., Bhattacharya, R., Vinje, J., Kobayashi, N., Sarkar, M.C., Bhattacharya, M.K. and Krishnan, T. 2008. Detection of a novel intergenogroup recombinant norovirus from Kolkata, India. Virology 377, 117–123. Nordgren, J., Bucardo, F., Dienus, O., Svensson, L. and Lindgren, P.E. 2008. Novel light-upon-extension real-time PCR assays for detection and quantification of genogroup I and II noroviruses in clinical specimens. J. Clin. Microbiol. 46, 164–170. Ntafis, V., Xylouri, E., Radogna, A., Buonavoglia, C. and Martella, V. 2010. Outbreak of canine norovirus infection in young dogs. J. Clin. Microbiol. 48, 2605–2608. Oka, T., Wang, Q.H., Katayama, K. and Saif, L.J. 2015. Comprehensive review of human sapoviruses. Clin. Microbiol. Rev. 28, 32–53. Oliver, S.L., Dastjerdi, A.M., Wong, S., El-Attar, L., Gallimore, C., Brown, D.W. and Bridger, J.C. 2003. Molecular characterization of bovine enteric caliciviruses: a distinct third genogroup of noroviruses (Norwalk-Like viruses) unlikely to be of risk to humans. J. Virol. 77, 2789–2798. Parra, G.I. 2019. Emergence of norovirus strains: a tale of two genes. Virus Evolution 5, vez048. Reuter, G., Zimsek-Mijovski, J., Poljsak-Prijatel, M., Di Bartolo, I., Ruggeri, F.M., Kantala, T., Maunula, L., Kiss, I., Kecskeméti, S., Halaihel, N., Buesa, J., Johnsen, C., Hjulsager, C.K., Larsen, L.E., Koopmans, M. and Böttiger, B. 2010. Incidence, diversity, and molecular epidemiology of sapoviruses in swine across Europe. J. Clin. Microbiol. 48, 363–368. Saitou, N. and Nei, M. 1987. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 4, 406–425. Sato, H., Yokoyama, M., Nakamura, H., Oka, T., Katayama, K., Takeda, N., Noda, M., Tanaka, T. and Motomura, K. 2017. Evolutionary constraints on the norovirus pandemic variant GII.4_2006b over the five-year persistence in Japan. Front. Microbiol. 8, 410. Scipioni, A., Mauroy, A., Vinjé, J. and Thiry, E. 2008. Animal noroviruses. Vet. J. 178, 32–45. Shibata, S., Sekizuka, T., Kodaira, A., Kuroda, M., Haga, K., Doan, Y.H., Takai-Todaka, R., Katayama, K., Wakita, T., Oka, T. and Hirata, H. 2015. Complete genome sequence of a novel GV.2 sapovirus strain, NGY-1, detected from a suspected foodborne gastroenteritis outbreak. Genome Announc. 3, e01553–14. Smiley, J.R., Hoet, A.E., Travén, M., Tsunemitsu, H. and Saif, L.J. 2003. Reverse transcription-PCR assays for detection of bovine enteric caliciviruses (BEC) and analysis of the genetic relationships among BEC and human caliciviruses. J. Clin. Microbiol. 41, 3089–3099. Sokel, S. and Kale, M. 2019. Investigation of norovirus genogroups (GI, GII and GIV) in stool of pet dogs with diarrhea. Pesq. Vet. Bras. 39, 402–408. Soma, T., Nakagomi, O., Nakagomi, T. and Mochizuki, M. 2015. Detection of norovirus and sapovirus from diarrheic dogs and cats in Japan. Microbiol. Immunol. 59, 123–128. Souza, M., Azevedo, M.S., Jung, K., Cheetham, S. and Saif, L.J. 2008. Pathogenesis and immune responses in gnotobiotic calves after infection with the genogroup II.4-HS66 strain of human norovirus. J. Virol. 82, 1777–1786. Summa, M., von Bonsdorff, C-H. and Maunula, L. 2012. Pet dogs: A transmission route for human noroviruses. J. Clin. Virol. 53, 244–247. Tamura, K., Stecher, G., Peterson, D., Filipski, A. and Kumar, S. 2013. MEGA6: molecular evolutionary genetics analysis version 6.0. Mol. Biol. Evol. 30, 2725–2729. Varela, M.F., Rivadulla, E., Lema, A. and Romalde, J.L. 2019. Human sapovirus among outpatients with acute gastroenteritis in Spain: a one-year study. Viruses 11, E144. Villabruna, N., Koopmans, M.P.G. and De Graaf, M. 2019. Animals as reservoir for human norovirus. Viruses 11, E478. Vinjé, J., Deijl, H., van der Heide, R., Lewis, D., Hedlund, K-O., Svensson, L. and Koopmans, M.P.G. 2000. Molecular detection and epidemiology of "sapporo-like viruses." J. Clin. Microbiol. 38, 530–536. Vinjé, J. and Koopmans, M.P. 2000. Simultaneous detection and genotyping of "Norwalk-Like viruses" by oligonucleotide array in a reverse line blot hybridization format. J. Clin. Microbiol. 38, 2595–2601. White, P.A. 2014. Evolution of norovirus. Clin. Microbiol. Infect. 20, 741–745. Wang, Q-H., Han, M.G., Cheetham, S., Souza, M., Funk, J.A. and Saif, L.J. 2005. Porcine noroviruses related to human noroviruses. Emerg. Infect. Dis. 11, 1874–1881. Wang, Q-H., Costantini, V. and Saif, L.J. 2007. Porcine enteric caliciviruses: genetic and antigenic relatedness to human caliciviruses, diagnosis and epidemiology. Vaccine 25, 5453–5466. Wang, Q-H., Souza, M., Funk, J.A., Zhang, W. and Saif, L.J. 2006. Prevalence of noroviruses and sapoviruses in swine of various ages determined by reverse transcription-PCR and microwell hybridization assays. J. Clin. Microbiol. 44, 2057–2062. Woode, G.N. and Bridger, J.C. 1978. Isolation of small viruses resembling astroviruses and caliciviruses from acute enteritis of calves. J. Med. Microbiol. 11, 441–452. Xue, L., Cai, W., Gao, J., Jiang, Y., Wu, H., Zhang, L., Zuo, Y., Dong, R., Pang, R., Zeng, H., Wu, S., Wang, J., Zhang, J. and Wu, Q. 2019. Genome characteristics and molecular evolution of the human sapovirus variant GII.8. Infect. Genet. Evol. 73, 362–367. Zheng, D-P., Ando, T., Fankhauser, R.L., Beard, R.S., Glass, R.I. and Monroe, S.S. 2006. Norovirus classification and proposed strain nomenclature. Virology 346, 312–323. | ||
| How to Cite this Article |
| Pubmed Style Matamoros DJP, Worsfold CS, Campos RC, Acuña HMB, Chacón EC, Sánchez CFJ. Molecular characterization of norovirus and sapovirus detected in animals and humans in Costa Rica: Zoo-anthropozoonotic potential of human norovirus GII.4. Open Vet J. 2023; 13(1): 74-89. doi:10.5455/OVJ.2023.v13.i1.8 Web Style Matamoros DJP, Worsfold CS, Campos RC, Acuña HMB, Chacón EC, Sánchez CFJ. Molecular characterization of norovirus and sapovirus detected in animals and humans in Costa Rica: Zoo-anthropozoonotic potential of human norovirus GII.4. https://www.openveterinaryjournal.com/?mno=53859 [Access: September 01, 2024]. doi:10.5455/OVJ.2023.v13.i1.8 AMA (American Medical Association) Style Matamoros DJP, Worsfold CS, Campos RC, Acuña HMB, Chacón EC, Sánchez CFJ. Molecular characterization of norovirus and sapovirus detected in animals and humans in Costa Rica: Zoo-anthropozoonotic potential of human norovirus GII.4. Open Vet J. 2023; 13(1): 74-89. doi:10.5455/OVJ.2023.v13.i1.8 Vancouver/ICMJE Style Matamoros DJP, Worsfold CS, Campos RC, Acuña HMB, Chacón EC, Sánchez CFJ. Molecular characterization of norovirus and sapovirus detected in animals and humans in Costa Rica: Zoo-anthropozoonotic potential of human norovirus GII.4. Open Vet J. (2023), [cited September 01, 2024]; 13(1): 74-89. doi:10.5455/OVJ.2023.v13.i1.8 Harvard Style Matamoros, D. J. P., Worsfold, . C. S., Campos, . R. C., Acuña, . H. M. B., Chacón, . E. C. & Sánchez, . C. F. J. (2023) Molecular characterization of norovirus and sapovirus detected in animals and humans in Costa Rica: Zoo-anthropozoonotic potential of human norovirus GII.4. Open Vet J, 13 (1), 74-89. doi:10.5455/OVJ.2023.v13.i1.8 Turabian Style Matamoros, Derling José Pichardo, Cristina Solís Worsfold, Rocío Cortés Campos, Hilda María Bolaños Acuña, Elena Campos Chacón, and Carlos Francisco Jiménez Sánchez. 2023. Molecular characterization of norovirus and sapovirus detected in animals and humans in Costa Rica: Zoo-anthropozoonotic potential of human norovirus GII.4. Open Veterinary Journal, 13 (1), 74-89. doi:10.5455/OVJ.2023.v13.i1.8 Chicago Style Matamoros, Derling José Pichardo, Cristina Solís Worsfold, Rocío Cortés Campos, Hilda María Bolaños Acuña, Elena Campos Chacón, and Carlos Francisco Jiménez Sánchez. "Molecular characterization of norovirus and sapovirus detected in animals and humans in Costa Rica: Zoo-anthropozoonotic potential of human norovirus GII.4." Open Veterinary Journal 13 (2023), 74-89. doi:10.5455/OVJ.2023.v13.i1.8 MLA (The Modern Language Association) Style Matamoros, Derling José Pichardo, Cristina Solís Worsfold, Rocío Cortés Campos, Hilda María Bolaños Acuña, Elena Campos Chacón, and Carlos Francisco Jiménez Sánchez. "Molecular characterization of norovirus and sapovirus detected in animals and humans in Costa Rica: Zoo-anthropozoonotic potential of human norovirus GII.4." Open Veterinary Journal 13.1 (2023), 74-89. Print. doi:10.5455/OVJ.2023.v13.i1.8 APA (American Psychological Association) Style Matamoros, D. J. P., Worsfold, . C. S., Campos, . R. C., Acuña, . H. M. B., Chacón, . E. C. & Sánchez, . C. F. J. (2023) Molecular characterization of norovirus and sapovirus detected in animals and humans in Costa Rica: Zoo-anthropozoonotic potential of human norovirus GII.4. Open Veterinary Journal, 13 (1), 74-89. doi:10.5455/OVJ.2023.v13.i1.8 |