| Research Article | ||
Open Vet J. 2023; 13(9): 1106-1115 Open Veterinary Journal, (2023), Vol. 13(9): 1106–1115 Original Research Colibacillosis in lambs and kids in Egypt: Prevalence, serogroups, antibiogram profile, virulence genes distribution and antimicrobial resistance genesHeba Hassan El-Nady, Mohamed Ibrahim Eissa, Naser Zeidan Abou-Zeid, Eman Beshry Abd-Elfatah, Ayman Ahmed Shehata and Elshaima Mohamed Fawzi*Infectious Diseases, Department of Animal Medicine, Faculty of Veterinary Medicine, Zagazig University, Zagazig, Egypt *Corresponding Author: Elshaima Mohamed Fawzi. Infectious Diseases, Faculty of Veterinary Medicine, Zagazig University, Zagazig, Egypt. Email: a.fawzy [at] zu.edu.eg; elshaimafawzi [at] yahoo.es Submitted: 07/06/2023 Accepted: 07/08/2023 Published: 30/09/2023 © 2023 Open Veterinary Journal
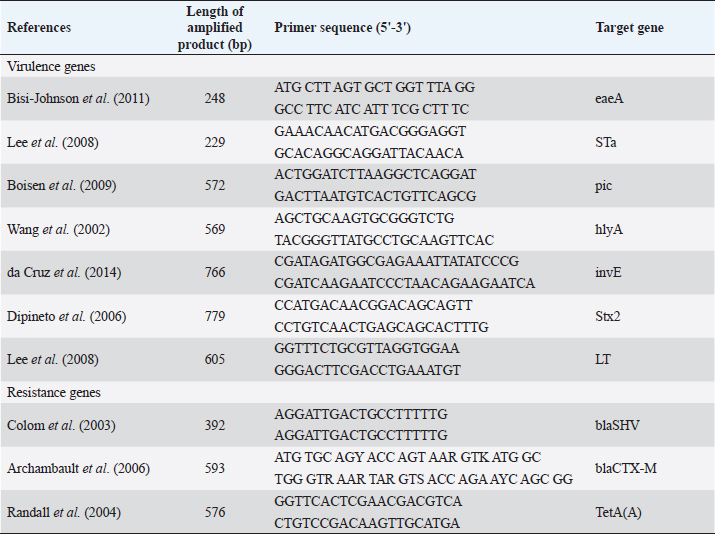
AbstractBackground: Small ruminants have a socioeconomic impact on Egypt’s production of meat, milk, and wool. Hence, every effort should be taken to prevent infections. Aim: To elucidate the prevalence and serogrouping of Escherichia coli (E. coli) strains from diarrheic lambs and kids, determine their antibiotic susceptibility and associated risk factors affecting the occurrence of the disease, and establish the most common virulence genes marker and major antimicrobial resistance genes. Methods: A total of 150 diarrheic animals (95 lambs and 55 kids) at different ages and seasons were subjected to clinical examination. Rectal swabs were collected from 150 diarrheic animals for isolation and biochemical identification of E. coli. Results: The bacteriological examination revealed that 62/95 lambs and 26/55 kids with percentages of 65% and 47%, respectively, showed infection with E. coli. Serotyping of 88 isolates of E. coli revealed the strains belonging to O2(8), O55(17), O84(5), O17(4), O6(8), O91(17), O26(9), O103(5), O126(5), O124(6), and O159(4). A total of 21 isolates were examined by multiplex polymerase chain reaction assay for detection of virulence and resistance genes. All examined isolates possessed a combination between intimin gene and heat-stable toxin (100%), the serine protease (pic) gene on 8/21 isolates of O55, O2, O6 (38%), and α-hemolysin gene on 8/21 isolates of O26, O91(38%) while adherent invasive gene (invA) gene on 3/21 isolates of O124, O159 (14%) which divided diarrheagenic E. coli into four types assigned to be atypical enteropathogenic E. coli (48%), atypical enterohemorrhagic E. coli 35%), atypical enterotoxigenic E. coli (6%), and atypical enteroinvasive E. coli (11%). On the other hand, the results of antimicrobial susceptibility testing revealed high resistance to ampicillin, erythromycin, and tetracycline (100%) and amoxicillin/clavulanic acid (92%) but were highly sensitive to gentamicin, imipenem, norfloxacin, ciprofloxacin, chloramphenicol, and amikacin (100%). Concerning to ß lactams antibiotic resistance genes of examined isolates had blaSHV (100%) and blaCTX-M (43%). For tetracycline, we detected the tetA in all examined isolates. Conclusion: The wide spread of atypical E. coli strains among diarrheic lambs and kids with marked resistance to several antibiotics of interest and the detection of major resistance genes assess the potential risk of this pathogen to animal and public health. Keywords: Escherichia coli, Lambs, Kids, Antibiogram profile, Virulence genes. IntroductionIn Egypt, small ruminant products are the second source in human diet consumption after bovine, it is a popular home-raised species in small-scale farmers or village flocks (Abd-Allah et al., 2019). Moreover, it could be a reservoir of many microorganisms (mo) that represent an economic and public health risk (Thomas et al., 2020). Neonatal diarrhea is one of the health problems leading to economic losses and mortalities in small ruminants’ flocks (Reidy et al., 2006). Colibacillosis is caused by pathogenic Escherichia coli, represented as one of the infectious agents causing neonatal diarrhea in lambs and kids that possesses a few concentrations of circulating immunoglobulins (Constable et al., 2017). Taxonomically, E. coli is categorized in the Enterobacteriaceae family. Six pathogenic strains of E. coli as enteropathogenic E. coli (EPEC), enteroaggregative E. coli, Shiga toxin-producing E. coli (STEC), diffusely adherents E. coli, enteroinvasive E. coli (EIEC), and enterotoxigenic E. coli (ETEC) are producing septicemia and diarrhea in human, animals and avian (Wani et al., 2013; Bashahun and Amina, 2017). Heat stable and heat labile toxins, are produced by ETEC and are encoded by the genes est and elt, respectively (Weiglmeier et al., 2010). Moreover, the outer membrane protein of pathogenic E. coli strains (EHEC and EPEC) known as intimin (eae) that encoded by intimin gene (eaeA) (Omerovic et al., 2017). Aumental use of antibiotics resulted in an increase in antimicrobial resistance (AMR) in both human health problems and veterinary medicine which become more prevalent worldwide (Blanco Crivelli et al., 2021). Many complex mechanisms are associated with the emergence of plasmid-mediated AMR of mo as metallo ß lactams and extended-spectrum beta-lactamses (ESBLs), active efflux pumps and plasmid-mediated quinolone resistance genes (Bhattacharyya et al., 2022). Assessment and diversity of gene resistance among bacterial isolates are essential to diagnose and understand the epidemiology of AMR spread among humans and animals (Boerlin et al., 2005). Abundant use of a wide variety of antibiotics such as aminoglycoside, penicillin, streptomycin, and sulfonamide without control from the authorities especially in developing countries leading to elevate the pathogenicity of m.o and constitute a major health concern to human and animal health (Wang et al., 2016; El-Adawy et al., 2018). Antibiotic resistance genes have been widely identified using polymerase chain reaction (PCR) (Momtaz et al., 2012). The wide spread of tetracycline resistance among m.o is due to the localization of tet genes on plasmids, transposons, and integrons (Roberts, 2003). TetA, blaCTX, and blaSHV genes were predominant in antibiotic-resistant E. coli mainly leading to antimicrobial treatment failure (Gozi et al., 2019). This study was performed to determine the prevalence rate and its associated risk factors that provoke diarrhea in lambs and kids, determine the actual incriminated virulence genes, detect the actual level of resistance of antimicrobial agents against E. coli isolated from diarrheic lambs and kids, and evaluate the distribution of major resistance genes in these isolates to detect the level of resistance in the study region. Material and MethodsAnimals and E. coli strains identificationA total of 150 diarrheic rectal swab samples were collected from 95 lambs and 55 kids aged from 1 day up to 4 months during the period from October 2021 to December 2022 from Al-Sharkia and Al-Ismailia Governorates. The data on prospective risk factors was related to a questionnaire of owners and direct observations of diarrheic lambs and kids. The questionnaire incorporated season, age, sex, breed, and other hygienic conditions were performed on each animal. Briefly, rectal sterile cotton swabs were collected from diarrheic animals in sterile MacConkey broth (Oxoid Ltd., Basingstoke, UK) in an ice box to the Infectious Diseases Laboratory, Faculty of Veterinary Medicine, Zagazig University, Egypt, for bacteriological examination. Rectal swabs were incubated aerobically at 37°C/24 hour to improve the probability of isolation. According to Quinn et al. (2011), a loopful of MacConkey broth was added to MacConkey’s agar and cultured for 37°C/24 hour to isolate E. coli strains, then a streak of lactose fermenter (pink) colonies were over Eosin Methylene Blue agar. Escherichia coli strains were identified using different specific biochemical tests according to Collee et al. (1996). Escherichia coli isolates serogroupsEscherichia coli serogroups were identified serologically using slide agglutination test according to Kok et al. (1996), by standard polyvalent and monovalent E. coli antisera. Antimicrobial sensitivity test (AST)AST was applied by disc diffusion method on E. coli isolates on Mueller–Hinton agar plates following 0.5 McFarland standards as per Clinical and Laboratory Standards Institute (CLSI, 2020) against thirteen antimicrobial agents (Oxoid, Basingstoke, UK): amoxicillin/clavulanic acid (AMC), amikacin (AK), norfloxacin (NOR), erythromycin (E), ciprofloxacin (CIP), gentamicin (CN), ceftazidime (CAZ), cefotaxime (CTX), trimethoprim-sulfamethoxazole (STX), chloramphenicol (C), ampicillin (AMP), imipenem (IMP), and tetracyclin (TE). The studied antimicrobial drugs characterized the E. coli isolates as being either susceptible, intermediate, or resistant. Detection of virulence and resistant genes by PCR techniqueFollowing the instructions on the QIAamp DNA Mini kit (Qiagen, Germany, GmbH), DNA was extracted from pure E. coli colonies. In a nutshell, 200 μl of the sample suspension was treated with 10 μl of proteinase K and 200 μl of lysis buffer at 56°C/10 minutes. After incubation, 200 μl of 100% ethanol was added, and nucleic acid was eluted using 100 μl of the kit’s provided elution buffer. Multiplex PCR techniqueDNA extract of E. coli isolates was exposed to multiplex PCR to recognize eaeA, heat-stable toxin (STa), serine protease (pic), α-hemolysin (hlyA), invE, Stx2, and (heat-labile toxin) LT virulence genes and blaSHV, blaCTX-M, and tetA resistant genes with specific oligonucleotide primers (Metabion, Germany) (Table 1). A 50 μl reaction containing 25 μl of Emerald Amp Max PCR Master Mix (Takara, Japan), 1 μl of each primer, 13 μl of water, and 8 μl of DNA template was used for the final PCR amplification. Initial denaturation took place during the amplification process at 94°C for 5 minutes. The annealing temperature was 58°C for (Stx2), 57°C for (Sta and LT), 64°C for (hlyA), 59°C for (invE), 58°C for (pic), 51°C for (eaeA), 54°C for (blaSHV), 54°C for (blaCTX-M), and 50°C for (TetA) and extension at 72°C for 40 seconds. At room temperature, the PCR products were separated on a 1% agarose gel (Applichem, Germany, GmbH). Before gel analysis, each gel slot was filled with 40 μl of the duplex PCR products and 20 μl of the uniplex PCR products. Thermo’s Fermentas 100 bp ladder was used to calculate the sizes of the fragments. A gel documentation system (Alpha Innotech, Biometra) was used to visualize the gel, and computer software was used to analyze the data. Table 1. Oligonucleotide primers sequence of virulence and resistance genes E. coli strain.
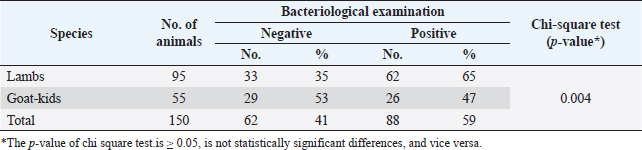
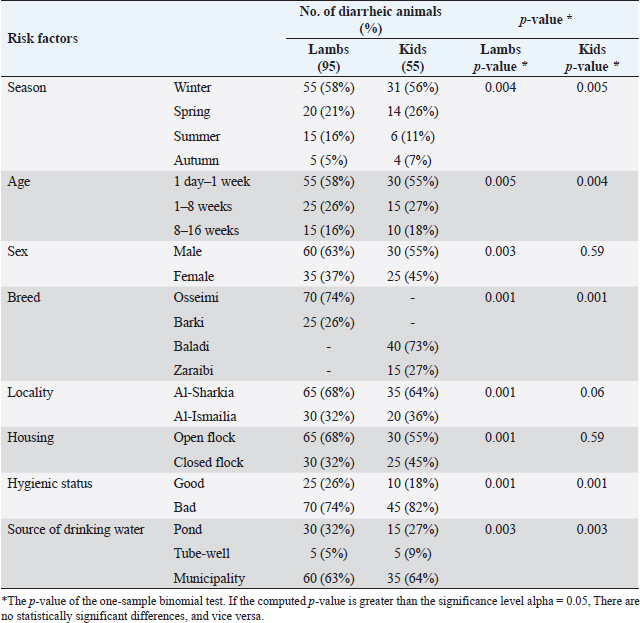
Statistical analysisUsing SPSS version 25 (SPSS Inc., Chicago, IL), the data were examined. One-sample chi-square and binomial tests were run, with p ≤ 0.05 considered statistically significant. Ethical approvalThe Zagazig University Institutional Animal Care and Use Committee (ZU-IACUC) reviewed and authorized this study with permission N. (ZU-IACUC/2/F/116/2022). ResultsThe colibacillosis-infected lambs and kids displayed a variety of clinical signs, including mild to profuse watery white diarrhea, minor dehydration, tachycardia, pale mucous membranes, severe weakness, a little rise in rectal temperature, and elevated respiratory rate. The bacteriological and biochemical identification of 150 rectal swabs of diarrheic lambs and kids (95 lambs and 55 kids aged from 1 day up to 4 months) illustrated a percentage of 65% (62/95) and 47% (26/55) E. coli infection in lambs and kids, respectively. The statistical analysis revealed a significant difference (p = 0.004) among lambs and kids (Table 2). As depicted in Table 3, all lambs and kids were diarrheic at the time of sample collection with the high risk recorded in winter (58% lambs, 56% kids) and spring (21% lambs, 26% kids) seasons. The highest prevalence rate was demonstrated significantly in lambs and kids at age 1 day-1 week (58% and 55%, respectively). A significant difference among diarrheic male and female lambs (p=0.003) as male lambs (63%) were more susceptible than females (37%), in contrast, no statistical significance among male and female kids (p=0.59) was recorded. lambs and kids breeds had a role in the prevalence of diarrhea, Osseimi lambs breed (74%)were more vulnerable than the Barki lambs breed (26%) and the Baladi kids breed (73%) were more vulnerable than the Zaraibi breed (27%) (p=0.001). Moreover, diarrheic lambs were more prevalent in Al-sharkia (68%) than Al-Ismailia (32%) while there is not statistical difference between Al-sharkia (64%) and Al-Ismailia (36%) localities in the prevalence of diarrhea in kids (p=0.06). Furthermore, the prevalence of diarrheic lambs housed in open flocks (68%) was higher than in closed flocks (32%) (p=0.001), while no statistical significance between open housed (55%) and closed housed (45%) concerning diarrheic kids flock (p=0.59). A significant association (p =0.001) in a hygienic condition, poor hygienic status showed a higher diarrheic rate of 74% in lambs and 82% in kids than good status. Source water supplied for animals had an effect on the prevalence rate of diarrhea among lambs and kids as in this study, pond and municipality water were highly significant (p =0.003). Table 2. The prevalence of E. coli between lambs and kids using chi-square test.
Table 3. Risk factors associated with colibacillosis in lambs and kids using one sample binomial test.
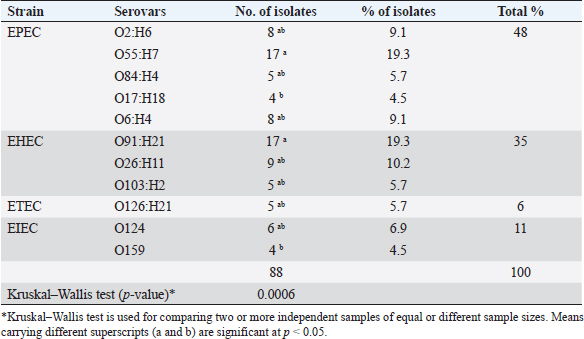
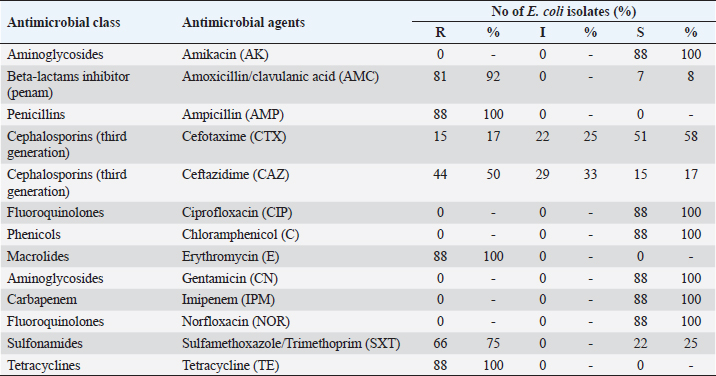
Serogrouping of E. coli isolates revealed 11 different E. coli serogroups, belonged to O2(8), O55(17), O84(5), O17(4), and O6(8) represented as atypical enteropathogenic E. coli (48%), O91(17), O26(9), and O103(5) represented as atypical enterohemorrhagic E. coli (35%), O126(5) represented as atypical enterotoxigenic E. coli (6%), and O124(6) and O159(4) represented as atypical enteroinvasive E. coli (11%). The highest prevalent serotypes were O55 and O91 (19.3%) and the lowest ones were O17 and O159 (4.5%). A significant difference between O55 and O91 in comparison with both O17 and O159 ( p= 0.0006) is illustrated in Table 4. The 88 E. coli isolates were tested against 13 different antimicrobial agents, and exhibited high resistance to ampicillin, erythromycin and tetracycline (100%), amoxicillin/clavulanic acid (92%), sulfamethoxazole/trimethoprim (75%), and ceftazidime (50%) but were sensitive to gentamicin, imipenem, norfloxacin, ciprofloxacin, chloramphenicol, amikacin (100%), and cefotaxime (58%) as showed in Table 5. Out of 88 E. coli isolates, 21 isolates represented for detection of virulence genes using multiplex PCR. In this study, eaeA and STa (100%) were the most prevalent virulence genes represented, the pic gene for EPEC represented on 8/21 isolates of O55, O2, O6 (38%), and hlyA gene for EHEC also represented on 8/21 isolates of O26, O91(38%) while invA gene for EIEC represented only on 3/21 isolates of O124, O159 (14%) but Stx2 and LT did not detect. Upon virulence gene characterization, E. coli isolates were alienated into typical ETEC (10%) and atypical combinations of ETEC/EHEC (38%), ETEC/EPEC (38%), and ETEC/EIEC (14%). All O55, O2, and O6 serotypes possessed the pic that represented an additional factor with eaeA and STa leading to the E. coli as aEPEC [A-typical EPEC]. Molecular diagnosis of E. coli AMR genes exhibited: blaSHV, blaCTX-M for (amoxicillin/clavulanic acid, cefotaxime, ampicillin, and ceftazidime) and tetA (for tetracycline). For amoxicillin/clavulanic acid, ampicillin, cefotaxime, and ceftazidime; blaSHV 21/21 (100%) showed a higher frequency than blaCTX-M 9/21 (43%). About 12/21 (57%) had a single gene, either blaCTX-M or blaSHV while 9/21 (43%) had both genes. For tetracycline, we detected the tetA in 21/21 (100%) of the isolates. DiscussionNeonatal diarrhea is a worldwide infectious disease causing financial losses in the livestock industry mainly in the first few weeks of life (Croxen et al., 2013). The same clinical signs of colibacillosis observed during this investigation were mentioned by Hassan et al. (2014) and Constable et al. (2017). A variation in the prevalence of colibacillosis among lambs and kids, as lambs 65% were more susceptible to infection by E. coli than kids 47%, might be due to the difference in number of examined animals. The prevalence of colibacillosis in lambs was 65% which exceeded the level mentioned by Ahmed et al. (2010) in Nigeria (36.84%). The current study’s high incidence could be related to improper preventative and control methods, untaken colostrum on the first days of lamb life, and unclean sheep housing. The prevalence of colibacillosis in kids was 47% which was lower than mentioned by Islam et al. (2016) in Bangladesh (52%) but higher than the recorded in Turkey (36.4%) by Türkyılmaz et al. (2014), in Saudia Arabia (30.8%) by Shabana et al. (2017), and in Rajasthan (31.43%) by Sharma et al. (2020). This dissimilarity might have been due to hygienic measures, geographic location, virulence, and strain of E. coli. Table 4. Serovars distribution of E. coli isolated from diarrheic lambs and kids.
Table 5. Antimicrobial sensitivity testing of E. coli isolated from diarrheic lambs and kids.
Colibacillosis was more common in the winter season followed by the spring season. Similar to Sonawane et al. (2012) but inconsistent with Islam et al. (2016) demonstrated that the diarrhea prevalence did not show any seasonal variation and Abdou et al. (2021) stated that diarrhea was more common in the dry season than the wet one. In this scenario, small ruminants aged from 1 day to 1 week were more likely to have a greater prevalence of infection, which matched the finding of Sharma et al. (2020) and Aklilu et al. (2013) reported that diarrhea was common in lambs and goat-kids of age less than 1 month. On the contrary, Shabana et al. (2017) noticed that sheeps and goats with age up to 12 months were highly susceptible to diarrhea. The prevalence of diarrhea in lambs was more common in open flocks (68%) than closed ones (32%) might be due to exposure to extreme cold conditions. This disagreed with Nasr et al. (2014) stated that diarrhea was higher in the closed system (83.70%) than in the open system (46.80%) and Abdou et al. (2021) noticed that diarrhea occurred in both open and closed flocks equally. The hygienic state of the area where lambs and kids were raised was shown to be significantly correlated, and low hygienic status exhibited a higher prevalence than good hygienic status. As detected by Islam et al. (2016) stated that poor hygiene has a main role in E. coli spread. Water sources for animal drinking from a pond and municipality had a high E. coli prevalence. Environmental contamination of water sources may be the main accusatory factor (Rashid et al., 2015). The 88 E. coli isolates were belonging to O2, O55, O84, O17, O6, O91, O26, O103, O126, O124, and O159 serotypes. The highest prevalent serotypes were O55 and O91 (19.3%) and the lowest ones were O17 and O159 (4.5%). This was in line with Ruchi and Kataria (2012) found that O55 was the most frequently isolated serogroup from diarrheic lambs and kids. In addition, Nasr et al. (2014) isolated the serogroups O55, O78, O125, O101, and O22 from diarrheic lambs. Hence, the analysis of our results proved that small ruminants in Egypt could be a potential source of infection in humans. Antimicrobial drugs are frequently misused in Egypt because their usage for feed efficiency and growth promotion is not properly restricted. Farmers typically administer excessive doses of antibiotics to sick animals based exclusively on their own experience and without a veterinarian’s prescription, supervision, or lab diagnosis, this creates selective pressure on the growth and distribution of infections that are resistant to antibiotics, including those that affect humans and animals (El-Twab et al., 2016). The antimicrobials with high resistance were erythromycin, ampicillin, and tetracycline (100%), followed by AMC (92%), SXT (75%), CAZ (50%), and CTX (17%) with (100%) sensitivity to amikacin, ciprofloxacin, chloramphenicol, gentamicin, imipenem, and norfloxacin which considered specific drugs for treatment of E. coli infection in lambs and kids. These findings were in line with Croxen et al. (2013) who mentioned that E. coli isolates were resistant to ampicillin and tetracycline and sensitive to chloramphenicol, gentamicin, and norfloxacin. While disagreed with Imre et al. (2022) who reported that E. coli isolates were less resistant to ceftazidime (3.6%, 1/28). Based on genotypic and phenotypic characteristics, most of the E. coli isolates (8/21, 38%) in this study were classified as aEPEC, which was in line with prior investigations (Vettorato et al., 2009; Maluta et al., 2014) that discovered few tEPEC strains in sheep and other animal species. Monitoring aEPEC in farm animals, such as sheep, became especially crucial since pathogenic strains can be transferred from animals to humans through food or direct contact with animals and their natural habitats (Brandal et al., 2012; Otero et al., 2013). In the actual result, observed that aEPEC was seriously found in lambs and kids. Similar findings in Spanish sheep and goats had been reported as well by Cortés et al. (2005). ETEC is one of the most significant pathogens that causes diarrhea in young small ruminants (Pourtaghi and Sodagari, 2016). About 10% of the isolates in this investigation carried specific ETEC genes mainly STa. According to the most recent research from India and Turkey, lamb fecal isolates included 9% and 11.2% ETEC (Bandyopadhyay et al., 2011; Türkyılmaz et al., 2014), respectively. In this study, the absence of LT in the isolates is not surprising as LT is considered atypical in ruminant isolates (Türkyılmaz et al., 2014). Eight (38%) of the E. coli isolates in the current investigation showed mixed combinations of ETEC and EPEC. The current results supported those of other research that discovered mixed combinations of several E. coli pathotypes in calves (Sharma et al., 2017; Aref et al., 2018). Other unusual combinations were also found, including ETEC/EHEC (38%) and ETEC/EIEC (14%). The emergence of novel pathotypes that are more pathogenic and cause severe diarrhea in young small ruminants may result from these unusual combinations. These isolates were thought to pose a risk to Egypt’s public health and were the cause of newborn diarrhea (Aref et al., 2018). This happens when virulence genes are added or removed, altering the lineage and promoting the formation of novel variants with distinct characteristics and pathogenicity (Ahmed et al., 2008). In this study, blaCTX-M and blaSHV AMR genes were responsible for developing the resistance against amoxicillin/clavulanic acid, ampicillin, cefotaxime, ceftazidime where blaSHV gene was predominant among ESBL genotypes and recorded in 21/21 (100%), blaCTX gene detected in 9/21 (43%). These results disagreed with (Abdallah et al., 2022; Mahmood et al., 2022) reported that the blaCTX gene was the most prevalent among E. coli isolates. The most common tet genes discovered in enterobacteria were tetA and tetB (Van et al., 2008). However, tetA markers are more prevalent than tetB markers (67%) compared to 31% (Karczmarczyk et al., 2011). In this study, all isolates carried the tetracycline efflux gene tetA. These findings agreed with Gozi et al. (2019) noted that among the tested E. coli isolates, tetA was found in 54 (69.2%) of the samples. ConclusionThis research proves the wide distribution of diarrheal E. coli pathotypes in lambs (65%) and kids (47%), with atypical EPEC (48%) being the most predominant pathotypes. The presence of eaeA and STa genes in all isolates created a typical virulent combination that threatened the public health hazard. Consequently, with the growing resistance of mostly used antibiotics [ampicillin, erythromycin, and tetracycline (100%), amoxicillin/clavulanic acid (92%), saulfamethoxazole/ trimethoprim (75%) and ceftazidime (50%)], isolated E. coli carried the most important antibiotic-resistant genes, as all were revealed to be positive for the blaSHV and tetA genes (100%) while 43% were positive to blaCTX-M. Thus, there is a need to implement an accurate surveillance program and exact control measures for drug administration in the veterinary field to minimize the dissemination of these virulent, resistant, and atypical strains to humans. AcknowledgmentThe owners of the cases who were admitted to the Zagazig Veterinary Clinic are thanked by the authors for using their animals and providing the samples. The staff from Zagazig University in Egypt’s Department of Animal Medicine are also acknowledged by the authors for their assistance with sample collecting. Author contributionsHHN, MIE, NZA, and EMF participated in the concept development, and study execution. While HHN, EBA, and AAS conducted data analysis and interpretation processes. The first draught of the manuscript was written by HHN and EMF. The manuscript revision and editing were done by HHN, EBA, and EMF. All of the study’s data were completely accessible to the authors, who also accepted responsibility for the accuracy of the data analysis and final submission. Conflict of interestThe authors declare that there is no conflict of interest. FundingNo organization provided funding for the completion of this work. Necessary facilities of the Department of Animal Medicine, Faculty of Veterinary Medicine, Zagazig University were used. Data availabilityThe manuscript data are included within the manuscript. ReferencesAbdallah, H.M., Al Naiemi, N., Elsohaby, I., Mahmoud, A.F., Salem, G.A. and Vandenbroucke-Grauls, C.M.J.E. 2022. Prevalence of extended-spectrum β-lactamase-producing Enterobacterales in retail sheep meat from Zagazig city, Egypt. BMC Vet. Res. 18, 191. Abd-Allah, S., Mohamed, M.I., Shoukry, M.M., Salman, F.M. and Rahman, A.E. 2019. Assessment of the traditional goat production systems in rural areas of the Nile Delta in Egypt. Bull. Natl. Res. Cent. 43, 114. Abdou, N.E., Majeed, Q.A.H., El-Azazy, O.M.E., Tahrani, L.M.A., AlAzemi, M.S. and Alajmi, A. 2021. Risk factors of diarrhea in small ruminants in Kuwait. Iran. J. Vet. Res. 22, 146–149. Ahmed, A., Egwu, G.O., Garba, H.S. and Magaji, A.A. 2010. Prevalence of bacterial pathogens and serotyping of E. coli isolates from diarrheic lambs in Sokoto state, Nigeria. Sokoto J. Vet. Sci. 8, 42–45. Ahmed, N., Dobrindt, U., Hacker, J. and Hasnain, S.E. 2008. Genomic fluidity and pathogenic bacteria: applications in diagnostics, epidemiology and intervention. Nat. Rev. Microbiol. 6, 387–394. Aklilu, M., Sisay, T., Tefera, G. and Tekalign, B. 2013. Identification and biotyping of Escherichia coli from diarrheic lambs in and around Debre Birhan town, Ethiopia. J. Environ. Anal. Toxicol. 3, 6. Archambault, M., Petrov, P., Hendriksen, R.S., Asseva, G., Bangtrakulnonth, A., Hasman, H. and Aarestrup, F.M. 2006. Molecular characterization and occurrence of extended-spectrum beta-lactamase resistance genes among Salmonella enterica serovar Corvallis from Thailand, Bulgaria, and Denmark. Microb. Drug Resist. 12, 192–198. Aref, N.E.M., Abdel-Raheem, A.R.A., Kamaly, H.F. and Hussien, S.Z. 2018. Clinical and sero-molecular characterization of Escherichia coli with an emphasis on hybrid strain in healthy and diarrheic neonatal calves in Egypt. Open Vet. J. 8, 351–359. Bandyopadhyay, S., Mahanti, A., Samanta, I., Dutta, T.K., Ghosh, M.K., Bera, A.K., Bandyopadhyay, S. and Bhattacharya, D. 2011. Virulence repertoire of Shiga toxin-producing Escherichia coli (STEC) and enterotoxigenic Escherichia coli (ETEC) from diarrhoeic lambs of Arunachal Pradesh, India. Trop. Anim. Health Prod. 43, 705–710. Bashahun, G.M. and Amina, A. 2017. Colibacillosis in calves: a review of literature. J. Anim. Sci. Vet. Med. 2, 62–71. Bhattacharyya, D., Banerjee, J., Habib, M., Thapa, G., Samanta, I., Nanda, P.K., Dutt, T., Sarkar, K. and Bandyopadhyay, S. 2022. Elucidating the resistance repertoire, biofilm production, and phylogenetic characteristics of multidrug-resistant Escherichia coli isolated from community ponds: a study from West Bengal, India. Water Environ. Res. 94, e1678. Bisi-Johnson, M.A., Obi, C.L., Vasaikar, S.D.,Baba, K.A. and Hattori, T. 2011. Molecular basis of virulence in clinical isolates of Escherichia coli and Salmonella species from a tertiary hospital in the Eastern Cape, South Africa. Gut Pathog. 3, 9. Blanco Crivelli, X., Bonino, M.P., Sanin, M.S., Petrina, J.F., Disalvo, V.N., Massa, R., Miliwebsky, E., Navarro, A., Chinen, I. and Bentancor, A. 2021. Potential zoonotic pathovars of diarrheagenic Escherichia coli detected in lambs for human consumption from Tierra del Fuego, Argentina. Microorganisms 9, 1710. Boerlin, P., Travis, R., Gyles, C.L., Reid-Smith, R., Janecko, N., Lim, H., Nicholson, V., McEwen, S.A., Friendship, R. and Archambault, M. 2005. Antimicrobial resistance and virulence genes of Escherichia coli isolates from swine in Ontario. Appl. Environ. Microbiol. 71, 6753–6761. Boisen, N., Ruiz-Perez, F., Scheutz, F., Krogfelt, K.A. and Nataro, J.P. 2009. High prevalence of serine protease autotransporter cytotoxins among strains of enteroaggregative Escherichia coli. Am. J. Trop. Med. Hyg. 80, 294–301. Brandal, L.T., Sekse, C., Lindstedt, B.A., Sunde, M., Løbersli, I., Urdahl, A.M. and Kapperud, G. 2012. Norwegian sheep are an important reservoir for human-pathogenic Escherichia coli O26: H11. Appl. Environ. Microbiol. 78, 4083–4091. CLSI. 2020. Performance standards for antimicrobial susceptibility testing, 30th ed. CLSI supplement M100. Wayne, PA: Clinical and Laboratory Standard Institute. Collee, J.G., Marmion, B.P. and Fraser, A.G. 1996. Mackie & McCartney practical medical microbiology, 14th ed. Edinburgh, UK: Churchill Livingstone, p: 486. Colom, K., Pèrez, J., Alonso, R., Fernández-Aranguiz, A., Lariňo, E. and Cisterna, R. 2003. Simple and reliable multiplex PCR assay for detection of blaTEM, blaSHV and blaOXA-1 genes in Enterobacteriaceae. FEMS Microbiol. Lett. 223, 147–151. Constable, P.D., Hinchcliff, K.W., Done, S.H. and Grünberg, W. 2017. Veterinary medicine: a textbook of the diseases of cattle, horses, sheep, pigs and goats, 11th ed. St. Louis, MO: Elsevier. Cortés, C., De la Fuente, R., Blanco, J., Blanco, M., Blanco, J.E., Dhabi, G., Mora, A., Justel, P., Contreras, A., Sánchez, A., Corrales, J.C. and Orden, J.A. 2005. Serotypes, virulence genes and intimin types of verotoxin-producing Escherichia coli and enteropathogenic E. coli isolated from healthy dairy goats in Spain. Vet. Microbiol. 110, 67–76. Croxen, M.A., Law, R.J., Scholz, R., Keeney, K.M., Wlodarska, M. and Finlay, B.B. 2013. Recent advances in understanding enteric pathogenic Escherichia coli. Clin. Microbiol. Rev. 26, 822–880. da Cruz, C.B., de Souza, M.C., Serra, P.T., Santos, I., Balieiro, A., Pieri, F.A., Nogueira, P.A. and Orlandi, P.P. 2014. Virulence factors associated with pediatric shigellosis in Brazilian Amazon. Biomed. Res. Int. 2014, 539697. Dipineto, L., Santaniello, A., Fontanella, M., Lagos, K., Fioretti, A. and Menna, L.F. 2006. Presence of Shiga toxin-producing Escherichia coli O157:H7 in living layer hens. Lett. App. Microbiol. 43, 293–295. El-Adawy, M., El-Aziz, M.A., El-Shazly, K., Ali, N.G. and El-Magd, M.A. 2018. Dietary propionic acid enhances antibacterial and immunomodulatory effects of oxytetracycline on Nile tilapia, Oreochromis niloticus. Environ. Sci. Pollut. Res. 25, 34200–34211. El-Twab, A., Ashraf, A., El-Hofy, F.I. and Rizk, A.M. 2016. Molecular characterization of quinolones and β-lactams resistant Salmonella serovars determinants in diarrheic calves, lambs and goats-kids in the Middle of Nile Delta, Egypt. Benha Vet. Med. J. 30, 171–182. Gozi, K.S., Froes, J.R., Deus Ajude, L.P.T., da Silva, C.R., Baptista, R.S., Peiró, J.R., Marinho, M., Mendes, L.C.N., Nogueira, M.C.L. and Casella, T. 2019. Dissemination of multidrug-resistant commensal Escherichia coli in feedlot lambs in Southeastern Brazil. Front. Microbiol. 10, 1394. Hassan, N., Sheikh, G., Hussain, S.A. and Nazir, G. 2014. Variation in clinical findings associated with neonatal colibacillosis in lambs before and after treatment. Vet. World 7, 262–265. Imre, K., Ban-Cucerzan, A., Herman, V., Sallam, K.I., Cristina, R.T., Abd-Elghany, S.M., Morar, D., Popa, S.A. and Imre, M. and Morar, A. 2022. Occurrence, pathogenic potential and antimicrobial resistance of Escherichia coli isolated from raw milk cheese commercialized in Banat Region, Romania. Antibiotics (Basel) 11(6), 721. Islam, K., Ahad, A., Barua, M., Islam, A., Chakma, S., Dorji, C., Uddin, M.A., Islam, S. and Ahasan, A.S.M.L. 2016. Isolation and epidemiology of multidrug resistant Escherichia coli from goats in Cox’s Bazar, Bangladesh. J. Adv. Vet. Anim. Res. 3, 166–172. Karczmarczyk, M., Walsh, C., Slowey, R., Leonard, N. and Fanning, S. 2011. Molecular characterization of multidrug-resistant Escherichia coli isolates from Irish cattle farms. App. Environ. Microbiol. 77, 7121–7127. Kok, T., Worswich, D. and Gowans, E. 1996. Some serological techniques for microbial and viral infections. In Practical medical microbiology. Eds., Collee, J., Fraser, A., Marmion, B. and Simmons, A., 14th ed. Edinburgh, UK: Churchill Livingstone, p: 978. Lee, S.I., Kang, S.G., Kang, M.L. and Yoo, H.S. 2008. Development of multiplex polymerase chain reaction assays for detecting enterotoxigenic Escherichia coli and their application to field isolates from piglets with diarrhea. J. Vet. Diagn. Invest. 20, 492–496. Mahmood, F.R.R. and Ahmed, I.M. 2022. Molecular detection of ESBL/AmpC ß-Lactamase Escherichia coli isolated from sheep in Mosul city. Iraqi J. Vet. Sci. 36, 387–392. Maluta, R.P., Fairbrother, J.M., Stella, A.E., Rigobelo, E.C., Martinez, R. and de Ávila, F.A. 2014. Potentially pathogenic Escherichia coli in healthy, pasture-raised sheep on farms and at the abattoir in Brazil. Vet. Microbiol. 169, 89–95. Momtaz, H., Rahimi, E. and Moshkelani, S. 2012. Molecular detection of antimicrobial resistance genes in E. coli isolated from slaughtered commercial chickens in Iran. Vet. Med. 57, 193–197. Nasr, M., Nabil, M.B., Hammouda, H.A. and Alaa, A.O. 2014. Epidemiological, clinical and bacteriological studies on bacterial lamb enteritis at Behera Province, Egypt. Alex. J. Vet. Sci. 43, 8–16. Omerovic, M., Müştak, H.K. and Kaya, I.B. 2017. Escherichia coli patotiplerinin virülens faktörleri. Etlik. Vet. Mikrobiyol. Derg. 28, 1–6. Otero, V., Rodríguez-Calleja, J.M., Otero, A., García-López, M.L. and Santos, J.A. 2013. Genetic characterization of atypical enteropathogenic Escherichia coli isolates from ewes’ milk, sheep farm environments, and humans by multilocus sequence typing and pulsed-field gel electrophoresis. App. Environ. Microbiol. 79, 5864–5869. Pourtaghi, H. and Sodagari, H.R. 2016. Antimicrobial resistance of entrotoxigenic and non-entrotoxigenic Escherichia coli isolated from diarrheic calves in Iran. Inter. J. Enteric Pathog. 4, e34557. Quinn, P.J., Markey, B.K., Leonard, F.C. 2011. Veterinary microbiology and microbial diseases, 2nd ed. Hoboke, NJ: Wiley-Blackwell, pp: 84–96. Randall, L.P., Cooles, S.W., Osborn, M.K., Piddock, L.J.V. and Woodward, M.J. 2004. Antibiotic resistance genes, integrons and multiple antibiotic resistance in thirty-five serotypes of Salmonella enterica isolated from humans and animals in the UK. J. Antimicrob. Chemother. 53, 208–216. Rashid, M., Rakib, M.M. and Hasan, B. 2015. Antimicrobial-resistant and ESBL-producing Escherichia coli in different ecological niches in Bangladesh. Infect. Ecol. Epidemiol. 5, 26712. Reidy, N., Lennon, G., Fanning, S., Power, E. and O’shea, H. 2006. Molecular characterisation and analysis of bovine rotavirus strains circulating in Ireland 2002–2004. Vet. Microbiol. 117, 242–247. Roberts, M.C. 2003. Tetracycline therapy: update. Clin. Infect. Dis. 36, 462–467. Ruchi, P. and Kataria, A.K. 2012. Serogrouping and colicin-related properties of Escherichia coli isolated from neonate diarrhoeic ruminants. Ruminant Sci. 1, 9–13. Shabana, I.I., Bouqellah, N.A. and Zaraket, H. 2017. Investigation of viral and bacterial enteropathogens of diarrheic sheep and goats in Medina, Saudi Arabia. Trop. Biomed. 34, 944–955. Sharma, R.K., Taku, A.K., Malik, A., Bhat, M.A., Javed, R., Badroo, G.A. and Kour, A. 2017. Molecular characterization and antimicrobial profiling of Escherichia coli isolates from diarrheic calves. Indian J. Anim. Sci. 87, 1467–1471. Sharma, S.K., Manat, N. and Joshi, M. 2020. Prevalence of colibacillosis in goat kids in Udaipur district of Rajasthan. Ind. J. Vet. Sci. Biotechnol. 16, 98–100. Sonawane, G.G., Singh, F., Tripathi, B.N., Dixit, S.K., Kumar, J. and Khan, A. 2012. Investigation of an outbreak in lambs associated with Escherichia coli O95 septicaemia. Vet. Pract. 13, 72–75. Thomas, K.M., de Glanville, W.A., Barker, G.C., Benschop, J., Buza, J.J., Cleaveland, S. and Crump, J.A. 2020. Prevalence of Campylobacter and Salmonella in African food animals and meat: a systematic review and meta-analysis. Int. J. Food Microbiol. 315, 108382. Türkyılmaz, S., Eskiizmirliler, S., Tunaligil, S. and Bozdogan, B. 2014. Identification, characterization and molecular epidemiology of Escherichia coli isolated from lamb and goat kids with diarrhoea. Acta Vet. Br. 82, 357–362. Van, T.T.H., Chin, J., Chapman, T., Tran, L.T. and Coloe, P.J. 2008. Safety of raw meat and shellfish in Vietnam: an analysis of Escherichia coli isolations for antibiotic resistance and virulence genes. Int. J. Food Microbiol. 124, 217–223. Vettorato, M.P., De Castro, A.F.P., Cergole-Novella, M.C., Camargo, F.L.L., Irino, K. and Guth, B.E.C. 2009. Shiga toxin-producing Escherichia coli and atypical enteropathogenic Escherichia coli strains isolated from healthy sheep of different populations in São Paulo, Brazil. Lett. Appl. Microbiol. 49, 53–59. Wang, G., Clark, C.G. and Rodgers, F.G. 2002. Detection in Escherichia coli of the genes encoding the major virulence factors, the genes defining the O157: H7 serotype, and components of the type 2 Shiga toxin family by multiplex PCR. J. Clin. Microbiol. 40, 3613–3619. Wang, J., Stanford, K., McAllister, T.A., Johnson, R.P., Chen, J., Hou, H., Zhang, G. and Niu, Y.D. 2016. Biofilm formation, virulence gene profiles, and antimicrobial resistance of nine serogroups of non-O157 Shiga toxin–producing Escherichia coli. Foodborne Pathog. Dis. 13, 316–324. Wani, S.A., Hussain, I., Beg, S.A., Rather, M.A., Kabli, Z.A., Mir, M.A. and Nishikawa, Y. 2013. Diarrhoeagennic Escherichia coli and salmonellae in calves and lambs in Kashmir: absence, prevalence and antibiogram. Rev. Sci. Tech. 32, 833–840. Weiglmeier, P.R., Rösch, P. and Berkner, H. 2010. Cure and curse: E. coli heat-stable enterotoxin and its receptor guanylyl cyclase C. Toxins J. 2, 2213–2229. | ||
| How to Cite this Article |
| Pubmed Style El-Nady HH, MIE, NZA, Abd-Elfatah EB, Shehata AA, Fawzi EM. Colibacillosis in lambs and kids in Egypt: Prevalence, serogroups, antibiogram profile, virulence genes distribution and antimicrobial resistance genes. Open Vet J. 2023; 13(9): 1106-1115. doi:10.5455/OVJ.2023.v13.i9.6 Web Style El-Nady HH, MIE, NZA, Abd-Elfatah EB, Shehata AA, Fawzi EM. Colibacillosis in lambs and kids in Egypt: Prevalence, serogroups, antibiogram profile, virulence genes distribution and antimicrobial resistance genes. https://www.openveterinaryjournal.com/?mno=154770 [Access: May 19, 2024]. doi:10.5455/OVJ.2023.v13.i9.6 AMA (American Medical Association) Style El-Nady HH, MIE, NZA, Abd-Elfatah EB, Shehata AA, Fawzi EM. Colibacillosis in lambs and kids in Egypt: Prevalence, serogroups, antibiogram profile, virulence genes distribution and antimicrobial resistance genes. Open Vet J. 2023; 13(9): 1106-1115. doi:10.5455/OVJ.2023.v13.i9.6 Vancouver/ICMJE Style El-Nady HH, MIE, NZA, Abd-Elfatah EB, Shehata AA, Fawzi EM. Colibacillosis in lambs and kids in Egypt: Prevalence, serogroups, antibiogram profile, virulence genes distribution and antimicrobial resistance genes. Open Vet J. (2023), [cited May 19, 2024]; 13(9): 1106-1115. doi:10.5455/OVJ.2023.v13.i9.6 Harvard Style El-Nady, H. H., , . M. I. E., , . N. Z. A., Abd-Elfatah, . E. B., Shehata, . A. A. & Fawzi, . E. M. (2023) Colibacillosis in lambs and kids in Egypt: Prevalence, serogroups, antibiogram profile, virulence genes distribution and antimicrobial resistance genes. Open Vet J, 13 (9), 1106-1115. doi:10.5455/OVJ.2023.v13.i9.6 Turabian Style El-Nady, Heba Hassan, Mohamed Ibrahim Eissa, Naser Zeidan Abo-Zeid, Eman Beshry Abd-Elfatah, Ayman Ahmed Shehata, and Elshaima Mohamed Fawzi. 2023. Colibacillosis in lambs and kids in Egypt: Prevalence, serogroups, antibiogram profile, virulence genes distribution and antimicrobial resistance genes. Open Veterinary Journal, 13 (9), 1106-1115. doi:10.5455/OVJ.2023.v13.i9.6 Chicago Style El-Nady, Heba Hassan, Mohamed Ibrahim Eissa, Naser Zeidan Abo-Zeid, Eman Beshry Abd-Elfatah, Ayman Ahmed Shehata, and Elshaima Mohamed Fawzi. "Colibacillosis in lambs and kids in Egypt: Prevalence, serogroups, antibiogram profile, virulence genes distribution and antimicrobial resistance genes." Open Veterinary Journal 13 (2023), 1106-1115. doi:10.5455/OVJ.2023.v13.i9.6 MLA (The Modern Language Association) Style El-Nady, Heba Hassan, Mohamed Ibrahim Eissa, Naser Zeidan Abo-Zeid, Eman Beshry Abd-Elfatah, Ayman Ahmed Shehata, and Elshaima Mohamed Fawzi. "Colibacillosis in lambs and kids in Egypt: Prevalence, serogroups, antibiogram profile, virulence genes distribution and antimicrobial resistance genes." Open Veterinary Journal 13.9 (2023), 1106-1115. Print. doi:10.5455/OVJ.2023.v13.i9.6 APA (American Psychological Association) Style El-Nady, H. H., , . M. I. E., , . N. Z. A., Abd-Elfatah, . E. B., Shehata, . A. A. & Fawzi, . E. M. (2023) Colibacillosis in lambs and kids in Egypt: Prevalence, serogroups, antibiogram profile, virulence genes distribution and antimicrobial resistance genes. Open Veterinary Journal, 13 (9), 1106-1115. doi:10.5455/OVJ.2023.v13.i9.6 |