| Research Article | ||
Open Vet J. 2023; 13(9): 1099-1105 Open Veterinary Journal, (2023), Vol. 13(9): 1099–1105 Original Research Molecular identification and phylogenetic confirmation of Sarcocystis species in slaughtered camels in Al-Najif province, IraqOla A. Aggar* and Mohammed T. S. Al-ZubaidiDepartment of Parasitology, College of Veterinary Medicine, University of Baghdad, Baghdad, Iraq *Corresponding Author: Ola A. Aggar. Department of Parasitology, College of Veterinary Medicine, University of Baghdad, Baghdad, Iraq. Email: Ula.Abd1105a [at] covm.uobaghdad.edu.iq Submitted: 10/06/2023 Accepted: 06/08/2023 Published: 30/09/2023 © 2023 Open Veterinary Journal
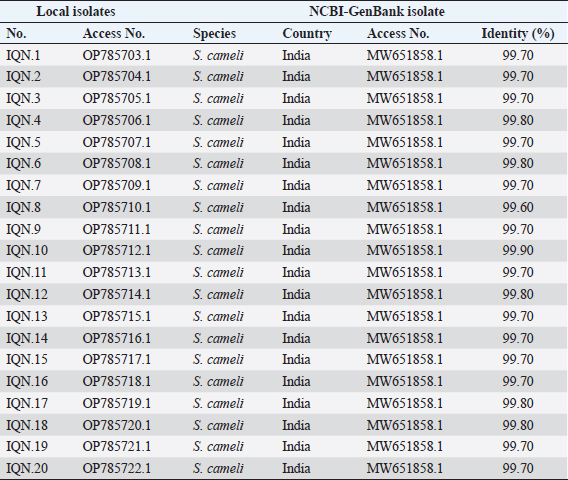
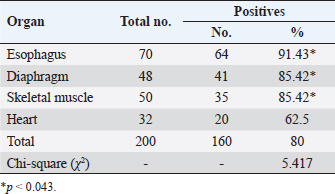
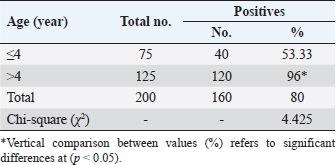
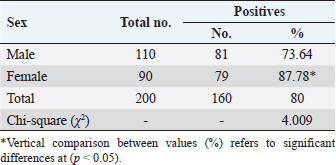
AbstractBackground: Sarcocystis is an intracellular parasite of particular importance as it infects many domestic animals as camels that play the role of intermediate host for the parasite. Aim: This study aimed to identify Sarcocystis species in camels by molecular assay with confirmation of local isolates by phylogenetic analysis. Methods: A total of 200 slaughtered camels (Camelus dromedarius) that were slaughtered in Al-Najaf province (Iraq) abattoirs from October (2021) to July (2022) were subjected to collect the fresh tissues from four organs (esophagus, diaphragm, skeletal muscle, and heart), to be tested later by the conventional polymerase chain reaction (PCR). Then, a total of 20 positive genomic DNA samples were sequenced, named, got specific access numbers (OP785703.1 to OP785722.1), and compared with the NCBI-GenBank isolates. Results: Targeting Cox1 gene, 80% of collected tissues were found positive by the conventional PCR assay. Phylogenetic tree analysis revealed that the local Sarcocystis isolates were identical to Indian S. cameli isolates at 99.70%–99.90%. Significantly, an increase in Sarcocystis infection was seen in the esophagus compared to the diaphragm, skeletal muscle, and heart; older (>4 years) than younger (≤4 years) camels, and in females more than males. Conclusion: To the best of our knowledge, this represents the first molecular study in Iraq that identifies Sarcocystis cameli in camels. However, additional epidemiological and molecular studies in camel populations as well as in other domestic and wild animals appeared to be necessary. Keywords: Sarcocystosis, S. cameli, Camelus dromedarius, PCR, Sequencing. IntroductionSarcocystis, first reported in 1843, is a genus of intracellular protozoan parasite which belongs to Sarcocystidae family, Eucoccidiorina order of Apicomplexa phylum. Worldwide, considerable production losses and health illness in animals have been recognized due to the presence of >200 species in the genus of Sarcocystis that infects a wide array of domestic and wild animals (Dubey et al., 2015a; Abbas et al., 2023). The life cycle of a parasite involves two hosts; definitive (predator) and intermediate (prey) hosts. However, Sarcocystis spp. having greater specificity to intermediate than definitive hosts (Prakas et al., 2023). In the intermediate host, the development of asexual stages of the parasite occurs usually in skeletal, cardiac, esophageal, and diaphragmatic muscles and to a lesser extent in CNS (Dubey et al., 2015a; Gajadhar et al., 2015; Prakas et al., 2021). The definitive host can be infected during ingestion of contaminated tissues with mature Sarcocystis that undergoes sexual reproduction to excrete the oocysts/sporocysts to the environment by feces (Lindsay and Dubey, 2020; Razooqi et al., 2022). One-humped or dromedary camels (Camelus dromedarius) reared widely in arid regions of many world countries (Burger et al., 2019; Zhu et al., 2019) has been used mainly for the production of meat that is characterized by the low percentages of fats and cholesterol when comparison to meats of other animal species (Faye, 2016; Kadim et al., 2018). However, the existence of parasite in camel meats might potentially downgrade their quality to be consumed by humans (Hamidinejat et al., 2013; Mohammadpour et al., 2020). Although Sarcocystis spp. can induce significant pathology or even mortality, most diseased camels generally exhibit subclinical illness (Metwally et al., 2020). Thus, the diagnosis of Sarcocystis spp. in camels using traditional tools represents a challenge for the researchers. Hence, molecular identification of Sarcocystis spp. in Iraqi camels with confirmation of local isolates by phylogeny was aimed in this study. Materials and MethodsStudy animals and samplingTotally, 200 slaughtered camels (C. dromedarius) of different ages (<2 to ≤5 years) and sexes (107 males and 93 females) were selected to the present study from the abattoir of Al-Najaf province (Iraq) during October (2021)–July (2022). Approximately, 100 g of fresh meat were obtained from the esophagus, diaphragm, skeletal muscle, and heart of each slaughtered camel, kept in individual plastic labeled containers, and saved frozen at −20C for later molecular examination. Molecular examinationConventional polymerase chain reaction (PCR) In accordance with the manufacturer instruction of the gSYAN DNA Extraction Kit (iNtRON Biotechnology, South Korea), the DNAs were extracted from collected samples and examined directly by the Nanodrop System (Thermo Scientific, UK) to estimate the purity and concentration of these DNAs samples (Gharban et al., 2023). According to the manufacturer instructions of GoTaqTM Green PCR MasterMix Kit (Bioneer, Korea), the Mastermix tubes were prepared at a modified final volume of 25 μl (5 μl of DNA template, 2 μl of forward primer, 2 μl of reverse primer, 4 μl of Green PCR Mastermix, and 12 l of nuclease-free water) using one set of primers [(F: 5′-ATG GCG TAC AAC AAT CAT AAA GAA-3′) and (R: 5-ATA TCC ATA CCG CCA TTG CCC AT-3′)] targeting Cox1 gene at a product size of 1,058 bp (Metwally et al., 2020). The PCR thermocycler was carried as follows: one cycle for initial denaturation (5 minutes/95°C); 35 cycles for denaturation (30 seconds/95°C), annealing (30 seconds/54°C) and extension (1 minute/72°C); and one cycle for final extension (5 minutes/72C). Agarose gel stained with 3 μl of Ethidium Bromide and Ladder Marker (100–2,000 bp) were analyzed by electrophoresis at 80 Volt, 80 Am for 90 minutes. PCR products of previously diagnosed and confirmed Sarcocystis spp. isolate was used as a positive control; while nuclease-free water was used as a negative control. Finally, the DNA fragments were visualized under the UV-Transilluminator (ATTA/Korea). Phylogenetic analysis Sequencing of some positive DNAs was performed to confirm the species of Sarcocystis, and study the genetic relationship between the local and NCBI-BLAST Sarcocystis isolates. Targeting the Cox1 gene, a total of 20 positive DNA samples in addition to forward primer were sent for the Macrogen Company (South Korea) to be sequenced by the AB DNA Sequencing System. First, the local isolates of Sarcocystis species were recorded in the National Center For Biotechnology Information (NCBI)-GenBank to obtain their access numbers; and then, the Phylogenetic tree analysis was performed by the MEGA 6.0 Software. Statistical analysisGraphPad Prism (GraphPad Software Inc., USA) Software was served to evaluate significant variation between results of this study using the one way-analysis of variance. Values were represented as percentages (%), and significant differences were p ˂ 0.05 (Gharban, 2023). Ethical approvalThe current study was licensed by the Scientific Committee of the Department of Parasitology in the College of Veterinary Medicine, University of Baghdad, Baghdad, Iraq. ResultsTargeting Cox1 gene, 80% (160/200) of meat samples were positive by the conventional PCR assay at a product size of 1,058 bp (Fig. 1). The genomic DNAs of 20 positive samples were named respectively as Sarcocystis spp. IQN.1 to IQN.20. Comparative analysis of local isolates with the NCBI-GenBank Sarcocystis isolates showed that there were nucleotide alignment similarities (*) and substitution mutations in Cox1 gene. Comparative homology sequence identity (%) to detect genetic variations between the local Sarcocystis spp. IQN-isolates (total no. 20) and the NCBI-BLAST Sarcocystis cameli Indian (MW651858.1) isolate of cox1 gene. This identity showed that there was 99% of similarity and 0.150%–0.50% of total genetic mutations/changes (Table 1, Fig. 2). According to organ, significantly higher increases (p < 0.05) in the prevalence of sarcocysts were detected in the esophagus (91.43%), while the lower one (p < 0.05) was seen in the heart (62.5%). However, insignificant variation (p > 0.05) in the prevalence of Sarcocystis spp. between diaphragm (85.42%) and skeletal muscle (85.42%) was reported (Table 2). According to the age of study camels, this study has seen that the parasite is more prevalent in camels aged >4 years (96%) than those aged ≤4 years (53.33%), (Table 3). Regarding the sex of study camels, the infection rate of sarcocystosis was increased significantly (p < 0.05) in females (87.78%) in comparison with males (73.64%), (Table 4). DiscussionSarcocystis infections have been demonstrated in different animal species such as cattle, buffalo, goats, and sheep (Dakhil et al., 2017; Al-Saadi et al., 2020; Swar and Shnawa, 2020; Abdullah, 2021). The molecular technique is one of the most useful diagnostic methods for the identification of many pathogens, but there is low molecular data about Sarcocystis infection in camels. In this study, the prevalence of Sarcocystis spp. in slaughtered camels was 80%. This agrees with the finding reported in camels (85.5%) in Egypt (Rabie et al., 2021). In this study, the Cox1 gene revealed a high efficacy in the detection of Sarcocystis spp. in tissue samples of slaughtered camels. Different studies reported that DNA barcoding system using the Cox1 gene has a high efficacy in discrimination between the invertebrate and vertebrate species (Rodrigues et al., 2017; Mioduchowska et al., 2018; Ahmed et al., 2022). Gjerde et al. (2015) concluded that the Cox1 gene can work better with DNA samples derived from Sarcocystis, and sequences of the Cox1 gene can vigorously distinguish between different species of Sarcocystis. Rodrigues et al. (2017) demonstrated the suitability of Cox1 as a gene of parasitic reorganization from those of highly related. Other studies reported that Cox1 has a well phylogenetic character than other genes (Stöger et al., 2016; Wang et al., 2017). Furthermore, some authors claimed that the evolution of the Cox1 gene enables researchers to distinguish between closely related species and investigate intraspecific variation (Changbunjong et al., 2016; Viricel and Rosel, 2022). The Cox1 has been demonstrated as the potential barcode marker for joint analysis of nuclear and mitochondrial markers of various species (Rodrigues et al., 2017; Ahmed et al., 2022).
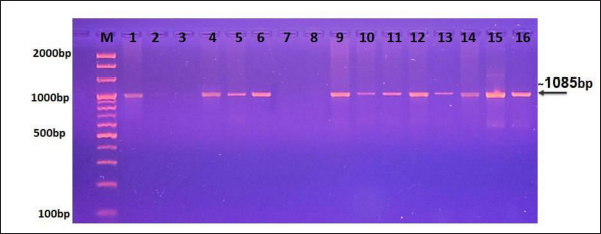
Fig. 1. Representative image shows the agarose-gel electrophoresis of PCR products targeting the Cox1 gene to detect the genus of Sarcocystis. Lane (M): Ladder marker (100–2,000 bp); Lane (1): positive control; Lane (2): negative control; Lanes (3, 7, 8): negative samples; Lanes (4–6 and 9–16): positive samples for Sarcocystis spp. at 1,058. Table 1. NCBI-BLAST Homology sequence identity between local and GenBank isolates.
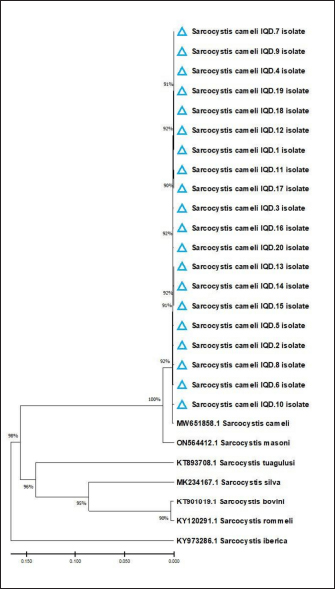
Fig. 2. Phylogenetic tree analysis based on Cox1 gene partial sequence in local IQN S. cameli isolates and NCBI-GenBank S. cameli isolates. Comparative identity for the genetic variations between the local isolates and NCBI-BLAST isolates showed that similarity was 98%; whereas, the total genetic mutations/changes were 0.2%–1.5%. Despite the discovery of five different Sarcocystis species in camels, multiple studies have shown that only S. cameli and S. ippeni are considered to be the real species infecting dromedary camels, namely S. cameli, S. ippeni, S. camelicanis, S. camelocanis, and S. miescheri (Dubey et al., 2015b; Omer et al., 2017; Rabie et al., 2021). Table 2. Total infection rate of sarcocystosis in camels according to organ.
Table 3. Total infection rate of sarcocystosis in camels according to age.
Table 4. Total infection rate of sarcocystosis in camels according to sex.
In this study, the phylogenetic analysis discovered that the local study isolates were identical to the global NCBI-GenBank Indian S. cameli isolate. This is in agreement with the results of other studies as S. cameli is the more prevalent species of Sarcocystis in camels to yet (Valinezhad et al., 2008; Dubey et al., 2015b; Omer et al., 2017; Asopa et al., 2023). For the first molecular proof of S. cameli in camels in Iran (Motamedi et al., 2011), the bradyzoite DNA was amplified from the 18S rRNA gene fragment using traditional PCR. Sarcocystis species can frequently infect the muscular tissue of the heart, tongue, esophagus, and diaphragm (Wahba et al., 2014; Ahmed et al., 2016). Our findings revealed that Sarcocystis spp. has existed in all examined organs but more commonly in the esophagus. Some research suggested that sarcocystosis is more frequent in the cremaster muscle (Bucca et al., 2011; Sağlam and Keleş, 2016). Shekarforoush et al. (2006) demonstrated that the heart is the most infected organ; while Al-Ani and Amr (2017) noted that camel’s diaphragm was the most frequently affected organ. According to Oryan et al. (2010), Sarcocystis appears to prefer the esophagus, tongue, and heart. In contrast, Gareh et al. (2020) reported that Sarcocystis can spread in various organs, mostly in the esophagus with a prevalence rate of 49%. Rabie et al. (2021) reported Sarcocystis in the esophageal, heart, and ocular muscles in addition to other organs, with a higher infection rate in the esophagus (85.5%). Asopa et al. (2023) showed that Sarcocystis spp. was prevalent in the tongue and esophagus of dromedary camels from Rajasthan’s Bikaner area. This variance in the distribution of S. cameli strains may be accounted for differences in the prevalence of Sarcocystis among camel organs (Hamidinejat et al., 2013). Another substantial risk factor for infection was age. In this study, the infection rate was considerably higher in older camels (>4 years) than in younger camels (<4 years). Other reports in Iran (Hamidinejat et al., 2013), Saudi Arabia (Omer et al., 2017), and Egypt (El-Bahy et al., 2019) have revealed similar results. The slower development of detectable cysts may account for the low prevalence of Sarcocystis in young camels. Another explanation for increasing the rate of infection in older camels is that the older animals are slaughtered at a higher rate than younger ones. In addition, keeping young camels indoors may reduce their exposure to disease (Valinezhad, et al., 2008, Hamidinejat et al., 2013, Omer et al., 2017). Sex was discovered as a crucial factor linked to infection. Our findings were consistent with numerous studies conducted in southern Ethiopia (Woldemeskel and Gumi, 2001), Iran (Valinezhad et al., 2008), and Egypt (Rabie et al., 2021) which found that male camels were more susceptible to infection than females. Omer et al. (2017) showed that the slaughtered female camels were usually over 4 years old, whereas male camels were under 2 years old. This disparity might be explained by the fact that most males are grazed outside for hard work, while most females are kept indoors for breeding under good and hygienic management. This might make males more susceptible to infection than females (Romero et al., 2017). An insignificant difference in the frequency of sarcocystosis between male and female camels was identified by Hamidinejat et al. (2013). A lack of relationship between sex and infection rates has been shown in similar studies on camels (Woldemeskel and Gumi, 2001; Shekarforoush et al., 2006; Valinezhad et al., 2008). ConclusionTo the best of our knowledge, this represents the first molecular study in Iraq that identifies Sarcocystis cameli in camels. However, additional epidemiological and molecular studies in camel populations as well as in other domestic and wild animals appeared to be necessary. AcknowledgmentsThe authors are thankful to the Colleges of Veterinary Medicine, University of Baghdad for providing the necessary facilities for the study. However, all materials used in this study were purchased by the authors and no external funds were received for this study. Conflict of interestThe authors declare that there is no conflict of interest. Author’s contributionOAA: Collection of tissue samples, extraction of DNAs, and preparation of Mastermix tubes; MTSA: PCR analysis, sequencing, and statistical analysis of obtained results. All authors have written the manuscript and approved the final copy of it. FundingThere is no external fund (private funding). Data availabilityAll data supporting the findings of this study are available within the manuscript. ReferencesAbbas, I., Baghdadi, H.B., Rizk, M.A., El-Alfy, E.S., Elmishmishy, B. and Gwida, M. 2023. Gastrointestinal parasites of dogs in Egypt: an update on the prevalence in Dakahlia Governorate and a meta-analysis for the published data from the country. Animals (Basel) 13(3), 1–21. Abdullah, S.H. 2021. Investigation of Sarcocystis spp. in slaughtered cattle and sheep by peptic digestion and histological examination in Sulaimani Province, Iraq. Vet. World 14(2), 468–477. Ahmed, A.M., Elshraway, N.T. and Youssef, A.I. 2016. Survey on Sarcocystis in bovine carcasses slaughtered at the municipal abattoir of El-Kharga, Egypt. Vet. World 9(12), 1461–1469. Ahmed, S., Ibrahim, M., Nantasenamat, C., Nisar, M.F., Malik, A.A., Waheed, R. and Alam, M.K. 2022. Pragmatic applications and universality of DNA barcoding for substantial organisms at species level: a review to explore a way forward. BioMed. Res. Int. 2022, 1–15. Al-Ani, F.K. and Amr, Z. 2017. Sarcocystis spp. prevalence in camel meat in Jordan. J. Dairy. Vet. Sci. 4, 1–3. Al-Saadi, S.A., Al-Mussawi, K.A. and Muhammed, H.A. 2020. Molecular identification of Sarcocystis species infection in sheep in Karbala Governorate–Iraq. Med. Legal. Update 20, 889–895. Asopa, S., Joshi, A., Dadhich, H. and Vyas, I. 2023. Prevalence of sarcocysts in tongue and oesophagus of dromedary camels from Bikaner district of Rajasthan. Ind. J. Vet. Sci. Biotechnol. 19(1), 96–98. Bucca, M., Brianti, E., Giuffrida, A., Ziino, G., Cicciari, S. and Panebianco, A. 2011. Prevalence and distribution of Sarcocystis spp. cysts in several muscles of cattle slaughtered in Sicily, Southern Italy. Food Control 22(1), 105–108. Burger, P.A., Ciani, E. and Faye, B. 2019. Old world camels in a modern world—a balancing act between conservation and genetic improvement. Anim. Gen. 50(6), 598–612. Changbunjong, T., Weluwanarak, T., Samung, Y. and Ruangsittichai, J. 2016. Molecular identification and genetic variation of hematophagous flies, (Diptera: Muscidae: Stomoxyinae) in Thailand based on cox1 barcodes. J. Asia Pac. Entomol. 19 (4), 1117–1123. Dakhil, H.G., Abdallah, B.H. and Abdallah, F.A. 2017. Molecular identification of Sarcocystis fusiformis and S. moulei infecting water buffaloes (Bubalus bubalis) in southern Iraq. World J. Pharm. Res. 6(3), 215–229. Dubey, J.P., Calero-Bernal, R., Rosenthal, B.M., Speer, C.A. and Fayer, R. 2015a. Sarcocystosis of animals and humans, 2nd ed. Boca Raton, FL: CRC Press, pp: 235–238. Dubey, J.P., Hilali, M., Van Wilpe, E., Calero-Bernal, R., Verma, S.K. and Abbas, I.E. 2015b. A review of sarcocystosis in camels and redescription of Sarcocystis cameli and Sarcocystis ippeni sarcocysts from the one-humped camel (Camelus dromedarius). Parasitology 142(12), 1481–1492. El-Bahy, N., El-Bagory, A.E.R., AbouLaila, M., Elkhatam, A. and Mady, H.M. 2019. Prevalence of Sarcocystis fusiformis and hydatid cyst among different ruminants at Menofia Governorate, Egypt. J. Curr. Vet. Res. 1(1), 1–10. Faye, B. 2016. The camel, new challenges for a sustainable development. Trop. Anim. Health. Prod. 48(4), 689–692. Gajadhar, A.A., Lalonde, L.F., Al-Adhami, B., Singh, B.B. and Lobanov, V. 2015. Foodborne apicomplexan protozoa: Coccidia. In Foodborne parasites in the food supply web. Sawston, UK: Woodhead Publishing, pp: 101–147. Gareh, A., Soliman, M., Saleh, A.A., El-Gohary, F.A., El-Sherbiny, H.M., Mohamed, R.H. and Elmahallawy, E.K. 2020. Epidemiological and histopathological investigation of Sarcocystis spp. in slaughtered dromedary camels (Camelus dromedarius) in Egypt. Vet. Sci. 7(4), 1–10. Gharban, H.A. (2023). Molecular prevalence and phylogenetic confirmation of bovine trichomoniasis in aborted cows in Iraq. Vet. World 16(3), 580–587. Gharban, H.A., Al-Shaeli, S.J. and Hussen, T.J. 2023. Molecular genotyping, histopathological and immunohistochemical studies of bovine papillomatosis. Open Vet. J. 13(1), 26–41. Gjerde, B., Hilali, M. and Mawgood, S.A. 2015. Molecular characterisation of three regions of the nuclear ribosomal DNA unit and the mitochondrial cox1 gene of Sarcocystis fusiformis from water buffaloes (Bubalus bubalis) in Egypt. Parasitol. Res. 114, 3401–3413. Hamidinejat, H., Hekmatimoghaddam, S., Jafari, H., Sazmand, A., Haddad Molayan, P., Derakhshan, L. and Mirabdollahi, S. 2013. Prevalence and distribution patterns of Sarcocystis in camels (Camelus dromedarius) in Yazd province, Iran. J. Parasit. Dis. 37, 163–165. Kadim, I.T., Al-Amri, I.S., Al Kindi, A.Y. and Mbaga, M. 2018. Camel meat production and quality: a review. J. Camel Pract. Res. 25(1), 9–23. Lindsay, D.S. and Dubey, J.P. 2020. Neosporosis, toxoplasmosis, and sarcocystosis in ruminants: an update. Vet. Clin. Food. Anim. Pract. 36(1), 205–222. Metwally, D.M., Al-Otaibi, T.T., Al-Turaiki, I.M., El-Khadragy, M. F. and Alajmi, R.A. 2020. Identification of Sarcocystis spp. in one-humped camels (Camelus dromedarius) from Riyadh and Dammam, Saudi Arabia, via histological and phylogenetic approaches. Animals (Basels) 10(7), 1108. Mioduchowska, M., Czyż, M.J., Gołdyn, B., Kur, J. and Sell, J. 2018. Instances of erroneous DNA barcoding of metazoan invertebrates: Are universal cox1 gene primers too “universal”? PLoS One 13(6), 1–13. Mohammadpour, R., Champour, M., Tuteja, F. and Mostafavi, E. 2020. Zoonotic implications of camel diseases in Iran. Vet. Med. Sci. 6(3), 359–381. Motamedi, G.R., Dalimi, A., Nouri, A. and Aghaeipour, K. 2011. Ultrastructural and molecular characterization of Sarcocystis isolated from camel (Camelus dromedarius) in Iran. Parasitol. Res. 108, 949–954. Omer, S.A., Alzuraiq, A.A. and Mohammed, O.B. 2017. Prevalence and molecular detection of Sarcocystis spp. infection in the dromedary camel (Camelus dromedarius) in Riyadh city, Saudi Arabia. Biomed. Res. 28(11), 1–10. Oryan, A., Ahmadi, N. and Mousavi, S.M.M. 2010. Prevalence, biology, and distribution pattern of Sarcocystis infection in water buffalo (Bubalus bubalis) in Iran. Trop. Anim. Health Prod. 42, 1513–1518. Prakas, P., Bea, A., Juozaitytė-Ngugu, E., Olano, I., Villanúa, D., Švažas, S. and Butkauskas, D. 2021. Molecular identification of Sarcocystis halieti in the muscles of two species of birds of prey from Spain. Parasites Vect. 14, 1–7. Prakas, P., Moskaliova, D., Šneideris, D., Juozaitytė-Ngugu, E., Maziliauskaitė, E., Švažas, S. and Butkauskas, D. 2023. Molecular identification of Sarcocystis rileyi and Sarcocystis sp. (closely related to Sarcocystis wenzeli) in intestines of mustelids from Lithuania. Animals (Basels)13(3), 467. Rabie, S., Hassanine, R.M.E.S., A Hassan, A., Abo Elhussien, O. and BM EL-Mahdi, M. 2021. Morphological and molecular characterization of Sarcocystis cameli and Sarcocystis ippeni from the muscles of one-humped camel (Camelus dromedarius) in New valley Governorate, Egypt. J. Vet. Sci. 4(3), 103–118. Razooqi, M.A., Gharban, H.A. and Al-Kaabi, M.A. 2022. Molecular and seroprevalence of toxoplasmosis in goats’ blood and milk in Iraq. Arch. Razi. Instit. 77(5), 1749–1755. Rodrigues, M.S., Morelli, K.A. and Jansen, A.M. 2017. Cytochrome c oxidase subunit 1 gene as a DNA barcode for discriminating Trypanosoma cruzi DTUs and closely related species. Parasites Vect. 10(1), 1–18. Romero, S., Carletti, T., Franco, C.D., Moré, G., Schnittger, L. and Florin-Christensen, M. 2017. Seropositivity to Sarcocystis infection of llamas correlates with breeding practices. Vet. Parasitol. Reg. Stud. Reports 10, 65–70. Sağlam, K. and Keleş, H. 2016. Sarcocystosis in the cremaster muscle of an infertile bull, spermiostasis and orchitis. Kocatepe Vet. J. 9(3), 252–254. Shekarforoush, S.S., Shakerian, A. and Hasanpoor, M.M. 2006. Prevalence of Sarcocystis in slaughtered one-humped camels (Camelus dromedarius) in Iran. Trop. Anim. Health Prod. 38(4), 301–303. Stöger, I., Kocot, K.M., Poustka, A.J., Wilson, N.G., Ivanov, D., Halanych, K.M. and Schrödl, M. 2016. Monoplacophoran mitochondrial genomes: convergent gene arrangements and little phylogenetic signal. BMC Evolut. Biol. 16(1), 1–18. Swar, S.O. and Shnawa, B.H. 2020. Ultrastructural and molecular characterization of Sarcocystis species derived from macroscopic sarcocysts of domestic sheep and goats in Soran City, Erbil, Iraq. World Vet. J. 10(4), 540–550. Valinezhad, A., Oryan, A. and Ahmadi, N. 2008. Sarcocystis and its complications in camels (Camelus dromedarius) of eastern provinces of Iran. Korean J. Parasitol. 46(4), 229–234. Viricel, A. and Rosel, P.E. 2022. Evaluating the utility of cox1 for cetacean species identification. Marine Mam. Sci. 28(1), 37–62. Wahba, A., Ayoub, M. and Soliman, K. 2014. Light and ultrastructure of Sarcocystis spp. of camels and associated pathological changes. Anim. Health Res. J. 2, 143–158. Wang, J., Zhang, L., Zhang, Q.L., Zhou, M.Q., Wang, X.T., Yang, X.Z. and Yuan, M.L. 2017. Comparative mitogenomic analysis of mirid bugs (Hemiptera: Miridae) and evaluation of potential DNA barcoding markers. Peer J. 5, e3661. Woldemeskel, M. and Gumi, B. 2001. Prevalence of sarcocysts in one-humped camel (Camelus dromedarius) from Southern Ethiopia. J. Vet. Med. 48(3), 223–226. Zhu, S., Zimmerman, D. and Deem, S.L. 2019. A review of zoonotic pathogens of dromedary camels. Ecohealth 16, 356–377. | ||
| How to Cite this Article |
| Pubmed Style Aggar OA, Al-Zubaidi MT. Molecular identification and phylogenetic confirmation of Sarcocystis species in slaughtered camels in Al-Najif province, Iraq. Open Vet J. 2023; 13(9): 1099-1105. doi:10.5455/OVJ.2023.v13.i9.5 Web Style Aggar OA, Al-Zubaidi MT. Molecular identification and phylogenetic confirmation of Sarcocystis species in slaughtered camels in Al-Najif province, Iraq. https://www.openveterinaryjournal.com/?mno=157039 [Access: May 19, 2024]. doi:10.5455/OVJ.2023.v13.i9.5 AMA (American Medical Association) Style Aggar OA, Al-Zubaidi MT. Molecular identification and phylogenetic confirmation of Sarcocystis species in slaughtered camels in Al-Najif province, Iraq. Open Vet J. 2023; 13(9): 1099-1105. doi:10.5455/OVJ.2023.v13.i9.5 Vancouver/ICMJE Style Aggar OA, Al-Zubaidi MT. Molecular identification and phylogenetic confirmation of Sarcocystis species in slaughtered camels in Al-Najif province, Iraq. Open Vet J. (2023), [cited May 19, 2024]; 13(9): 1099-1105. doi:10.5455/OVJ.2023.v13.i9.5 Harvard Style Aggar, O. A. & Al-Zubaidi, . M. T. (2023) Molecular identification and phylogenetic confirmation of Sarcocystis species in slaughtered camels in Al-Najif province, Iraq. Open Vet J, 13 (9), 1099-1105. doi:10.5455/OVJ.2023.v13.i9.5 Turabian Style Aggar, Ola A., and Mohammed T.S. Al-Zubaidi. 2023. Molecular identification and phylogenetic confirmation of Sarcocystis species in slaughtered camels in Al-Najif province, Iraq. Open Veterinary Journal, 13 (9), 1099-1105. doi:10.5455/OVJ.2023.v13.i9.5 Chicago Style Aggar, Ola A., and Mohammed T.S. Al-Zubaidi. "Molecular identification and phylogenetic confirmation of Sarcocystis species in slaughtered camels in Al-Najif province, Iraq." Open Veterinary Journal 13 (2023), 1099-1105. doi:10.5455/OVJ.2023.v13.i9.5 MLA (The Modern Language Association) Style Aggar, Ola A., and Mohammed T.S. Al-Zubaidi. "Molecular identification and phylogenetic confirmation of Sarcocystis species in slaughtered camels in Al-Najif province, Iraq." Open Veterinary Journal 13.9 (2023), 1099-1105. Print. doi:10.5455/OVJ.2023.v13.i9.5 APA (American Psychological Association) Style Aggar, O. A. & Al-Zubaidi, . M. T. (2023) Molecular identification and phylogenetic confirmation of Sarcocystis species in slaughtered camels in Al-Najif province, Iraq. Open Veterinary Journal, 13 (9), 1099-1105. doi:10.5455/OVJ.2023.v13.i9.5 |