| Research Article | ||
Open Vet J. 2024; 14(2): 604-616 Open Veterinary Journal, (2024), Vol. 14(2): 604–616 Original Research Prevalence and antimicrobial resistance of Enterobacteriaceae in the north of KazakhstanYuliya Aleshina1, Anara Yeleussizova2, Anara Mendybayeva1, Pavel Shevchenko1 and Raushan Rychshanova1*1Research Institute of Applied Biotechnology, A. Baitursynov Kostanay Regional University, Kostanay, Republic of Kazakhstan 2Department of Veterinary Sanitation, A. Baitursynov Kostanay Regional University, Kostanay, Republic of Kazakhstan *Corresponding Author: Raushan Rychshanova. Research Institute of Applied Biotechnology, A. Baitursynov Kostanay Regional University, Kostanay, Republic of Kazakhstan. Email: raushan.rychshanova [at] gmail.com Submitted: 05/09/2023 Accepted: 15/01/2024 Published: 29/02/2024 © 2024 Open Veterinary Journal
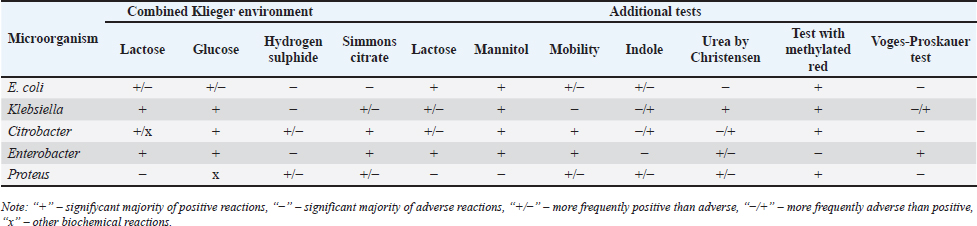
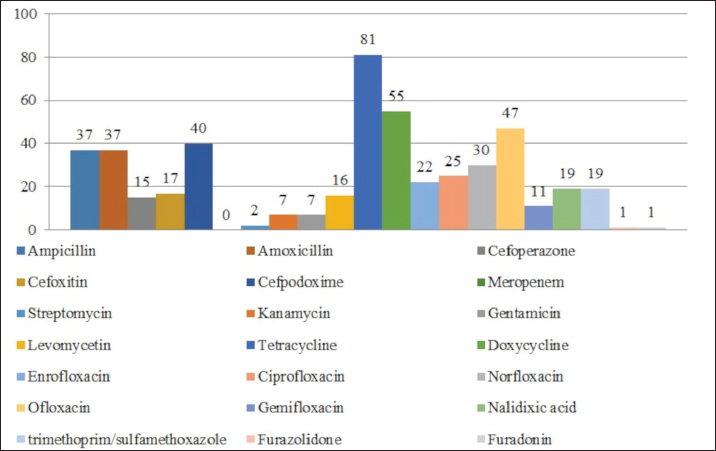
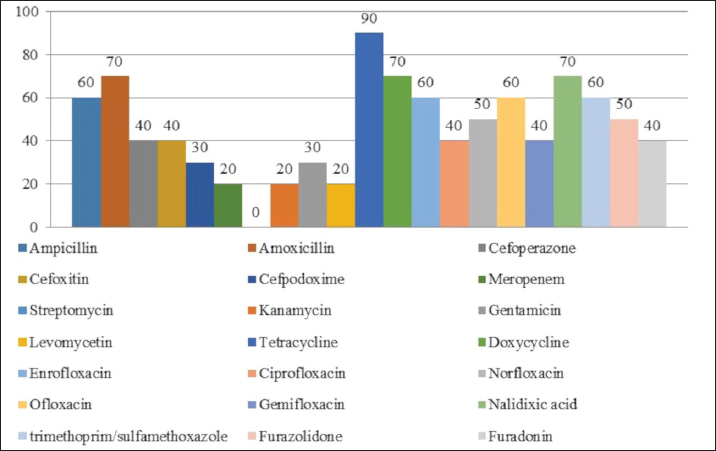
AbstractBackground: An increasing number of drugs are used each year in the treatment of small pets (cats and dogs), including medicines (cephalosporins and fluoroquinolones) used in human therapy. Aim: The purpose of this study was to isolate and explore the antibiotic resistance of opportunistic Enterobacteriaceae (Escherichia coli, Klebsiella, Proteus, Ci trobacter, Enterobacter) from cats and dogs, and to isolate resistance genes in the microorganisms. Methods: In 2021, 808 samples of biological material from small domestic animals were collected in veterinary clinics in Kostanay. From these, 210 microorganisms were isolated and identified. Results: A large majority of the strains sampled belonged to E. coli—149 (70.9%), Enterobacter—11 (5.2%), Klebsiella—28 (13.3%), Proteus—12 (5.7%) and 10 Citrobacter isolates (4.8%). In all isolates identified, antibiotic resistance/sensitivity was determined by disc-diffusion method to ampicillin, cefoxitin, gentamicin, levomycetin, tetracycline, ciprofloxacin, norfloxacin, ofloxacin, cefoperazone, cefpodoxime, streptomycin, kanamycin, doxycycline, gemifloxacin, nalidixic acid, furazolidone, furadonine, amoxicillin, and enrofloxacin. Conclusion: The study has demonstrated that the greatest number of Enterobacteriaceae were sensitive to the action of meropenem, which belongs to the group of beta-lactam antibiotics; resistance was demonstrated against tetracycline, doxycycline, ampicillin, amoxicillin, ofloxacin, and cefpodoxime. The most common genes encoding antimicrobial resistance were as follows: BlaTEM and OXA in 41 and 28 isolates, respectively, encoding resistance to beta-lactams; StrA and StrB in 45 and 48 isolates encoding aminoglycosides; and tetA and tetB in 43 and 28 isolates encoding tetracyclines. Obtained data demonstrate that uncontrolled and frequent use of beta-lactam and tetracycline antibacterials, in cats and dogs, results in the spread of genotypic resistance among micro-organisms of the family Enterobacteriaceae. Keywords: Antibiotic sensitivity, Citrobacter, Klebsiella, Microorganism, Proteus. IntroductionAn increasing number of drugs are used each year in the treatment of small pets (cats and dogs), including medicines (cephalosporins and fluoroquinolones) used in human therapy. Studies demonstrate that antimicrobial resistance is increasing among microorganisms that cause infections in animals. Pets live in close proximity to humans, increasing the risk of transmission of resistant microorganisms between animals and humans (Biyashev et al., 2019). Moreover, the widespread use of antimicrobials in veterinary medicine contributes to the selection and dissemination of resistant strains, as these animals often receive the same antibiotics used in human medicine (Yespembetov et al., 2019). Examining resistance in opportunistic microorganisms like Enterobacteriaceae allows one to estimate the risks of resistance spread from pets’ commensal microflora. In general, this study addresses an underexplored aspect of the global antibiotic resistance problem associated with small domestic animals. This topic has been the subject of research by many scientists. For example, Vasaikar et al. (2017) investigated the resistance genes responsible for extended-spectrum beta-lactamase-producing (ESBL) and carbapenemase-producing (CRE) Klebsiella species isolated in Mt. The study also aimed to understand their epidemiology (Vasaikar et al., 2017). Umeda et al. (2019) aimed to investigate the prevalence and characteristics of cephalosporin-resistant Enterobacteriaceae in sheltered dogs and cats. They collected fecal samples or rectal swabs from 151 dogs and 182 cats, screening for antimicrobial-resistant bacteria and conducting phenotypic and genotypic analyses. Genotyping revealed high genetic diversity among the isolates, with no significant correlation found between the history of human association and the presence of the bacteria in either dogs or cats. The study concluded that companion animals might play a role in the circulation of antimicrobial-resistant bacteria, with a variety of individual attributes and histories observed among the animals in Osaka, Japan. Karkaba et al. (2019) in a study conducted in Auckland, New Zealand, fecal swabs from 586 pets (225 cats and 361 dogs) were analyzed to detect the presence of Escherichia coli extended-spectrum beta-lactamase-producing bacteria (ESBL-E). Surveys were also conducted among pet owners. The aim of the study was to determine the prevalence of ESBL-E in pets and identify factors associated with it. The overall prevalence of ESBL-E was 6.4%. The identified beta-lactamase genes included blaCTX-M-14 and blaCMY-2. Many isolates had genetic types associated with infections in humans and animals. Variables found to be independently associated with an increased risk of carrying ESBL-E included the treatment of animals with antimicrobials, family members working in veterinary clinics, and family members traveling abroad (Tusupbekova et al., 2022). The study concluded that pets can be colonized by ESBL-E, which are genotypically similar to bacteria that infect humans and animals, and that veterinary clinics may be hotbeds of antimicrobial resistance. The results showed that Klebsiella pneumoniae were the majority species isolated, followed by K. oxytoca. The prevalence of ESBL production in all Klebsiella species was significant. There was a notable increase in the rate of ESBL genes over the five-year study period. The researchers found high antibiotic resistance in decreasing order to various antibiotics. Carbapenems, amikacin, tigecycline, cefoxitin, levofloxacin, pip/tazo, ciprofloxacin, and fosfomycin were suggested for the treatment of drug-resistant Klebsiella. Methicillin-resistant Staphylococcus aureus, methicillin-resistant Staphylococcus pseudintermedius, vancomycin-resistant enterococci, ESBL or CRE Enterobacteriaceae and gram-negative bacteria are the main microbiological hazards directly or indirectly affecting human health, and their sources are pets (Pomba et al., 2017). The transmission of such microorganisms from dogs poses a potential public health risk, as pets can serve as reservoirs of resistant bacteria to which humans are exposed (Radzykhovskyi et al., 2022). The potential for transmission of E. coli between humans and pets is illustrated, for example, by the isolation of an identical clone of E. coli from a dog and from the feces of family members where it lives. This study is extremely relevant as it examines the prevalence of antibiotic-resistant strains of opportunistic Enterobacteriaceae among domestic animals—cats and dogs. The study of antibiotic resistance in animals is of great importance for assessing the risks of spreading antibiotic-resistant bacteria and resistance genes from animals to humans. The results of the study allow to assess the extent of this problem in a particular region and develop effective strategies to control the situation. The data obtained is a valuable contribution to the study of the global problem of antibiotic resistance. Contact with pets increases the risk of E. coli colonization with ESBL by a factor of 7. Currently, there is little information available about antibiotic resistance in cats and dogs, but several studies indicate a situation identical to the clinical situation in humans. Enterobacteriaceae cause various infections in cats and dogs, with urinary tract infections (UTIs) occurring in a large majority of cases. Enterobacteriaceae, such as K. pneumoniae and Proteus mirabilis, are regularly isolated from pets, but E. coli is the most common pathogen (Lord et al., 2021). Antimicrobials, particularly β-lactams, are frequently required to treat these infections successfully, but antimicrobial resistance is increasing. Resistance to β-lactams is mainly due to the production of β-lactamases, a variant of which can hydrolyze and inactivate extended-spectrum cephalosporins. It is particularly important as cephalosporins are important antimicrobials for human and animal medicine (OIE List of Antimicrobials of Veterinary Importance, 2007; Critically Important Antimicrobials for Human Medicine, 2019). Better knowledge in this area would facilitate using antimicrobials, especially since therapy is usually initiated before the results of sensitivity testing and some treatment recommendations suggest that if resistance to a particular antimicrobial exceeds 10%, it should not be used as the first choice for treatment. In addition, UTIs are frequently caused by Enterobacteriaceae from the host’s fecal flora, and there is evidence that genes can be passed from commensal to pathogenic Enterobacteriaceae. The transfer of antibiotic-resistant strains from animals to humans is still being explored around the world. Currently, there is no data on the prevalence of both resistant strains and resistance genes among domestic cats and dogs in Kazakhstan. Exploring the determinants of antimicrobial resistance in small domestic animals (cats and dogs) will allow an assessment of the current pattern of resistance prevalence at the genetic level (Antimicrobial resistance, 2021). There is a gap in the data on the prevalence of resistant strains in the microflora of domestic animals in certain regions and comprehensive studies of phenotypic and genotypic mechanisms of resistance in these microorganisms. The specifics of resistance transmission between animals and humans are also not well understood. This paper fills these gaps by providing information on the prevalence and mechanisms of resistance of Enterobacteriaceae in northern Kazakhstan and demonstrating the link to antibiotic use in veterinary medicine, allowing for a better assessment of the risks of transmission to humans. The study of antibiotic resistance among commensal Enterobacteriaceae in domestic animals is extremely important for science, as it allows for a better understanding of the epidemiology and mechanisms of antibiotic resistance. Commensal bacteria from animals can serve as a reservoir of resistance genes and a source of infection for humans. Studying the characteristics of resistance in different microorganisms helps to understand the ways of transmission and the formation of resistant forms. Data on the prevalence of resistant strains in animal populations are important for monitoring the situation and developing control strategies. Such research contributes to the study of the global threat of antibiotic resistance and is of great practical importance for public health. The purpose of these studies was to isolate and explore the antibiotic resistance of opportunistic Enterobacteriaceae (E. coli, Klebsiella, Proteus, Citrobacter, Enterobacter) from cats and dogs, and to isolate resistance genes in the microorganisms. Materials and MethodsThe work was performed in the departments of microbiological and molecular genetic analysis of the Research Institute of Applied Biotechnology of A. Baitursynov Kostanay Regional University. The study analyzed biological material (n=808), from clinically healthy, and with various abnormalities, isolated in veterinary clinics in Kostanay. The authors chose a comprehensive approach that includes standard microbiological methods for identifying resistance genes. This made it possible to obtain a representative sample of microorganisms from animals, determine their species, investigate antibiotic susceptibility profiles, and identify genetic determinants of resistance. The chosen methodology is optimal for a comprehensive study of the problem of antibiotic resistance in animal microflora from both phenotypic and genotypic perspectives. Samples were extracted by dry, sterile swabs into clean, sterile, disposable tubes, observing all aseptic rules. Microbiological tests were performed according to Bergey’s bacterial identifier and the approved guidelines for the microbiological diagnosis of diseases caused by Enterobacteriaceae. Upon receipt of the material in the laboratory, initial cultures were made on meat-peptone broth and then transferred to plates with CHROMagar™ Orientation and CHROMagar™ E. coli universal chromogenic, differential diagnostic media. After culturing, if colonies characteristic of E. coli, Klebsiella, Citrobacter, Enterobacter, and Proteus growth were identified, smears were prepared and Gram stained, and then their biochemical properties were explored (Biyashev et al., 2016). Antimicrobial sensitivity tests on isolated bacterial cultures were performed by applying standard antibiotic disks to a freshly sown lawn and recording the results on the presence of microbial growth retardation around the disks. Each of the isolated microbial cultures was tested for susceptibility to levomycetin (30 μg), cefoxitin (30 μg), ampicillin (10 μg), gentamicin (10 μg), ciprofloxacin (5 μg), norfloxacin (10 μg), tetracycline (30 μg), ofloxacin (5 μg), trimethoprim (1.25 μg), interpretation was done according to the current guidelines of the European Committee on Antimicrobial Susceptibility Testing (2022); resistance to cefoperazone (75 μg), streptomycin (10 μg), doxycycline (30 μg), cefpodoxime (10 μg), kanamycin (30 μg), nalidixic acid (30 μg), furazolidone (300 μg), gemifloxacin (5 μg), furadonine (300 μg)—as recommended by the Clinical and Laboratory Standards Institute (2019); resistance to enrofloxacin (5 μg) and amoxicillin (25 μg) according to the MUK 4.2.1890-04 “Determination of the sensitivity of microorganisms to antibacterial drugs” (2004) (FSR, 2009). The presence and identification of genes encoding resistance of opportunistic pathogens to antimicrobial agents were performed by polymerase chain reaction with detection of results in agarose gel (Tyliszczak et al., 2018). Ethical approvalAll sampling was conducted according to the European Convention for the Protection of Pets Animals (1987) and the Universal Declaration on Animal Welfare (2007). ResultsDuring the clinical examination and history, 452 animals with obvious symptoms of gastrointestinal diseases (gastrointestinal congestion, diarrhea, bloating, vomiting) were identified out of 808 examined animals. Biomaterial (anal, oral, and vaginal wipes, urine and feces) was collected from each animal presenting symptoms of gastrointestinal damage. Of 452 samples of biological material, 210 strains of opportunistic pathogens were analyzed for biochemical properties. Among them: 149 strains of E. coli, 28 strains of Klebsiella, 10 strains of Citrobacter, 11 strains of Enterobacter and 12 strains of Proteus (Table 1). Each of the microbial isolates isolated corresponded to its family and genus in terms of tinctorial, cultural, and morphological properties. Escherichia coli grew on combined Kliegler’s medium with gas emission and with complete or partial staining of the column and bevel from red to yellow. In each of the isolated E. coli specimens, fermentation of mannitol and lactose and no growth on Christensen’s medium with urea and Simons’ medium were observed. The test with methylated red can be called positive, while the Foges-Proskauer test—negative. The growth of Klebsiella microorganisms on Kliegler’s medium is characterized by a complete discoloration of the medium, without the development of hydrogen sulfide. The oxidation of mannitol was similarly complete with lactose. On Simmons’ medium, it can be observed both a complete color change in the medium and a partial color change. Reaction with methylene red was positive, indole and Foges-Proskauer reactions were absent rather than positive Enterobacteriaceae of the genus Citrobacter grew with complete or partial yellow staining of the medium and the production of gas on Klieger’s medium. Positive reactions are characterized by growth on Simons citrate, fermentation of lactose and mannitol, and a test with methylated red. Growth in urea by Christensen and the development of indole can rather be classified as negative. There was no reaction from Vorges-Proskauer. Enterobacter bacteria distinguished themselves by their characteristic growth, yellow staining, and gas emission. The fermentation of glucose and manatee was complete, and growth on Simons’ medium was characterized by complete staining of the medium, and on Christens’ medium—more positive than negative. The reaction with Foges-Proskauer was positive and the test with methylated red—negative. The biochemical properties of Proteus indicate their inability to ferment lactose, respectively the growth on the Klieger medium occurred with an initial yellow discoloration of the slant and a subsequent change of color to red. Reactions with Simons’ medium, indole development, and growth in Cristens’ urine can be regarded as rather positive. Fermentation of glucose and manatee was not observed. Forges-Proskauer test—no reaction, test with methylated red—positive. Each of the microbial isolates can be attributed to the corresponding biochemical properties typical of bacteria of the E. coli and the genera Klebsiella, Citrobacter, Enterobacter, and Proteus. The next step was to record the results of the distribution of symptomatic and quantitative manifestations of the different genera of Enterobacteriaceae (Table 2). Vomiting was reported in 83% of cats and dogs, of which 144 cases were due to viral diseases (29.9% of all symptomatic animals). Intestinal and gastric inflammation were identified in 91 animals (43.3%). In addition, the results presented indicate a preponderance of infectious diseases (67.4%) in dogs, and non-infectious diseases (86.7%) in cats, both in diseases not related to the influence of Enterobacteriaceae and in cases where they are present. Out of 210 isolates of opportunistic pathogens, five major genera were explored. Among them: 149 strains of E. coli (70.0%), 28 strains of Klebsiella (13.3%), 10 strains of Citrobacter (4.7%), 11 strains of Enterobacter (5.3%), and 12 strains of Proteus (5.7%). The third stage involved establishing the antimicrobial vulnerability of the previously named strains and identifying their resistance genes. Of the 149 strains of E. coli strains were resistant to tetracycline 81% (120 strains), doxycycline 55% (82 strains), ofloxacin 47% (70 strains), cefpodoxime 40% (59 strains), ampicillin and amoxicillin 37% (55 strains to each antibiotic), norfloxacin 30% (45 strains), ciprofloxacin 25% (37 strains), enrofloxacin 22% (32 strains), trimethoprim and nalidixic acid 19% (28 strains), cefoxitin 17% (25 strains), levomycetine 16% (24 strains), cefoperazone 15% (23 strains), gemifloxacin 11% (16 strains), kanamycin and gentamicin 7% (11 strains), streptomycin 2% (three strains), furazolidone and furadonine 1% (one strain). No resistant strains were identified to meropenem (Fig. 1). Table 1. Biochemical properties of selected strains of opportunistic Enterobacteriaceae.
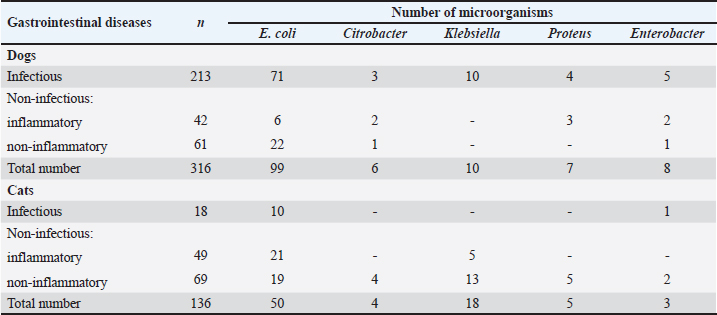
Table 2. Bacterial culture isolates obtained from animals with gastrointestinal diseases.
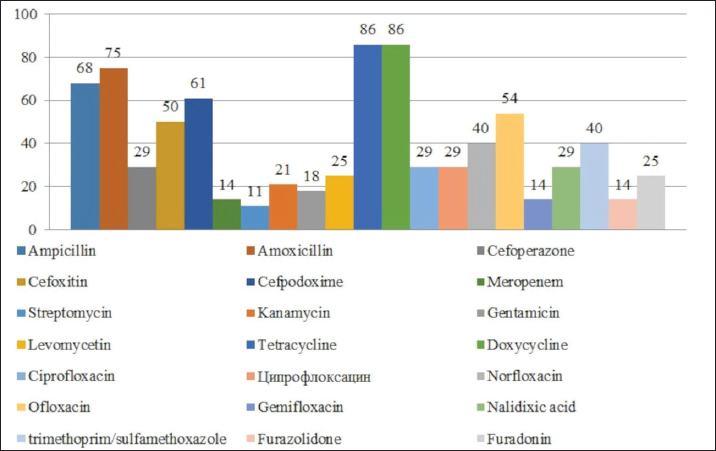
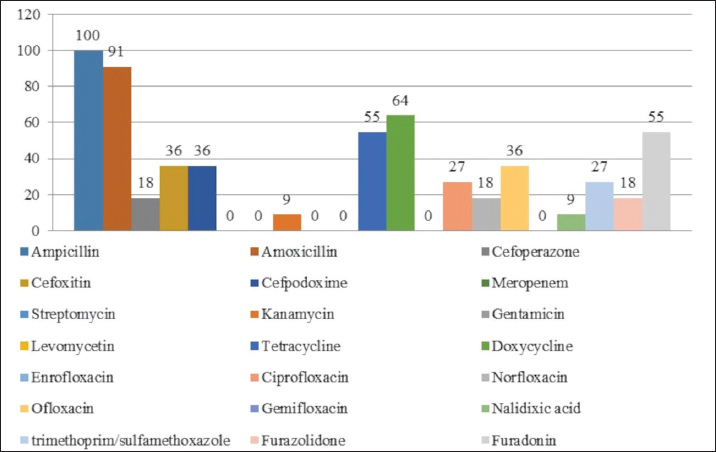
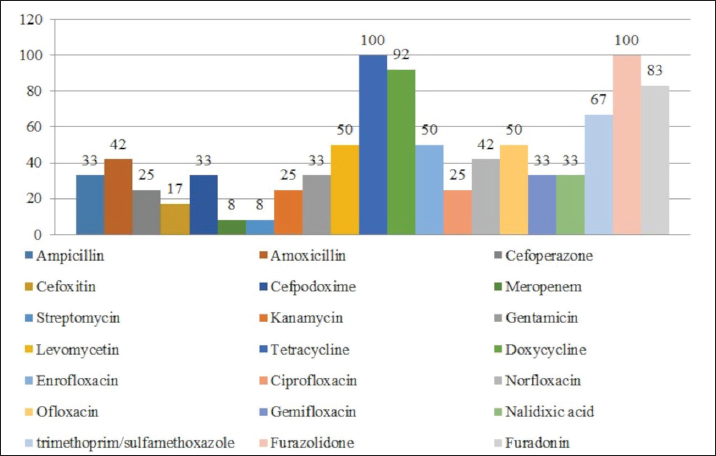
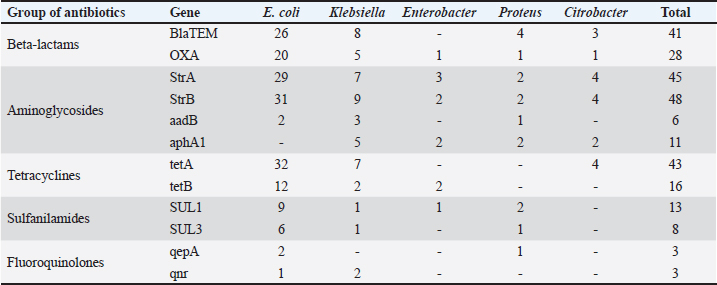
Fig. 1. Antibiotic resistance of E. coli isolates (%). Of the 28 Klebsiella strains examined, 86% (24 strains) of the microorganisms were resistant to tetracycline and doxycycline, 75% (21 strains) to amoxicillin, 68% (19 strains) to ampicillin, to cefpodoxime 61% (17 strains), to ofloxacin 54% (15 strains), to cefoxitin 50% (14 strains), to norfloxacin and trimethoprim 40% each (11 strains), to nalidixic acid cefoperazone enrofloxacin and ciprofloxacin 29% (eight strains each), to levomycetin and furadonine 25% (seven strains each) of microorganisms, to kanamycin 21% (six strains), to gentamicin 18% (five strains), to meropenem, gemifloxacin and furazolidone, 14% each (four strains), and the lowest number of organisms were resistant to streptomycin, 11% (three strains) (Fig. 2). Antibiotic resistance testing of 10 Citrobacter isolates resulted in all microorganisms being 90% resistant to tetracycline, amoxicillin, doxycycline, and nalidixic acid was resistant to 70% of the microorganisms, ampicillin, enrofloxacin Ofloxacin trimethoprim 60% of strains, norfloxacin, furazolidone 50%, cefoperazone, cefoxitin, ciprofloxacin, gemifloxacin, and furodonin 40% each, cefpodoxime and gentamicin 30%, meropenem, kanamycin and levomycetine 20% of organisms. No streptomycin-resistant strains were identified (Fig. 3). Of the 11 Enterobacter strains isolated, the microorganisms demonstrated 100% resistance to ampicillin, 91% to amoxicillin, 64% to doxycycline, 55% to tetracycline, and furadonine, 36% for cefoxitin, cefpodoxime, ofloxacin, 27% for ciprofloxacin and trimethoprim, 18% for cefoperazone, norfloxacin, and furazolidone, and 9% for kanamycin and nalidixic acid. Meropenem, streptomycin, gentamicin, levomycetin, enrofloxacin, and gemifloxacin were not identified as resistant to the isolates of Enterobacteriaceae (Fig. 4). A study of 12 microorganisms of the genus Proteus for antibiotic resistance demonstrated that 100% of the isolates were resistant to tetracycline and furazolidone; 92% of isolates were resistant to doxycycline, 83% of microorganisms were resistant to furadonine, 67% of isolates were resistant to trimethoprim, 50% of isolates were resistant to levomycetin, enrofloxacin, and ofloxacin, 42% of microorganisms were resistant to amoxicillin and norfloxacin, to ampicillin, cefpodoxime, gentamicin, gemifloxacin, and nalidixic acid 33%, to cefoperazone, kanamycin, enrofloxacin 25%, 17% were resistant to cefoxitin. The strains demonstrated the least resistance (8%) to meropenem and streptomycin (Fig. 5). The final stage of the work was to perform research to determine the resistance genes of the organisms. The results of these studies, using the polymerase chain reaction technique, from 149 samples of E. coli DNA (deoxyribonucleic acid), 46 samples were obtained that have genes encoding resistance to beta-lactase antibiotics (BlaTEM—26, OXA—20), aminoglycosides—62 (StrA—29, StrB—31, aadB—2), tetracyclines—44 (tetA—32 and tetB—12), sulfonamides—15 (SUL1—9, SUL3—6), fluoroquinolones—3 (qepA—2, qnr—1).
Fig. 2. Antibiotic resistance of Klebsiella isolates (%).
Fig. 3. Antibiotic resistance of Citrobacter isolates (%).
Fig. 4. Antibiotic resistance of Enterobacter isolates (%).
Fig. 5. Antibiotic resistance of Proteus isolates (%). Table 3. Resistance genes of microorganisms.
In 28 Klebsiella samples, genes encoding resistance to beta-lactams—13 (BlaTEM—8, OXA—5), aminoglycosides—24 (StrA—7, StrB—9, aadB—3, aphA1—5), tetracyclines—9 (tetA—7 and tetB—2), sulfonamides—2 (SUL1—1, SUL3—1), fluoroquinolones—2 (gene qnr) (Table 3). In 11 Enterobacter DNA samples, the OXA gene encoding resistance to beta-lactamases was identified in 1, the aminoglycosides in 7 (StrA—3, StrB—2, aphA1—2), the tetB gene in 2, and the SUL1 gene to sulfonamides in 1. No fluoroquinolone-resistant ones have been identified. In 12 samples of Proteus DNA, genes coding for resistance to antibiotic groups were selected: beta-lactams—5 (BlaTEM—4, OXA—1), aminoglycosides—7 (StrA—2, StrB—2, aadB—1 and aphA1—2), sulphonamides—3 (SUL1—2, SUL3—1) and fluoroquinolones—1 (qepA). No tetracycline-resistant drugs have been isolated. A study of 10 samples of Citrobacter DNA revealed 4 genes encoding beta-lactam antibiotics (BlaTEM—3, OXA—1), 10 genes encoding aminoglycosides (StrA—4, StrB—4, aphA1—2) and 4 tetA genes encoding tetracyclines. No sulfanilamide- and fluoroquinolone-resistant ones have been identified. DiscussionAs a result of this research, 808 samples of biological material (urine, feces, wipes from the rectum, mouth, nose, and vagina) were collected from dogs and cats in Kostanay veterinary clinics. Among these, 210 (26%) micro-organisms were isolated and identified. These findings correlate with several studies in the Netherlands, where the prevalence of Enterobacteriaceae colonization in dogs and cats (including healthy and sick animals) ranged from 3.1% to 55% (van den Bunt et al., 2020). The vast majority of the strains sampled belonged to E. coli—149 (70.9%), Klebsiella—28 (13.3%), Enterobacter—11 (5.2%), Proteus—12 (5.7%), and 10 Citrobacter isolates (4.8%). In each of the isolates identified, antibiotic resistance/susceptibility was determined using the disc-diffusion method. The results with high resistance in E. coli strains to tetracycline are consistent with several studies from the United Kingdom and Portugal, which had a high percentage of resistant strains (40%) (Wedley et al., 2017). Other recent clinical trials in Belgium, however, report 12% resistance to ampicillin and 17% resistance to tetracycline, but very low or undetectable levels of resistance were to enrofloxacin and gentamicin, which is consistent with the research data. Studies in Australia have reported increased resistance of E. coli isolates to ampicillin, amoxicillin, cefpodoxime, and no resistance to carbapenems, which is consistent with the research results (Saputra et al., 2017). A study of Klebsiella strains identified that the greatest number of isolates demonstrated resistance to tetracycline, doxycycline, amoxicillin, and ampicillin, while those resistant to meropenem were not detected. Studies conducted in 2017–2019 in China, Thailand, and Portugal confirm the high prevalence of resistance to these antibacterials (Marques et al., 2019; Phoo et al., 2020; Zhang et al., 2021). Notably, these studies demonstrate the link between antibiotic resistance in Klebsiella strains isolated from domestic animals and their hosts, thus confirming the potential for the transmission of resistant strains from animals to humans. There are few studies on the prevalence of antibiotic resistance in Citrobacter isolates from small domestic animals (cats and dogs) worldwide. The results demonstrate that the maximum number of micro-organisms were resistant to tetracycline, amoxicillin, doxycycline, ampicillin, enrofloxacin, trimethoprim/sulfamethoxazole, norfloxacin, cefoperazone, cefoxitin, and ciprofloxacin. Sensitivity was demonstrated for streptomycin. Studies by Harada et al. (2019) demonstrated that Citrobacter strains isolated from cats and dogs presented resistance to ciprofloxacin, trimethoprim/sulfamethoxazole, while they were sensitive to meropenem. In a study by Ramona et al. (2020), microorganisms of the genus Citrobacter were sensitive to meropenem and resistant to ampicillin and amoxicillin. In Nigeria, the data were partly similar, with Citrobacter indicating resistance to amoxicillin, streptomycin, gentamicin, and ciprofloxacin (Babatunde et al., 2017; Mocherniuk et al., 2022). Enterobacter strains have been identified as being most resistant to ampicillin, amoxicillin, tetracycline, cefoxitin, and trimethoprim while being sensitive to the action of gentamicin, enrofloxacin, meropenem, streptomycin. These findings are partly consistent with those obtained in Japan and Italy, the difference is the list of antibacterials to which sensitivity in the microorganisms was explored (Mezzatesta et al., 2012; Harada et al., 2014, 2017). In addition, a study by Li et al. (2021) mentions high resistance in Enterobacter isolates to β-lactam antibacterials in both dogs and cats. The micro-organisms of the genus Proteus are established to have reduced resistance to tetracycline, doxycycline, ampicillin, amoxicillin, levomycetine, enrofloxacin, trimethoprim/sulfamethoxazole, and nitrofurans. Similar results have been reported in Japan and Denmark, where there were high numbers of strains resistant to both enrofloxacin and ampicillin, but strains were sensitive to gentamicin (Pedersen et al., 2007). In a Canadian study, ampicillin-resistant strains were identified while enrofloxacin-resistant strains demonstrated sensitivity; in a German study, on the other hand, the isolates were resistant to enrofloxacin (Grobbel et al., 2007). In Spain, tetracycline resistance and reduced resistance to cephalosporins were identified in 80% of Proteus strains, and sensitivity to gentamicin was identified (Awosile et al., 2018). In general, there are differences in the prevalence of resistance among opportunistic Enterobacteriaceae isolates in different countries. It could be explained by differences in the strains and resistance genes circulating in different territories. As a result of the studies, it was established that the largest number of Enterobacteriaceae was sensitive to the action of meropenem, belonging to the group of beta-lactam antibiotics, among E. coli and Enterobacter isolates there were no resistant strains, among Proteus strains one isolate was resistant, among Citrobacter—2. Among the microorganisms isolated 10 isolates (1 Escherichia, 4 Klebsiella, 3 Proteae, 2 Citrobacter) presented resistance to all examined groups of antibiotics, whereas 20 strains of E. coli were completely sensitive to the action of antibacterial agents and 13 isolates presented intermediate sensitivity. Among other microorganisms, both fully sensitive and intermediate strains to all antibiotics have not been identified. Most of the isolated microbial strains (88%) were resistant, except for 23 E. coli, 1 Klebsiella and 1 Citrobacter. Notably, in general, the highest number of all isolated microorganisms demonstrated resistance to the group of tetracyclines and beta-lactams, which, according to the state register of veterinary drugs and fodder additives of the Committee of Veterinary Control and Supervision of the Ministry of Agriculture of the Republic of Kazakhstan, are the drugs of choice in the treatment of infectious animal diseases. In a study conducted in Spain, beta-lactams were the antimicrobial drug most commonly prescribed in dogs (Gómez-Poveda and Moreno, 2018; Irgashev et al., 2023). Ampicillin is a good treatment for sporadic bacterial cystitis caused by E. coli in cats and dogs. Using this antimicrobial to treat infections caused by E. coli should be used with caution due to the rapid development of resistance caused by beta-lactamase production (Boehmer et al., 2018). Antibacterials of the tetracycline range are used in the treatment of wounds, skin, and digestive tract diseases in cats and dogs. Intensive use of antibiotics in these classes results in the spread of resistant microorganisms and reduces their effectiveness (Paliy et al., 2023). The results of these studies, using the polymerase chain reaction technique, are from 149 samples of E. coli DNA. 46 samples of E.coli, were obtained that have genes encoding resistance to beta-lactase antibiotics and 4 tetA genes encoding tetracyclines. The experiment demonstrates that 85 DNA samples characterized by genotypic antibiotic resistance are found to be associated with phenotypic profiles. Therewith, resistance genes to aminoglycoside antimicrobials (StrA, StrB, aadB, aphA1) were detected in 22 samples, whereas no phenotypic resistance to these drugs was identified in these microorganisms. Probably, these could be the so-called “silenced” genes, which are found in several studies but have been understudied thus far (Li et al., 2014; Wang et al., 2017). Phenotypic resistance to the fluoroquinolone group (enrofloxacin, ciprofloxacin, norfloxacin, ofloxacin, gemifloxacin) was present in 113 strains of microorganisms, whereas genotypic resistance, i.e. the presence of qepA, qnrA genes was absent. Probably, this is explained by differences in the mode of action of resistance, namely overactivity of the efflux pump and reduction of membrane permeability, a similar situation observed in studies by other scientists (Galal et al., 2019; Bardasheva et al., 2021). It should be noted that the study has certain limitations, such as: – The sample of animals was limited to the northern region of Kazakhstan. To obtain a more complete picture of the country, studies in other regions are needed. – The study covered only five genera of enterobacteria. For a comprehensive analysis, it would be advisable to expand the range of microorganisms studied. – Antibiotic susceptibility was determined only by the disc diffusion method. It is recommended to add minimal inhibitory concentrations. – Resistance studies were limited to genes of the main antibiotic groups. It is advisable to extend the PCR analysis to other known genes. – There is a lack of data on the molecular typing of strains to identify links between animal and human isolates. These limitations can be overcome in future studies by expanding the geography and sample size, increasing the range of microorganisms analyzed, using additional methods of antibiotic susceptibility testing, screening for other resistance genes and molecular typing of isolates. ConclusionThe study examined 808 samples of biomaterial from small domestic animals, resulting in the selection of microorganisms and analysis of their resistance to antimicrobial agents. It is established that 452 animals had gastrointestinal disease, and using antibiotics is widely exploited in veterinary practice. A high prevalence of Enterobacteriaceae strains has been detected in cats and dogs in the northern region of the Republic of Kazakhstan. The symptomatic and quantitative findings of the different Enterobacteriaceae genera were as follows: E. coli 149 (70.9%), Klebsiella 28 (13.3%), Enterobacter 11 (5.2%), Proteus 12 (5.7%) and 10 Citrobacter isolates (4.7%). The majority of the samples (88%) were characterized by the presence of multidrug resistance. Among isolated microorganisms ten isolates demonstrated resistance to all studied groups of antibiotics, moreover, 20 E. coli strains were fully sensitive to the action of antibacterial agents and 13 isolates demonstrated intermediate sensitivity. The study of antibiotic resistance of isolated strains demonstrated high resistance to beta-lactams and tetracyclines. Resistance to these groups of antimicrobials is conditioned upon the presence of the resistance genes BlaTEM, OXA, tetA, and tetB in the microorganisms. The Enterobacteriaceae examined were least resistant to meropenem. The result of the experiment presented that 85 DNA samples characterized by genotypic antibiotic resistance were found to be associated with phenotypic profiles. Therewith, resistance genes to aminoglycoside antimicrobials were detected in 22 samples, whereas no phenotypic resistance to these drugs was detected in these microorganisms. Phenotypic resistance to the fluoroquinolone group was present in 113 microbial strains, whereas genotypic resistance, i.e., the presence of the qepA, qnrA genes, was absent. The authors established that uncontrolled and frequent use of beta-lactam and tetracycline antibacterials in cats and dogs’ results in the spread of genotypic resistance among Enterobacteriaceae microorganisms. Based on the above, the results allowed for estimating the current prevalence of antibiotic-resistant forms of opportunistic pathogens detected from cats and dogs in the northern region of Kazakhstan and determining their phenotypic and genotypic profile, which will better control treatment standards of diseases caused by opportunistic Enterobacteriaceae. The rational use of antibiotics in veterinary medicine is an extremely important strategy to preserve the effectiveness of these medicines and prevent the spread of antibiotic-resistant microorganisms in the environment, including pets and humans. The prospects for future research are to explore the features and modes of transmission of these opportunistic Enterobacteriaceae between domestic animals and from animals to humans and to consider the reasons for preventing the development of these microorganisms. AcknowledgmentsThe authors are grateful to the Ministry of Science and Higher Education of the Republic of Kazakhstan, for providing the fund for this study. Author contributionsThis project was conceived and designed by YA and RR, with data acquisition by YA, AY, AM, PS and RR. Analysis and interpretation of the data were performed by YA, RR and AY. The article was drafted by YA and RR, with the revision for intellectual content performed by YA, AY, AM, PS and RR. All authors gave final approval for article submission. Conflict of interestThe authors declare that there is no conflict of interest. FundingThe research was funded by the Ministry of Science and Higher Education of the Republic of Kazakhstan, project “Prevalence of Determinants of Antibacterial Drug Resistance” (Grant No. AP09058122). Data availabilityThe data used to support the primary findings of this study are included in the article. Complete datasets generated and analyzed during the study are available from the corresponding author upon reasonable request. ReferencesAntimicrobial Resistance. (2021). Available via https://www.who.int/news-room/fact-sheets/detail/antimicrobial-resistance (Accessed 10 July 2023). Awosile, B.B., Mcclure, J.T., Saab, M.E. and Heider, L.C. 2018. Antimicrobial resistance in bacteria isolated from cats and dogs from the Atlantic Provinces, Canada from 1994–2013. Canadian Vet. J. 59, 885–893. Babatunde, D.O., Adesola, A.E. and Oladimeji, O.D. 2017. Antibiotic resistance profiling and microbiota of the upper respiratory tract of apparently healthy dogs in Ibadan, South West Nigeria. African J. Infect. Dis. 17, 11–18. Bardasheva, A.V., Fomenko, N.V., Kalymbetova, T.V., Babkin, I.V., Chretien, S.O., Zhirakovskaya, E.V., Tikunova, N.V. and Morozova, V.V. 2021. Genetic characteristics of the growth of Klebsiella isolates circulating in Novosibirsk. Vavilovskii Zhurnal Genet. Selektsii. 25(2), 234–245. Biyashev, K.B., Biyashev, B.K. and Saribayeva, D.A. 2016. Persistence of the Escherichia coli 64G-probiotic strain in the intestine of calves. Biol. Med. 8(2), 2–3. Biyashev, K.B., Kirkimbaeva, Z.S., Biyashev, B.K., Makbuz, A.Z. and Bulegenova, M.D. 2019. Determination of the level of resistance of probiotic strain Escherichia coli 64 g to hydrochloric acid, bile and antimicrobial agents. Ecol. Environ. Conserv. 25(4), 1930–1933. Boehmer, T., Vogler, A.J., Thomas, A., Sauer, S., Hergenroether, M. and Straubinger, R.K. 2018. Phenotypic characterization and whole genome analysis of extended-spectrum betalactamase-producing bacteria isolated from dogs in Germany. PLoS One 13, e0206252. Clinical and Laboratory Standards Institute. 2019. M100-ED29:2019. Performance Standards for Antimicrobial Susceptibility Testing. 29th ed. Wayne, PA: Clinical and Laboratory Standards Institute. Critically Important Antimicrobials for Human Medicine. 2019. [Online] Available via https://www.who.int/publications/i/item/9789241515528 (Accessed 24 August 2023). European Committee on Antimicrobial Susceptibility Testing. 2022. [Online] Available via https://www.eucast.org/ (Accessed 23 August 2023). European Convention for the Protection of Pets Animals. 1987. [Online] Available via https://rm.coe.int/168007a67d (Accessed 24 August 2023). FSR. 2009. A set of discs for determining sensitivity to antimicrobial drugs-1 (ND-PMP-1) according to TU 9398-006-01967164-2009B. 2009. [Online] Available via https://nevacert.ru/reestry/med-reestr/fsr-2009-06290-o28446 (Accessed 25 August 2023). Galal, L., Abdel Aziz, N.A. and Hassan, W.M. 2019. Defining the relationship between phenotypic and genotypic resistance profiles of multidrugresistant enterobacterial clinical isolates. Adv. Exp. Med. Biol. 1214, 9–21. Gómez-Poveda, B. and Moreno, M.A. 2018. Antimicrobial prescriptions for dogs in the capital of Spain. Front. Vet. Sci. 5, 309. Grobbel, M., Lübke-Becker, A., Alesík, E., Schwarz, S., Wallmann, J., Werckenthin, C. and Wieler, L.H. 2007. Antimicrobial susceptiblity of Klebsiella spp. and Proteus spp. from various organ systems of horses, dogs and cats as determined in the BfT-GermVet monitoring program 2004–2006. Berl. Münchener tierärztliche Wochenschr. 120, 402–411. Harada, K., Ayaka, N., Takae, S., Yujiro, M., Ken, K., Tadashi, M. and Yasushi, K. 2014. Phenotypic and molecular characterisation of antimicrobial resistance in Proteus mirabilis isolates from dogs. J. Med. Microbiol. 63, 1561–1567. Harada, K., Shimizu, T., Mukai, Y., Kuwajima, K., Sato, T. and Kajino, A. 2017. Phenotypic and molecular characterization of antimicrobial resistance in Enterobacter spp. isolates from companion animals in Japan. PLoS One 12(3), e0174178. Harada, K., Takae, S., Hiroichi, O., Yui, K., Tadashi, M. and Yuzo, T. 2019. Characterization of antimicrobial resistance in Serratia spp. and Citrobacter spp. isolates from companion animals in Japan: nosocomial dissemination of extended-spectrum cephalosporin-resistant citrobacter freundii. Microorg. 7(3), 64–71. Irgashev, A., Nurgaziev, R., Nurmanov, Ch., Asanova, E. and Ishenbaeva, S. 2023. Respiratory form of infectious rhinotracheitis: analysis of immunomorphological reactions. Sci. Horiz. 26(10), 32–43. Karkaba, A., Hill, K., Benschop, J., Pleydell, E. and Grinberg, A. 2019. Carriage and population genetics of extended spectrum β-lactamase-producing Escherichia coli in cats and dogs in New Zealand. Vet. Microbiol. 233, 61–67. Li, X., Yao, Z., Yuan, L., Qi, W., Shuchang, A. and Jichao, C. 2014. Identification of Klebsiella pneumoniae strains harboring inactive extended-spectrum beta-lactamase antibiotic-resistance genes. Chin. Med. J. 127(17), 3051–3057. Li, Y., Fernández, R., Durán, I., Molina-López, R.A. and Darwich, L. 2021. Antimicrobial resistance in bacteria isolated from cats and dogs from the Iberian Peninsula. Front. Microbiol. 11, 621597. Lord, J., Gikonyo, A., Miwa, A. and Odoi, A. 2021. Antimicrobial resistance among Enterobacteriaceae, Staphylococcus aureus, and Pseudomonas spp. isolates from clinical specimens from a hospital in Nairobi, Kenya. Peer J. 9, e11958. Marques, C., Menezes, J., Belas, A., Aboim, C., Cavaco-Silva, P., Trigueiro, G., Gama, L.T. and Pomba, C. 2019. Klebsiella pneumoniae causing urinary tract infections in companion animals and humans: population structure, antimicrobial resistance and virulence genes. J. Antimicrob. Chemother. 74, 594–602. Mezzatesta, M.L., Gona, F. and Stefani, S. 2012. Enterobacter cloacae complex: clinical impact and emerging antibiotic resistance. Future Microbiol. 7, 887–902. Mocherniuk, M., Kukhtyn, M., Horiuk, Yu., Savchuk, L. and Mizyk, V. 2022. Identification of the bioaerosol microbiota in veterinary clinics as the key to preventing nosocomial infection. Sci. Horiz. 25(11), 31–40. MUK 4.2.1890-04 “Determination of the sensitivity of microorganisms to antibacterial drugs”. 2004. [Online] Available via https://files.stroyinf.ru/Index2/1/4293754/4293754463.htm (Accessed 26 August 2023) . OIE List of Antimicrobials of Veterinary Importance. 2007. [Online] Available via https://www.woah.org/app/uploads/2021/03/oie-list-antimicrobials.pdf (Accessed 24 August 2023). Paliy, A., Pavlichenko, O., Rodionova, K., Morozov, M. and Dankevych, N. 2023. Treatment of pets with the active substance dexpanthenol in wound processes. Sci. Horiz. 26(3), 9–23. Pedersen, K., Pedersen, K., Jensen, H., Finster, K., Jensen, V.F. and Heuer, O.E. 2007. Occurence of antimicrobial resistance in bacteria from diagnostic samples from dogs. J. Antimicrob. Chemother. 60, 775–781. Phoo, M., Ngasaman, R., Indoung, S., Naknaen, A., Chukamnerd, A. and Pomwised, R. 2020. Occurrence of ndm-5 and antibiotic resistance genes among Escherichia coli and Klebsiella pneumonia in companion animals in Thailand. Southeast Asian J. Trop. Med. Public Health 51(3), 391–405. Pomba, C., Rantala, M., Greko, C., Baptiste, K., Catry, B., Duijkeren, E., Mateus, A., Moreno, M., Pyörälä, S., Ružauskas, M., Sanders, P. and Teale, C. 2017. Public health risk of antimicrobial resistance transfer from companion animals. J. Antimicrob. Chemother. 72(4), 957–968. Radzykhovskyi, M., Dyshkant, O., Gutyj, B., Sachuk, R. and Palytsia, Y. 2022. Adenovirus infections in dogs: diagnostic features. Ukr. J. Vet. Sci. 13(4), 50–59. Ramona, I., Cerbo, A., Messi, P. and Sabia, C. 2020. antibiotic resistance and virulence traits in vancomycin-resistant enterococci (VRE) and extended-spectrum β-Lactamase/AmpC-producing (ESBL/AmpC) enterobacteriaceae from humans and pets. Antibiotics 9, 152–159. Saputra, S., Jordan, D., Mitchell, T., San Wong, H., Abraham, R.J. and Kidsley, A. 2017. Antimicrobial resistance in clinical Escherichia coli isolated from companion animals in Australia. Vet. Microbiol. 211, 43–50. Tusupbekova, M.M., Sharapatov, Y.A., Pronkin, E.A., Lavrinenko, A.V. and Turgunov, Y.M. 2022. Comparative study of morphological changes in the kidney and ureter of a rabbit with various methods of infection. Clin. Exp. Morphol. 11(1), 62–72. Tyliszczak, B., Drabczyk, A. and Kudłacik–Kramarczyk, S. 2018. Smart, self-repair polymers based on acryloyl-6-aminocaproic acid and modified with magnetic nanoparticles—preparation and characterization. Int. J. Polym. Anal. Charact. 23(3), 226–235. Umeda, K., Hase, A., Matsuo, M., Horimoto, T. and Ogasawara, J. 2019. Prevalence and genetic characterization of cephalosporinresistant Enterobacteriaceae among dogs and cats in an animal shelter. J. Med. Microbiol. 68(3), 339–345. Universal Declaration on Animal Welfare. 2007. [Online] Available via https://web.archive.org/web/20090219033045/http://animalsmatter.org/downloads/UDAW_Text_2005.pdf (Accessed 22 September 2023). van den Bunt, G., Fluit, A.C., Spaninks, M.P., Timmerman, A.J., Geurts, Y., Kant, A., Scharringa, J., Mevius, D., Wagenaar, J.A. and Bonten, M.J.M. 2020. Faecal carriage, risk factors, acquisition and persistence of ESBL-producing Enterobacteriaceae in dogs and cats and co-carriage with humans belonging to the same household. J. Antimicrob. Chemother. 75, 342–350. Vasaikar, S., Obi, L., Morobe, I. and Bisi-Johnson, M. 2017. Molecular characteristics and antibiotic resistance profiles of Klebsiella isolates in Mthatha, Eastern Cape Province, South Africa. Int. J. Microbiol. 17, 8486742. Wang, Y., Zhang, R. and Shen, J. 2017. Comprehensive resistome analysisreveals the prevalence of NDM and MCR-1in Chinese poultry production. Nat. Microbiol. 2, 16260. Wedley, A.L., Dawson, S., Maddox, T.W., Coyne, K.P., Pinchbeck, G.L. and Clegg, P. 2017. Carriage of antimicrobial resistant Escherichia coli in dogs: Prevalence, associated risk factors and molecular characteristics. Vet. Microbiol. 199, 23–30. Yespembetov, B.A., Syrym, N.S., Syzdykov, M.S., Kuznetsov, A.N., Koshemetov, Z.K., Mussayeva, A.K., Basybekov, S.Z., Kanatbayev, S.G., Mankibaev, A.T. and Romashev, C.M. 2019. Impact of geographical factors on the spread of animal brucellosis in the Republic of Kazakhstan. Comp. Immunol. Microbiol. Infect. Dis. 67, 101349. Zhang, Z., Lei, L., Zhang, H., Dai, H., Song, Y., Li, L., Wang, Y. and Xia, Z. 2021. Molecular investigation of Klebsiella pneumoniae from clinical companion animals in Beijing, China, 2017–2019. Pathog. 10, 271. | ||
| How to Cite this Article |
| Pubmed Style Aleshina Y, Yeleussizova A, Mendybayeva A, Shevchenko P, Rychshanova R. Prevalence and antimicrobial resistance of Enterobacteriaceae in the north of Kazakhstan. Open Vet J. 2024; 14(2): 604-616. doi:10.5455/OVJ.2024.v14.i2.1 Web Style Aleshina Y, Yeleussizova A, Mendybayeva A, Shevchenko P, Rychshanova R. Prevalence and antimicrobial resistance of Enterobacteriaceae in the north of Kazakhstan. https://www.openveterinaryjournal.com/?mno=168397 [Access: April 27, 2024]. doi:10.5455/OVJ.2024.v14.i2.1 AMA (American Medical Association) Style Aleshina Y, Yeleussizova A, Mendybayeva A, Shevchenko P, Rychshanova R. Prevalence and antimicrobial resistance of Enterobacteriaceae in the north of Kazakhstan. Open Vet J. 2024; 14(2): 604-616. doi:10.5455/OVJ.2024.v14.i2.1 Vancouver/ICMJE Style Aleshina Y, Yeleussizova A, Mendybayeva A, Shevchenko P, Rychshanova R. Prevalence and antimicrobial resistance of Enterobacteriaceae in the north of Kazakhstan. Open Vet J. (2024), [cited April 27, 2024]; 14(2): 604-616. doi:10.5455/OVJ.2024.v14.i2.1 Harvard Style Aleshina, Y., Yeleussizova, . A., Mendybayeva, . A., Shevchenko, . P. & Rychshanova, . R. (2024) Prevalence and antimicrobial resistance of Enterobacteriaceae in the north of Kazakhstan. Open Vet J, 14 (2), 604-616. doi:10.5455/OVJ.2024.v14.i2.1 Turabian Style Aleshina, Yuliya, Anara Yeleussizova, Anara Mendybayeva, Pavel Shevchenko, and Raushan Rychshanova. 2024. Prevalence and antimicrobial resistance of Enterobacteriaceae in the north of Kazakhstan. Open Veterinary Journal, 14 (2), 604-616. doi:10.5455/OVJ.2024.v14.i2.1 Chicago Style Aleshina, Yuliya, Anara Yeleussizova, Anara Mendybayeva, Pavel Shevchenko, and Raushan Rychshanova. "Prevalence and antimicrobial resistance of Enterobacteriaceae in the north of Kazakhstan." Open Veterinary Journal 14 (2024), 604-616. doi:10.5455/OVJ.2024.v14.i2.1 MLA (The Modern Language Association) Style Aleshina, Yuliya, Anara Yeleussizova, Anara Mendybayeva, Pavel Shevchenko, and Raushan Rychshanova. "Prevalence and antimicrobial resistance of Enterobacteriaceae in the north of Kazakhstan." Open Veterinary Journal 14.2 (2024), 604-616. Print. doi:10.5455/OVJ.2024.v14.i2.1 APA (American Psychological Association) Style Aleshina, Y., Yeleussizova, . A., Mendybayeva, . A., Shevchenko, . P. & Rychshanova, . R. (2024) Prevalence and antimicrobial resistance of Enterobacteriaceae in the north of Kazakhstan. Open Veterinary Journal, 14 (2), 604-616. doi:10.5455/OVJ.2024.v14.i2.1 |