| Research Article | ||
Open Veterinary Journal, (2024), Vol. 14(1): 164-175 Original Research Antibacterial effect of Moringa oleifera on Staphylococcus aureus and Pseudomonas aeruginosa isolated from raw milk and some dairy products with special reference to biofilm gene expressionRowyda Mohamed Yousry Elshazely1*, Ibrahim H. Amer2, Salah F. A. Abd-El Aal2 and Asmaa B. M. B. Tahoun21Dakahlia Veterinary Directorate, Mansoura City, Egypt 2Department of Food Hygiene, Safety and Technology, Faculty of Veterinary Medicine, Zagazig University, Zagazig City, Egypt *Corresponding Author: Rowyda Mohamed Yousry Elshazely. Dakahlia Veterinary Directorate, Mansoura City, Egypt. Email: rowyda.vet2021 [at] gmail.com Submitted: 01/10/2023 Accepted: 15/12/2023 Published: 31/01/2024 © 2024 Open Veterinary Journal
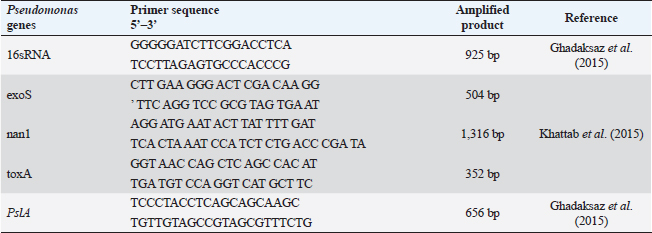
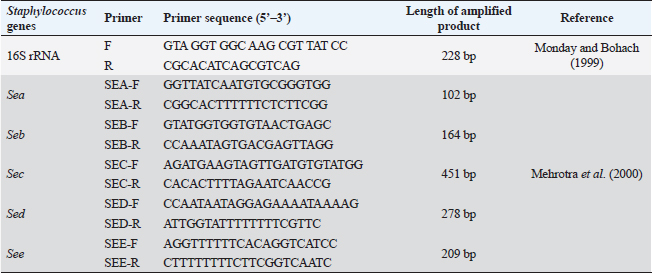
ABSTRACTBackground: Pseudomonas aeruginosa (P. aeruginosa) and Staphylococcus aureus (S. aureus) are well defined as food poisoning pathogens that are highly resistant and need continuous studies. Aim: The purpose of the work was to examine phenotypic and genotypic characteristics of both P. aeruginosa and S. aureus, and treatment trials with medicinal plants. Methods: Samples were examined for isolation of P. aeruginosa and S. aureus on selective media followed by biochemical confirmation, biofilm formation, genes detection, and expression of P. aeruginosa pslA biofilm gene was performed by quantitative real-time polymerase chain reaction after treatment with 0.312 mg/ml Moringa oleifera aqueous extract as a minimum inhibitory concentration. Results: The highest isolation rate of P. aeruginosa was 20% from both raw milk and Kariesh cheese, followed by 16% and 12% from ice cream and processed cheese, respectively, while the highest isolation rate of S. aureus was 36% from raw milk followed by 28% in ice cream and 16% in both Kariesh cheese and processed cheese. 30% of P. aeruginosa isolates were biofilm producers, while only 21% of S. aureus isolates were able to produce biofilm. The P. aeruginosa isolates harbor virulence-associated genes nan1, exoS, toxA, and pslA at 100%, 80%, 40%, and 40%, respectively. Staphylococcus aureus SEs genes were examined in S. aureus strains, where SEA and SEB genes were detected with 60%, but no isolate harbored SEC, SED, or SEE. The significant fold change of P. aeruginosa pslA expression was 0.40332 after treatment with M. oleifera aqueous extract. Conclusion: Pseudomonas aeruginosa and S. aureus harbor dangerous virulence genes that cause food poisoning, but M. oleifera extract could minimize their action. Keywords: Antibacterial, Moringa oleifera, Biofilm gene expression, Staphylococcus aureus, Pseudomonas aeruginosa. IntroductionFood-borne pathogens can easily take their way to milk and dairy products and contaminate them (Painter et al., 2013). Micro-organisms express their virulence by the act of some genes that encode products inside the host body (Muthu et al., 2014). Recently diseases caused by food-borne pathogens have a great attention globally (Bhunia, 2018). For this, regular checking of milk and dairy products’ microbial content and virulence genes became of high value and concern (Momtaz et al., 2012). Staphylococcus aureus has been proven to be the main cause of many food poisoning cases and outbreaks that take place when having contaminated meals that contain milk or dairy products (Vazquez-Sanchez et al., 2014), due to their high content of carbohydrates and protein (Fletcher et al., 2015). Gastro-enteric disturbances are induced after having meals containing staphylococcal heat-stable enterotoxins (SEs) (Mossong et al., 2015). Staphylococcus aureus can produce about 23 enterotoxins (Podkowik et al., 2013), where the most prominent ones are A (SEA) and D (SED), while B (SEB), C (SEC), and E (SEE) come after them (Balaban and Rasooly, 2000). A highly genetic plasticity Pseudomonas aeruginosa easily adapts to various environments, so it is easily isolated from soil and water and it has been found colonized in various living animals (Streeter and Katouli, 2016). Pathogenicity of P. aeruginosa is related to biofilm formation [where alginates enter the colonies composition to antagonist antibiotics medication and host immunity (Moradian et al., 2012)], numerous membranes existence, and extracellular virulence factors production (Khattab et al., 2015) mainly exotoxins which are extremely toxic causing tissue damage (Sato and Frank, 2004), acute infection and food poisoning (Nikbin et al., 2012). ExoS (exoenzyme S) is one of three ADP-ribosyltransferases (Wolska et al., 2012) that are produced by P. aeruginosa type III secretion system into the cytoplasmic matrix of epithelial cells directly (Fadhil et al., 2016). The role of nan1 (neuraminidase) gene is starting colonization and facilitating the pathogen adherence easily to the respiratory tract and buccal epithelium, as it is encoded to sialidase causing long term infection (Moradian et al., 2012). Biofilm initiation and then keeping it is done by the pslA gene (polysaccharide synthesis locus A) (Yang et al., 2011). Multidrug-resistant (MDR) pathogens are in continuous progress in reducing the effect of antibiotics (Brown and Wright, 2016) increasing health problems and deaths (Ali et al., 2018). Recently, alternative medicinal plants have been of great concern instead of antibiotics (Zuo et al., 2008) like Moringa oleifera plant which is called the miracle tree due to its role in nutrition and medicine (Oyeyinka and Oyeyinka, 2018), besides its leaves aqueous extract has been confirmed to be consumed orally safely in high doses (Awodele et al., 2012). It has a proven bactericidal effect and several medicinal uses (Suarez et al., 2005). Several health benefits that gained after having Moringa leaves besides its great nutritional value (Teixeira et al., 2014), which cannot be counted as it acts as a diuretic, anti-neoplastic, anti-diabetic, anti-inflammatory, antimicrobial, antioxidant, antihypertensive, anti-hyperlipidemia, antipyretic, antiulcer, cardio protectant, and hepato-protectant (Vongsak et al., 2014). In our study, we have examined the prevalence of food poisoning pathogens (S. aureus and P. aeruginosa), their biofilm reaction, and virulence genes in milk and some dairy products, then, we have examined M. oleifera leaves aqueous extract effect on the recovered isolated P. aeruginosa. Materials and MethodsSample collectionA 100 samples of raw milk, Kariesh cheese, processed cheese, and ice cream (25 each), were randomly purchased from small outlets, vendors, and markets in Sharkia and Dakahlia Provinces, Egypt, which were examined during the period from November 2021 to April 2022. Bacteriological phenotypic examinationBacteriological examinationOne milliliter of the examined sample was enriched in 9 ml of trypton soya (TS) broth, then a loopful was streaked onto both HiCrome™ Staph Selective Agar (Himedia, Mumbai) (M1913) (APHA, 2015) and Pseudomonas Agar Base (Himedia, Mumbai) (M085) with glycerol and CetriNix Supplement (Himedia, Mumbai) (FD029) (APHA, 2015). The suspected colonies were subcultured on trypticase soya broth and incubated at 30°C for 24 hours to obtain subculture. The suspected isolates were then biochemically examined via cystine-lactose-electrolyte-deficient (C.L.E.D) agar (Thomson and Miller, 2003). Congo red agar (CRA) test for biofilm productionCultures were streaked onto modified CRA medium plates (Mariana et al., 2009). Molecular examination of suspected isolatesMolecular identification of isolated pathogensIsolates which were biofilm producers (five isolates of each pathogen, for S. aureus two isolates were taken from raw milk samples, two from Kariesh cheese samples, and one from ice cream sample, while P. aeruginosa isolates were three from raw milk samples, one from processed cheese sample, and one from ice cream sample) were further examined in the Polymerase Chain Reaction (PCR) Unit, Animal Health Research Institute, Zagazig, Sharkia, Egypt. Extraction of bacterial DNA was done according to manufacturer’s guidelines. 16S rRNA for both S. aureus (Monday and Bohach, 1999) and P. aeruginosa isolates (Ghadaksaz et al., 2015). Table 1. Oligonucleotide primers, their specific sequence, and the specific amplified product of P. aeruginosa.
Table 2. Oligonucleotide primers, their specific sequence, and the specific amplified product of S. aureus.
Table 3. Cycling conditions of the different primers during PCR of P. aeruginosa.
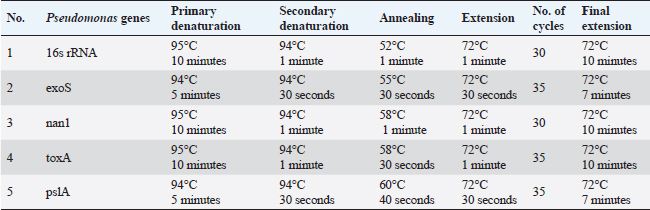
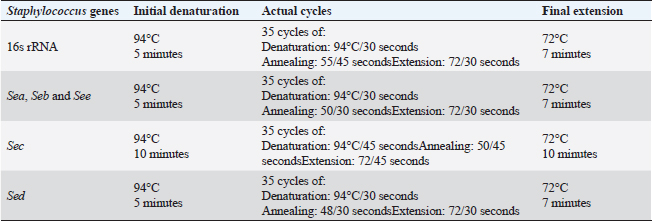
Virulence genes PCR amplificationSEs A to E virulence-associated genes were examined in S. aureus isolates (Mehrotra et al., 2000). Pseudomonas aeruginosa isolates were examined for toxA (exoenzyme A), exoS, nan1 virulence-associated genes (Khattab et al., 2015) and pslA biofilm gene (Ghadaksaz et al., 2015) (Tables 1–4). Moringa oleifera treatmentIt was done according to Shah et al. (2015). Extract preparation (0.5%)Preparation of P. aeruginosa isolatePseudomonas aeruginosa isolate used in this experiment is highly virulent and was isolated from raw milk during this study, confirmed biochemically and by the conventional method of PCR, in addition, it harbored exoS and nan1 virulence genes, moreover it reacted as a medium biofilm producer on CRA and was positive for pslA biofilm gene. The isolate was incubated in TS broth at 35°C overnight. Preparation of S. aureus isolateStaphylococcus aureus isolate examined is highly virulent, where it was isolated from a Kareish cheese sample, and biochemically confirmed, also, it was positive for 16S rRNA and SEB gene and it was a strong biofilm producer. That isolate was incubated in a TS broth tube and at 35°C overnight. Determination of antibacterial activity of aqueous extract of M. oleiferaMinimum inhibitory concentration (MIC) and sub inhibitory concentration (SIC) by micro-dilution methodSerial dilution of 100 μl of 0.5% M. oleifera aqueous extract was performed in 96 well plates (TechnoPlastic Products, Switzerland), then it was inoculated with 5 × 105 CFU/ml of P. aeruginosa and S. aureus apart and overnight incubation was performed at 37°C, inoculation of each well were performed on each specific media to determine MIC and SIC; the lowest concentration prevents the growth of pathogen and the concentration before it, respectively, then examined on CRA to determine MIC and SIC (CLSI, 2013). Cycling conditions of the different primers during PCR of S. aureus.
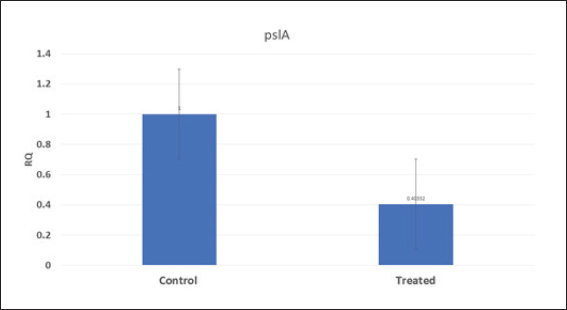
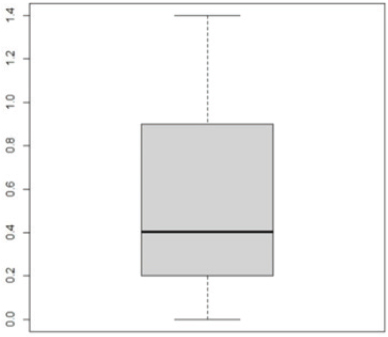
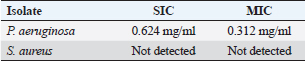
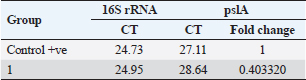
Pseudomonas aeruginosa pslA expression by quantitative real-time (qRT)-PCRThe next step was gene expression by qRT-PCR where the housekeeping gene used was 16S rRNA to find out the different levels of expression. The reaction was described by El-Demerdash and Bakry (2020). The gene expression variation was estimated by comparing the Ct of the examined sample and positive control based on “ΔΔCt” method (Yuan et al., 2006). Statistical analysisR studio was operated for both heatmap and box plot. Moreover, one-way analysis of variance SPSS version 22 for Windows was used for fold change analysis of M. oleifera aqueous extract against P. aeruginosa isolate. ResultsBacteriological phenotypic examinationFigure 1 expresses phenotypic bacteriological results, where P. aeruginosa was isolated from whole samples with 30 (30%). The highest isolation rate was seen in raw milk as 48%, followed by 36% in ice cream, lower results were 20% and 16% in Kariesh cheese and processed cheese, respectively. 17% of isolates were identified biochemically as 20%, for both raw milk and Kariesh cheese, and 12% and 16% in processed cheese and ice cream, respectively. About 30% of P. aeruginosa isolates were biofilm producers, where 12% were intermediate, 18% of isolates were weak, while (12/17) of isolates were negative biofilm producers with 70%. Staphylococcus aureus was isolated from 32 (32%) of the examined samples. The highest isolation rate was detected in 44% of raw milk samples, followed by 40% of ice cream samples, lower results were recorded as 28% in Kariesh cheese samples and 16% of processed cheese samples. 24% of isolates were identified biochemically as 36% in raw milk samples, 28% in ice cream isolates, and 16% in both Kariesh cheese and processed cheese. Five of 24 (21%) S. aureus isolates were biofilm producers as 8% were strong producers and 12% were intermediate, while 79% were negative. Molecular examination of suspected isolatesFigure 2 shows the results of molecular examination of suspected isolates, where all of the recovered P. aeruginosa isolates were positive for both 16S rRNA gene (a confirmatory target for P. aeruginosa) and nan1 gene, while 66.6%, 100%, and 100% of examined strains from raw milk, processed cheese, and ice cream samples, respectively, contained exoS gene, while both toxA and pslA genes were detected in isolates which were taken from raw milk and soft processed cheese samples with 33.3% and 100%, respectively (Fig. 3). All recovered S. aureus isolates were positive for 16S rRNA gene (a confirmatory target for S. aureus), S. aureus isolates were examined for SEs A to E, SEA was positive in 100% of examined strains from all raw milk and ice cream samples, while SEB was detected in 50% and 100% of examined strains from raw milk and Kariesh cheese samples, respectively (Fig. 4), none of SEC, SED, and SEE was detected in any S. aureus strain. Antibacterial activity of aqueous extract of M. oleiferaThere was no MIC detected on P. aeruginosa and S. aureus. Although a biofilm of S. aureus did not affect by M. oleifera aqueous extract but P. aeruginosa biofilm formation MIC was 0.312 mg/ml, SIC was 0.624 mg/ml (Table 5), while relative expression of pslA biofilm gene treatment with MIC (3.2%) of M. oleifera aqueous extract revealed significant decrease in the expression level (0.40332 fold change) as shown in Table 6 and Figures 5 and 6. DiscussionFoodborne diseases mostly affect developing countries due to bad food handling and poor sanitary (WHO, 2015). In our study, P. aeruginosa was isolated from 30 (30%) of the examined samples (Fig. 1). Pseudomonas was examined in a number of studies in Egypt, and raw milk and dairy products were reported to be contaminated with P. aeruginosa, with the prevalence of 48% raw milk samples, 18% soft cheese and 8% of Kareish cheese (Ibrahim et al., 2022), P. aeruginosa was present in 20%,16%, and 8%, respectively, in examined raw milk, Kareish cheese, and ice cream samples (Atia et al., 2022), lower results were recorded by Abdel-Hameed and Saleem (2019) who found P. aeruginosa in 11.6% of raw milk samples and 10% of ice cream samples. About 70% of psychrotrophic pathogens like Pseudomonas species can be easily recovered from chilled dairy products such as raw milk and fresh cheese (Langsrud et al., 2003), as these are the main reservoirs of Pseudomonas spp. (Quintieri et al., 2019). Pseudomonas species are able to form an extracellular matrix called biofilm (Brooks and Flint, 2008), thus making them highly resistant to detergents and cleaning operations (Giaouris et al., 2014). About 30% of the isolates that have been recovered from examined samples in this study were biofilm producers. Higher results were obtained by Radovanic et al. (2020) who found that 54 (90%) of Pseudomonas isolates of milk samples are biofilm producers, the most prominent group was moderately biofilm producers with (70%) 42 isolates, followed by 10 (17%) weak biofilm producers, then 2 isolates (3%) were strong biofilm producers, while 6 (10%) isolates did not show any biofilm production. Different results were obtained by El-Demerdash and Bakry (2020) who used CRA to classify P. aeruginosa isolates taken from food samples and found that 21 isolates (66.6%) were biofilm producers as 19% were strong and 28.6% were intermediate, while 19% was negative. Arslan et al. (2011) also used CRA for slime production in Pseudomonas isolates taken from homemade cheese and all of them were negative. Biofilm plays a great role in P. aeruginosa pathogenicity causing a chronic form of infection and also resisting antimicrobial agents (Olsen, 2015). Staphylococcus aureus was isolated as 32% (Fig. 1) of total samples. Lower results were obtained by Mirzaei et al. (2012) who found that the incidence of S. aureus in raw milk samples was 14% and 6% in ice cream samples, also lower results were reported in cheese samples by Baumgartner et al. (2014) that was (13%) and Hadi and Hassan (2013) who isolated S. aureus from raw milk samples with percentage (13.4%) and from white raw soft cheese samples with (20%) percentage. Higher results of S. aureus prevalence were 75%, 40%, and 50% in raw milk, Kareish cheese, and ice cream samples, respectively, which were obtained by Al-Ashmawy et al. (2016), also higher results of S. aureus were found by Fadel and Ismail (2015); the results were 63.3%, 40%, and 36.7% of Kareish cheese, raw buffaloes’ milk, and ice cream samples, respectively. The acidity of Kareish cheese may play a great role in decreasing the incidence and prevalence rates of S. aureus to lower rates than those in raw milk (Al-Ashmawy et al., 2016). Improper personal hygiene and bad hygienic practices during dairy product manufacturing are the main causes of milk and dairy S. aureus contamination (Chye et al., 2004). About 21% of the S. aureus isolates in our study were biofilm producers. Different results were recorded by Avila-Novoa et al. (2018) who isolated S. aureus from dairy industry surfaces and examined these isolates on CRA, the results were that 75% of isolates were biofilm producers, 16.6% were negative and 8.3% were non characteristic phenotype. The critical point of slime formation is that the effect of antibiotics is lowered by decreasing their dose when the biofilm prevents some from passing inside the pathogen (Singh et al., 2010).
Fig. 1. Heat map that shows the overall phenotypic examination: incidence of examined food poisoning pathogens P. aeruginosa and S. aureus and their biofilm formation profile. The X axis of the heat map resamples examined pathogens each on its selective media, their biochemical identification, and their ability to produce biofilm, while the Y axis resamples type of samples. The blue color of the map indicates positive results, while the white color indicates negative results. The numbers of examined samples are found horizontally on the map.
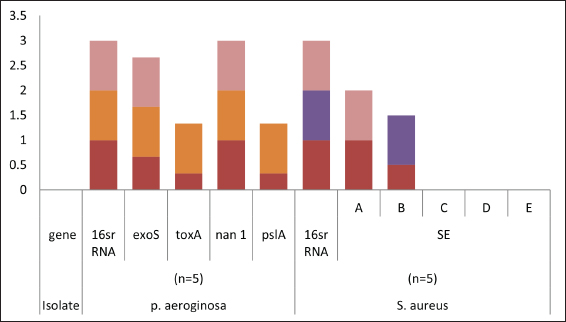
Fig. 2. Distribution of different genes in food poisoning isolates recovered from examined samples (red for raw milk, orange for processed cheese, purple for Kariesh cheese, and pink for ice cream).
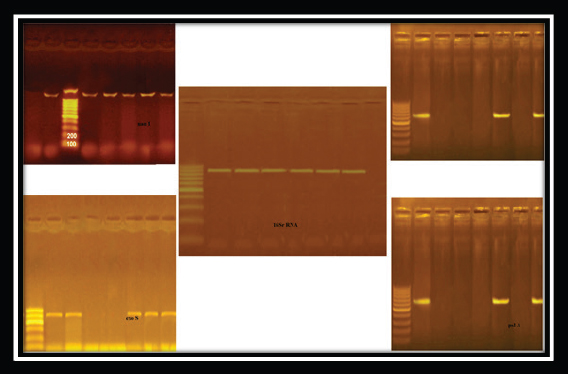
Fig. 3. Pseudomonas aeruginosa genes detected by conventional PCR.
Fig. 4. Staphylococcus aureus enterotoxin genes detected by conventional PCR.
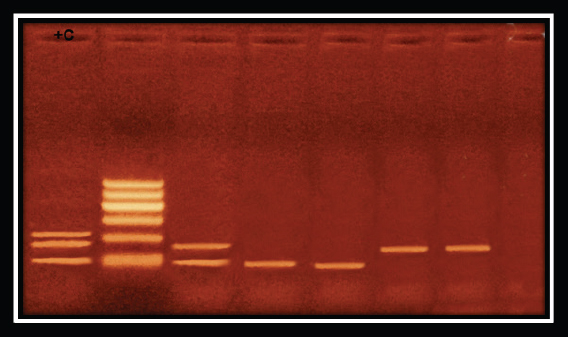
Fig. 5. Showing biofilm gene expression of P. aeruginosa using real time PCR after treatment with M. oleifera.
Fig. 6. Boxplot showing variation of biofilm gene expression of P. aeruginosa detected by using real time PCR after treatment with M. oleifera aquas extract. All recovered P. aeruginosa isolates were positive for 16S rRNA gene (a confirmatory specific target for P. aeruginosa genus) (Ghadaksaz et al., 2015), while 66.6%, 100%, and 100% of examined strains from raw milk, soft processed cheese, and ice cream samples, respectively, contained exoS gene, that was similar to Mohammadzadeh et al. (2017) who reported 100% P. aeruginosa isolates had gene exoA, nearly similar results were confirmed by Ameen et al. (2019) where 69.7% out of P. aeruginosa isolates had exoS genes. Other studies reported lower results as Younis et al. (2015) found out (17.14%) of isolates harbored exoS gene and (36.8%) of isolates carried exoS gene according to the study of Banerjee et al. (2017). In our study, both toxA and pslA genes were only detected in isolates that were taken from raw milk samples with 33.3% and 100% of soft processed cheese, while AL-Sheikhly et al. (2020) recorded higher results as all examined isolates harbored pslA gene. For the nan1 gene, it was detected in 100% of examined strains, lower results were detected by Tawfiq (2018) where only (41.2%) of isolates carried nan1 gene. Many reasons such as strain virulence, immunity, and contamination level explain the different results (Khan and Cerniglia, 1994). In our study, all recovered S. aureus isolates were positive for 16S rRNA gene (a confirmatory specific target for S. aureus genus) (Monday and Bohach, 1999), S. aureus isolates were examined for SEs A to E, SEA was positive in 100% of examined strains isolated from all raw milk and ice cream samples, while SEB was detected in 50% and 100% of examined strains from raw milk and Kariesh cheese samples, respectively. None of SEC, SED, and SEE was detected in any S. aureus strain. Although Al-Ashmawy et al. (2016) recorded the same results for SEa, higher rates for SEb and SEc were recorded by them as 100%. On the other hand, Oliveira et al. (2011) found a low percentage of the SEc gene in 6% of S. aureus isolates recovered from subclinical mastitic milk. Both Jørgensen et al. (2005) and Morandi et al. (2007) found no SEe in isolates recovered from milk samples which agrees with our study. These SEs pose a great hazard to health (WHO, 2007) as the low amount of these toxins could easily be a reason for food poisoning (Le Loir et al., 2003) moreover, the high heat stability of staphylococcal enterotoxin makes the incidence of food poisoning more risky (Mossong et al., 2015). In our study, M. oleifera aqueous extract could not act as the bactericidal for P. aeruginosa in the MIC test; this was similar to Abalaka et al. (2012) who explained that point due to the high resistance nature of this pathogen. On the other hand, 0.312 mg/ml of the aqueous leaves extract was detected as MIC on P. aeruginosa biofilm formation and SIC was 0.624 mg/ml, pslA biofilm gene expression of P. aeruginosa isolate was detected by qRT-PCR before and after treatment with the detected MIC (0.312 mg/ml) M. oleifera aqueous extract and we found out a revealed significant decrease in the expression level (Tables 5 and 6, Figs. 5 and 6) (0.40332-fold change). Phytochemicals inside M. oleifera leaves act as antibacterial materials (Bukar et al., 2010). Many studies examined pslA gene expression, where El-Demerdash and Bakry (2020) examined the effect of amikacin and cefotaxime on the pslA gene, the fold changes were between 0.1 and 0.7, and AL-Sheikhly et al. (2020) examined gentamicin at 512 μg/ml on pslA gene, but its effect was lower than expected according to them. Staphylococcus aureus isolate in our study was highly virulent, it was not affected by M. oleifera aqueous extract, which may be due to active ingredients in inadequate amounts (De Zoysa et al., 2019), as some of them were lost during evaporation at the step of preparation of aqueous extract (Ibrahim and Kebede, 2020). Table 5. Showing MIC and SIC of 0.5% M. oleifera aqueous extract against S. aureus and P. aeruginosa.
Table 6. Showing biofilm gene expression of P. aeruginosa using real time PCR after treatment with M. oleifera (16S rRNA as a housekeeping gene).
ConclusionTreatment with an aqueous extract of M. oleifera 0.5% showed a significant effect on P. aeruginosa pslA gene, although there was no change in the MIC test, this medicinal plant has a great antibacterial effect and needs further examinations. AcknowledgementsOur study had no specific grant. Author contributionsAll of the authors contributed to this study and approved the final manuscript. Conflict of interestThe authors declare that there is no conflict of interest. FundingNone. Data AvailabilityAll data supporting the findings of this study are available within the manuscript. ReferencesAbalaka, M.E., Daniyan, S.Y., Oyeleke, S.B. and Adeyemo, S.O. 2012. The antibacterial evaluation of Moringa oleifera leaf extracts on selected bacterial pathogens. J. Microbiol. Res. 2(2), 1–4. Abdel-Hameed, A.H.M. and Saleem, K.L. 2019. Biosensors and bio-based methods for the separation and detection of foodborne pathogens. Adv. Food Nut. Res. 54, 1–44. Al-Ashmawy, M.A., Ibrahim, K.S., Mohammed, S.A., Elhadidy, M. and Tamura, T. 2016. Prevalence, molecular characterization, and antimicrobial susceptibility of methicillin-resistant Staphylococcus aureus (MRSA) isolated from milk and dairy products. Foodborne Pathogens Dis. 13(3), 2038. Ali, J., Rafiq, Q.A. and Ratcliffe, E. 2018. Antimicrobial resistance mechanisms and potential synthetic treatments. Future Sci. OA 4, FSO290. AL-Sheikhly, M.A., Musleh, L.N. and Al-Mathkhury, H.J.F. 2020. Gene expression of pelA and pslA in Pseudomonas aeruginosa under gentamicin stress. Iraqi J. Sci. 61(2), 295–305. Ameen, F., Reda, S.A., El-Shatoury, S.A., Riad, E.M., Enany, M.E. and Alarfaj, A.A. 2019. Prevalence of antibiotic resistant mastitis pathogensin dairy cows in Egypt and potential biological control agents produced from plant endophytic actinobacteria. Saudi J. Biol. Sci. 26, 1492–1498. APHA. 2015. Compend ium of methods for the microbiological examination of foods, 5th ed. Eds., Salfinger, Y. and Tortorello, M.L. Washington, DC: USA. Arslan, S., Eyi, A. and Özdemir, F. 2011. Spoilage potentials and antimicrobial resistance of Pseudomonas spp. isolated from cheeses. J. Dairy Sci. 94, 5851–5856. Atia, R.M., Mohamed, H.A., Abo ElRoos, N.A. and Awad, D.A. 2022. Incidence of Pseudomonas species and effect of their virulence factors on milk and milk products. Benha Vet. Med. J. 42, 1–5. Avila-Novoa, M.G., Iñ´ıguez-Moreno, M., Sol´ıs-Vel´azquez, O.L., Gonz´alez-G´omez, O.J., Guerrero-Medina, P. and Guti´errez-Lomel´, M. 2018. Biofilm formation by Staphylococcus aureus isolated from food contact surfaces in the dairy industry of Jalisco, Mexico. J. Food Qual. 2018, 1746139. Awodele, O., Oreagbe, I.A., Odoma, S., da Silva, J.A.T. and Osunkalo, V.O. 2012. Toxicological evaluation of the aqueous leaf extract of Moringa oleifera Lam. (Moringaceae). J. Ethnopharmacol. 139, 300–306. Balaban, N. and Rasooly, A. 2000. Staphylococcal enterotoxins. Int. J. Food Microbiol. 61, 1–10. Banerjee, S., Batabyal, K., Joardar, S.N., Isore, D.P., Dey, S., Samanta, I., Samanta, T.K. and Murmu, S. 2017. Detection and characterization of pathogenic Pseudomonas aeruginosa from bovine subclinical mastitis in West Bengal, India. Vet. World 10, 738–742. Baumgartner, A., Niederhauser, I. and Johler, S. 2014. Virulence and resistance gene profiles of Staphylococcus aureus strains isolated from ready-to-eat foods. J. Food Protect. 77(7), 1232–1236. Bhunia, A.K. 2018. Foodborne microbial pathogens: mechanisms and pathogenesis. New York, NY: Springer, vol. 2, pp: 2319–3867. Brooks, J.D. and Flint, S.H. 2008. Biofilms in the food industry: problems and potential solutions. Int. J. Food Sci. Technol. 43(12), 2163–2176. Brown, E.D. and Wright, G.D. 2016. Antibacterial drug discovery in the resistance era. Nature 529, 336–343. Bukar, A., Uba, A. and Oyeyi, T.I. 2010. Antimicrobial profile of Moringa oleifera Lam. extracts against some foodborne microorganisms. Bayero J. Pure Appl. Sci. 3(1), 43–48. Chye, F., Abdullah, A. and Ayob, M.K. 2004. Bacteriological quality and safety of raw milk in Malaysia. Food Microbiol. 21, 535–554. CLSI. 2013. M100-S23 performance standards for antimicrobial. Wayne, PA: CLSI. De Zoysa, M.H.N., Rathnayake, H., Hewawasam, R.P. and Wijayaratne, W.M.D.G.B. 2019. Determination of in vitro antimicrobial activity of five Sri Lankan medicinal plants against selected human pathogenic bacteria. Int. J. Microbiol. 2019, 8. El-Demerdash, A.S. and Bakry, N.R. 2020. Evaluation of the synergistic effect of amikacin with cefotaxime against Pseudomonas aeruginosa and its biofilm genes expression. Gene expression and phenotypic traits. IntechOpen; doi:10.5772/intechopen.91146. Fadel, H.M. and Ismail, J. 2015. Occurrence and zoonotic importance of methicillin-resistant Staphylococcus aureus in raw milk and some dairy products at Ismailia City, Egypt. Zagazig Vet. J. 43(3), 95–104. Fadhil, L., Al-Marzoqi, A.H., Zahraa, M.A. and Shalan, A.A. 2016. Molecular and phenotypic study of virulence genes in a pathogenic strain of Pseudomonas aeruginosa isolated from various clinical origins by PCR: profiles of genes and toxins. Res. J. Pharm. Biol. Chem. Sci. 7, 590–598. Fletcher, S., Boonwaat, L., Moore, T., Chevada, R. and Conaty, S. 2015. Investigating an outbreak of staphylococcal food poisoning among travelers across two Australian states. Western Pac. Surveill. Res. J. 6, 17–21. Ghadaksaz, A., Fooladi, A.A.I., Hosseini, H.M. and Amin, M. 2015. The prevalence of some Pseudomonas virulence genes related to biofilm formation and alginate production among clinical isolates. J. Appl. Biomed. 13, 61–68. Giaouris, E., Heir, E., Hébraud, M., Chorianopoulos, N., Langsrud, S., Møretrø, T., Habimano, O., Desvaux, M., Renier, S. and Nychas, G.J. 2014. Attachment and biofilm formation by foodborne bacteria in meat processing environments: causes, implications, role of bacterial interactions and control by alternative novel methods. Meat Sci. 97, 289–309. Hadi, M.G.K. and Hassan A.A.A. 2013. Detection of methicillin or multidrug resistant Staphylococcus aureus (MRSA) in locally produced raw milk and soft cheese in Baghdad markets. Iraqi J. Vet. Med. 37(2), 226–231. Ibrahim, M.M., Elsaied, E.I., Abd El Aal, S.F. and Bayoumi, M.A. 2022. Prevalence of Pseudomonas aeruginosa in milk and some dairy products with reduction trials by some natural preservatives. J. Adv. Vet. Res. 12(4), 434–438. Ibrahim, N. and Kebede, A. 2020. In vitro antibacterial activities of methanol and aqueous leave extracts of selected medicinal plants against human pathogenic bacteria. Saudi J. Biol. Sci. 27, 2261–2268. Jørgensen, H., Mork, T., Hogasen, H. and Rovik, L. 2005. Enterotoxigenic Staphylococcus aureus in bulk milk in Norway. J. Appl. Microbiol. 99, 158–166. Khan, A.A. and Cerniglia, C.E. 1994. Detection of Pseudomonas aeruginosa from clinical and environmental samples by amplification of the exotoxin A gene using PCR. Appl. Environ. Microbiol. 60, 3739–3745. Khattab, M.A., Nour, M.S., and El-Sheshtawy, N.M. 2015. Genetic identification of Pseudomonas aeruginosa virulence genes among different isolates. J. Microb. Biochem. Technol. 7, 274–277. Langsrud, S., Sundheim, G., Borgmann-Strahsen, R. and Borgmann-Strahsen, R. 2003. Intrinsic and acquired resistance to quaternary ammonium compounds in food-related Pseudomonas spp. J. Appl. Microbiol. 95, 874–882. Le Loir, Y., Baron, F. and Gautier, M. 2003. Staphylococcus aureus and food poisoning. Genet. Mol. Res. 2, 63–76. Mariana, N.S., Salman, S.A., Neela, V. and Zamberi S. 2009. Evaluation of modified Congo red agar for detection of biofilm produced by clinical isolates of methicillin–resistance Staphylococcus aureus. African J. Microbiol. Res. 3(6), 330-338. Mehrotra, M., Wang, G. and Johnson, W.M. 2000. Multiplex PCR for detection of genes for Staphylococcus aureus enterotoxins, exfoliative toxins, toxic shock syndrome toxin 1, and methicillin resistance. J. Clin. Microbiol. 38(3), 1032–1035. Mirzaei, H., Farhoudi, H., Tavassoli, H., Farajli, M. and Monadi, A. 2012. Presence and antimicrobial susceptibility of methicillin resistant Staphylococcus aureus in raw and pasteurized milk and ice cream in Tabriz by culture and PCR techniques. Afr. J. Microbiol. Res. 6, 6224–6229. Mohammadzadeh, A., Mardaneh, J., Ahmadi, R. and Adabi J. 2017. Evaluation of the virulence features and antibiotic resistance patterns of pathogenic Pseudomonas aeruginosa strains isolated from hospitalized patients in Gonabad, Iran. Arch. Pediatr. Infect. Dis. 5(3), 41267. Momtaz, H., Farzan, R., Rahimi, E., Safarpoor Dehkordi, F. and Souod, N. 2012. Molecular characterization of Shiga toxin-producing Escherichia coli isolated from ruminant and donkey raw milk samples and traditional dairy products in Iran. Sci. World J. 2012, 231342. Monday, S.R. and Bohach, G.A. 1999. Use of multiplex PCR to detect classical and newly described pyrogenic toxin genes in staphylococcal isolates. J. Clin. Microbiol. 37(10), 3411–3414. Moradian, F., Perdosi, E. and Molana, Z. 2012. Molecular detection of integron genes and pattern of antibiotic in P. aeruginosa strains isolates from intensive unit. Iran. J. MAM 1(4), 424. Morandi, S., Brasca, M., Lodi, R., Cremonesi, P.B. and Castiglioni, B. 2007. Detection of classical enterotoxins and identification of enterotoxin genes in Staphylococcus aureus from milk and dairy products. Vet. Microbiol. 124, 66–72. Mossong, J., Decruyenaere, F., Moris, G., Ragimbeau, C., Olinger, C.M., Johler, S., Perrin, M., Hau, P. and Weicherderding, P. 2015. Investigation of a staphylococcal food poisoning outbreak combining case-control, traditional typing and whole genome sequencing methods, Luxembourg, June 2014. Euro. Surveill. 20, 30059. Muthu, G., Suresh, A., VishnuPrabu, D., Munirajan, A.K., Esther Mary, S., Sathishkumar, E., Gopinath, P. and Srivani, S. 2014. Detection of virulence genes from Salmonella species in Chennai, India, Centre for Info Bio Technology (CIB Technol). J. Microbiol. 3(1), 11–14. Nikbin, V., Aslani, M., Sharafi, Z., Hashemipour, M., Shahcheraghi, F. and Ebrahimipour, G. 2012. Molecular identification and detection of virulence genes among Pseudomonas aeruginosa isolated from different infectious origins. Iran. J. Microbiol. 4, 118–123. Oliveira, L., Rodrigues, A.C., Hulland, C. and Ruegg, P.L. 2011. Enterotoxin production, enterotoxin gene distribution, and genetic diversity of Staphylococcus aureus recovered from milk of cows with subclinical mastitis. Am. J. Vet. Res. 72, 1361–1368. Olsen, I. 2015. Biofilm-specific antibiotic tolerance and resistance. Euro. J. Clin. Microbiol. Infect. Dis. 34, 877–886. Oyeyinka, A.T. and Oyeyinka, S.A. 2018. Moringa oleifera as a food fortificant: recent trends and prospects. J. Saudi Soc. Agric. Sci. 17, 127–136. Painter, J.A., Hoekstra, R.M., Ayers, T., Tauxe, R.V., Braden, C.R., Angulo, F.J. and Griffin, P.M. 2013. Attribution of foodborne illnesses, hospitalizations, and deaths to food commodities by using outbreak data, United States, 1998–2008. Emerg. Infect. Dis. 19, 407–415. Podkowik, M., Park, J.Y., Seo, K.S., Bystron, J. and Bania, J. 2013. Enterotoxygenic potential of coagulase-negative Staphylococci. Int. J. Food Microbiol. 163, 34–40. Quintieri, L., Fanelli, F. and Caputo, L. 2019. Antibiotic resistant Pseudomonas spp. spoilers in fresh dairy products: an underestimated risk and the control strategies. Foods 8, 372. Radovanic, R.S., Rajic, N.S., Ranin, L.A.S., Vuckovic, N.O., Milosavljevic, A.T., Ranin, J. and Gajic, I. 2020. Biofilm production and antimicrobial resistance of clinical and food isolates of Pseudomonas spp. Curr. Microbiol. 77, 4045–4052. Sato, H. and Frank, D.W. 2004. ExoU is a potent intracellular phospholipase. Mol. Microbiol. 53(5), 1279–1290. Shah, M.A., Don Bosco, S.J. and Mir, S.A. 2015. Effect of Moringa oleifera leaf extract on the physicochemical properties of modified atmosphere packaged raw beef. J. Food Packaging Shelf Life. 3, 31–38. Singh, R., Ray, P., Das, A. and Sharma, M. 2010. Penetration of antibiotics through Staphylococcus aureus and Staphylococcus epidermidis biofilms. J. Antimicrob. Chemother. 65(9), 1955–1958. Streeter, K. and Katouli, M. 2016. Pseudomonas aeruginosa: a review of their pathogenesis and prevalence in clinical settings and the environment. Infect. Epidemiol. Med. 2, 25–32. Suarez, M., Haenni, M., Canarelli, S.; Fisch, F., Chodanowski, P., Servis, C., Michielin, O., Freitag, R., Moreillon, P. and Mermod, N. 2005. Structure-function characterization and optimization of a plant-derived antibacterial peptide. Antimicrob. Agents Chemother. 49, 3847–3857. Tawfiq, S.M. 2018. Bacteriological and genetic study of Pseudomonas aeruginosa isolates. Iraqi J. Agric. Sci. 44(1), 22–35. Teixeira, E.M.B., Carvalho, M.R.B., Neves, V.A., Silva, M.A. and Arantes-Pereira, L. 2014. Chemical characteristics and fractionation of proteins from Moringa oleifera Lam. leaves. Food Chem. 147, 51–54. Thomson, R.B. and Miller, J.M. 2003. Specimen collection, transport, and processing: bacteriology. In Manual of clinical microbiology, 8th ed. Eds., Murray, P.R., Baron, E.J., Jorgensen, J.H., Pfaller, M.A. and Yolken, R.H. Washington, DC: American Society for Microbiology. Vazquez-Sanchez, D., Cabo, M.L., Ibusquiza, P.S. and Rodriguez Herrera, J.J. 2014. Biofilm-forming ability and resistance to industrial disinfectants of Staphylococcus aureus isolated from fishery products. Food Control 39, 8–16. Vongsak, B., Sithisam, P. and Gritsanapan, W. 2014. Simultaneous HPLC quantitative analysis of active compounds in leaves of Moringa oleifera Lam. J. Chromatogr. Sci. 52, 641–645. WHO. 2007. Food safety, risk assessment. Available via http://www.who.int/foodsafety/micro/riskassessment/en/index.html (Accessed 30 April 2023). WHO. 2015. Estimates of the global burden of foodborne diseases: foodborne disease burden epidemiology reference group 2007-2015. Geneva, Switzerland: World Health Organization. Wolska, K., Koti, B., Mioduszewska, H., Sempruch, C., Borkowska, L. and Rymuza, K. 2012. Occurrence of the nan1 gene and adhesion of Pseudomonas aeruginosa isolates to human buccal epithelial cells. Biol. Lett. 49(1), 59–64. Yang, L., Hu, Y.F., Liu, Y., Zhang, J.D., Ulstrup, J. and Molin, S. 2011. Distinct roles of extracellular polymeric substances in Pseudomonas aeruginosa biofilm development. Environ. Microbiol. 13, 1705–1717. Younis, G., Awad, A., Maghawry, M. and Selim, F. 2015. Extracellular enzymes and toxins of Pseudomonas aeruginosa strains isolated from clinically diseased egyptian cows. Adv. Anim. Vet. 3, 522–526. Yuan, J.S., Reed, A., Chen, F. and Stewart C.N. 2006. Statistical analysis of realtime PCR data. BMC Bioinform. 7, 85. Zuo, G.Y., Wang, G.C., Zhao, Y.B., Xu, G.L., Hao, X.Y. and Han, J. 2008. Screening of Chinese medicinal plants for inhibition against clinical isolates of methicillin-resistant Staphylococcus aureus (MRSA). J. Ethnopharmacol. 120, 287–290. | ||
| How to Cite this Article |
| Pubmed Style Elshazely RM, Amer IH, Aal SFA, Tahoun AB, . Antibacterial effect of Moringa oleifera on Staphylococcus aureus and Pseudomonas aeruginosa isolated from raw milk and some dairy products with special reference to biofilm gene expression. Open Vet J. 2024; 14((1) (Zagazig Veterinary Conference)): 164 -175 . doi:10.5455/OVJ.2024.v14.i1.15 Web Style Elshazely RM, Amer IH, Aal SFA, Tahoun AB, . Antibacterial effect of Moringa oleifera on Staphylococcus aureus and Pseudomonas aeruginosa isolated from raw milk and some dairy products with special reference to biofilm gene expression. https://www.openveterinaryjournal.com/?mno=175314 [Access: May 08, 2024]. doi:10.5455/OVJ.2024.v14.i1.15 AMA (American Medical Association) Style Elshazely RM, Amer IH, Aal SFA, Tahoun AB, . Antibacterial effect of Moringa oleifera on Staphylococcus aureus and Pseudomonas aeruginosa isolated from raw milk and some dairy products with special reference to biofilm gene expression. Open Vet J. 2024; 14((1) (Zagazig Veterinary Conference)): 164 -175 . doi:10.5455/OVJ.2024.v14.i1.15 Vancouver/ICMJE Style Elshazely RM, Amer IH, Aal SFA, Tahoun AB, . Antibacterial effect of Moringa oleifera on Staphylococcus aureus and Pseudomonas aeruginosa isolated from raw milk and some dairy products with special reference to biofilm gene expression. Open Vet J. (2024), [cited May 08, 2024]; 14((1) (Zagazig Veterinary Conference)): 164 -175 . doi:10.5455/OVJ.2024.v14.i1.15 Harvard Style Elshazely, R. M., Amer, I. H., Aal, S. F. A., Tahoun, A. B. & (2024) Antibacterial effect of Moringa oleifera on Staphylococcus aureus and Pseudomonas aeruginosa isolated from raw milk and some dairy products with special reference to biofilm gene expression. Open Vet J, 14 ((1) (Zagazig Veterinary Conference)), 164 -175 . doi:10.5455/OVJ.2024.v14.i1.15 Turabian Style Elshazely, Rowyda M.Y., Ibrahim H. Amer, Salah F.A. Abd-El Aal, Asmaa B.M.B. Tahoun, and . 2024. Antibacterial effect of Moringa oleifera on Staphylococcus aureus and Pseudomonas aeruginosa isolated from raw milk and some dairy products with special reference to biofilm gene expression. Open Veterinary Journal, 14 ((1) (Zagazig Veterinary Conference)), 164 -175 . doi:10.5455/OVJ.2024.v14.i1.15 Chicago Style Elshazely, Rowyda M.Y., Ibrahim H. Amer, Salah F.A. Abd-El Aal, Asmaa B.M.B. Tahoun, and . "Antibacterial effect of Moringa oleifera on Staphylococcus aureus and Pseudomonas aeruginosa isolated from raw milk and some dairy products with special reference to biofilm gene expression." Open Veterinary Journal 14 (2024), 164 -175 . doi:10.5455/OVJ.2024.v14.i1.15 MLA (The Modern Language Association) Style Elshazely, Rowyda M.Y., Ibrahim H. Amer, Salah F.A. Abd-El Aal, Asmaa B.M.B. Tahoun, and . "Antibacterial effect of Moringa oleifera on Staphylococcus aureus and Pseudomonas aeruginosa isolated from raw milk and some dairy products with special reference to biofilm gene expression." Open Veterinary Journal 14.(1) (Zagazig Veterinary Conference) (2024), 164 -175 . Print. doi:10.5455/OVJ.2024.v14.i1.15 APA (American Psychological Association) Style Elshazely, R. M., Amer, I. H., Aal, S. F. A., Tahoun, A. B. & (2024) Antibacterial effect of Moringa oleifera on Staphylococcus aureus and Pseudomonas aeruginosa isolated from raw milk and some dairy products with special reference to biofilm gene expression. Open Veterinary Journal, 14 ((1) (Zagazig Veterinary Conference)), 164 -175 . doi:10.5455/OVJ.2024.v14.i1.15 |