| Research Article | ||
Open Veterinary Journal, (2024), Vol. 14(1): 370-388 Original Research Individual genomic loci, transcript level, and biochemical profile of immune and antioxidant markers associated with genetically identified bacterial mastitis in Shami goats in EgyptEman Ebissy1, Asmaa Darwish1, Amani A. Hafez1, Ahmed Ateya2*, and Ahmed El-Sayed11Department of Animal Health and Poultry, Animal and Poultry Production Division, Desert Research Center (DRC), Cairo, Egypt 2Department of Development of Animal Wealth, Faculty of Veterinary Medicine, Mansoura University, Mansoura, Egypt *Corresponding Author: Ahmed Ateya. Department of Development of Animal Wealth, Faculty of Veterinary Medicine, Mansoura University, Mansoura, Egypt. Email: ahmed_ismail888 [at] yahoo.com Submitted: 01/10/2023 Accepted: 15/12/2023 Published: 31/01/2024 © 2024 Open Veterinary Journal
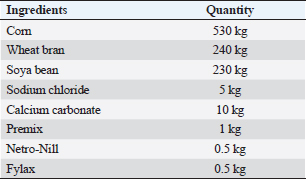
ABSTRACTBackground: Mastitis in goats is unquestionably a grave concern, with far-reaching implications for both animal well-being and productivity, while also presenting a potential threat to public health. Aim: The study aimed to compare culture methods and multiplex PCR (m-PCR) in the detection of the most three common mastitis-causing pathogens (Staphylococcus aureus, Escherichia coli, and Streptococcus spp.) and investigate the gene expression, single nucleotide polymorphisms (SNPs), serum concentrations of immunological and antioxidant indicators linked to mastitis in Shami goats. Methods: One hundred Shami do (50 Shami goats with clinical mastitis and 50 normal goats taken as control group). The culture methods and m-PCR analysis were used to find the bacteria in the milk samples. Blood samples were obtained to assess some hemato-biochemical parameters, detect SNPs, and determine the expression of certain immunological and antioxidant indicators in the genes. Results: The culture method detected the pathogens causing mastitis in 90% of the milk samples, but m-PCR detected them in 100% of the milk samples. SNPs linked to mastitis resistance/susceptibility in examined does were detected through DNA sequencing of immunological and antioxidant indicators. The magnitude of gene expression varied significantly between the resistant and mastitis-affected groups. Significant (P ˂ 0.05) elevations were noticed in WBCs count, mainly neutsrophils count, serum levels of BHB, NEFA, triglycerides, LDL-C, AST, ALT, ALP, creatinine, total protein, globulin, Ca, K, GPx, MDA, acute phase proteins, and cytokines in mastitis affected does as compared to control. While RBCs count, PCV%, lymphocytes count, serum concentration of glucose, cholesterol, HDL-C, albumin, Na, Cl, P, GSH, SOD, and catalase significantly (P ˂ 0.05) diminished in mastitis affected does compared to healthy ones. APPs and pro-inflammatory cytokines scored high sensitivities and NPVs but TNF-α and serum amyloid A (SAA) had the highest percentages of increase. Conclusion: The study confirmed that m-PCR is the most sensitive method for bacteria identification (S. aureus, E. coli, and Strept. spp.) while SNPs in antioxidant and immunological genes may be important genetic indicators for mastitis risk or resistance in Shami does. To establish an effective management plan and forecast the most sensitive risk time for illness onset, gene expression profiles of the tested genes may also be employed as proxy biomarkers. TNF-α and SAA may be precious indicators for the detection of caprine mastitis. Keywords: Shami does, Single nucleotide polymorphisms, Antioxidants, m-PCR, Mastitis. IntroductionIt is impossible to overstate the profitable significance of goats, particularly in underdeveloped nations. Worldwide, goats are an extremely useful source of meat, milk, fur, and skin (Wilson, 1987). Most developing nations’ livestock economies and cultures are heavily reliant on goats. The most economically significant part of animal production is goats. They endure harsh environmental conditions in semi-arid and desert regions. According to Joint (2011), Egypt has a total of 4.2 million goats. The Damascus or Shami goat is well regarded by farmers in the eastern Mediterranean region for its outstanding reproductive and productivity performance (Khazaal, 2009). The capacity of Damascus goats to adapt to high temperatures and their appropriateness for breeding in lowlands give them an advantage over sheep in terms of their ability to reproduce while benefiting from arid pastures (Barıtçı and Adıgüzel, 2017). Damascus goats are used extensively to increase the production of other breeds since they are regarded as a dual-purpose animal for the production of meat and milk with a high genetic level (Mavrogenis et al., 2006). Small ruminants frequently develop mastitis, which is characterized by alterations in the physical, chemical, and microbial characteristics of the milk as well as pathological lesions in the glandular tissue (Mishra et al., 2018). Constructed on etiopathological findings and inspections, mastitis is typically categorized as subclinical, acute, subacute, chronic, and gangrenous (Narenji Sani et al., 2015). Teat injuries, poor management and cleanliness, and malfunctioning milking equipment are examples of predisposing factors that have been shown to speed up the spread of infectious organisms and the progression of disease (Zenebe, 2014). The key factors relating to the economic losses of mastitis are milk production decline, deterioration in product quality, and compromised hygiene standards rendering the final product unsuitable for consumption (Mota, 2008). In addition, Staphylococcus species have emerged as the leading culprits behind subclinical or clinical mastitis in numerous countries (Schukken et al., 2009). Important Streptococcus species for bovine mastitis include Strept. agalactiae, Strept.dysgalactiae, and Strept.uberis (Dmitriev et al., 2006). According to Mohamed (2013), they are to blame for over 80% of all mastitis diagnosed in Egypt. It is challenging to distinguish between the many species within this vast group of pathogens (Koskinen et al., 2009). Furthermore, the absence of particular biochemical markers for species identification means that the pathogens causing mastitis cannot be reliably identified simply on phenotypic alterations (Capurro et al., 2009). DNA sequence-based molecular techniques have shown to be an effective means of species identification, rapid, accurate, and trustworthy identification techniques are still needed (Shome et al., 2011). Blood biochemical investigations give much information about animal’s nutritional status, health, and wellbeing; therefore, they can be used to assess the health of animals in general (Faraz, 2019). Based on our present comprehension, the information on immunological, antioxidant states, SNPs, and gene expression alterations associated with caprine mastitis is scarce. In addition, the absence of accurate diagnostic and predictive tools for these conditions hinders our ability to enhance preventative and therapeutic strategies for these disorders. The current aimed to investigate SNPs, the gene expression profile of immune and antioxidant bio-markers, besides identifying the most frequent pathogens causing mastitis in goats using multiplex PCR (m-PCR). Materials and MethodsAnimalsFifty Shami goats with clinical mastitis and 50 normal goats (the control group), 3–8 years (5.1 ± 1.7) and 27.5–45 kg (33.7 ± 5.8), were raised at Mariut Research Station, El-Amria, Alexandria, Desert Research Center (DRC), Egypt. Shami does do ranged in lactation from the first to the seventh. Water was accessible to the does at all times, and they were kept in semi-open shaded pens with 750 g of concentrate feed mixture (CFM) + 750 g of alfalfa hay/head/day for feeding. Table 1 displays the ingredients of the basal diet. When available, the natural pasture which consisted of grass, berseem, darawa, and green herbage was fed. The investigated were put through clinical examination in accordance with the pre-established standard protocols (Pugh, 2012), and the results were documented at the same time. Clinical mastitis was identified based on physical examination of the glands through inspection and palpation (Jackson and Cockcroft, 2002) and evaluation of produced milk for aberrant color and consistency. The processes were carried out, from September 2022 to June 2023. SamplingMilk sampleAccording to Hogan et al.(1999), teats were prepared aseptically before samples were taken manually. 70% ethanol was applied to the mammary teats after cleaning and drying them, and first milk streams were discarded. Then, milk samples (10–15 ml) from both mammary halves of each doe were collected in sterile tubes, labeled, and straight to the laboratory for California mastitis test (CMT) and somatic cell count (SCC) screening. Using a Bently Soma Count 150 (Bentley Instrument, Chaska, MN, USA), somatic cells were counted electronically. The CMT supplied by Taba Medical Pharma Company®, was performed using 3–5 ml of goat milk and an equivalent volume of CMT reagent, and they were quickly combined by swirling or rotating. The amount of gel that developed and the degree of color change were used to rate the response (Gebrewahid et al., 2012). The CMT core was given a value of 0, +1, +2, or +3 depending on the reaction’s severity, and milk samples with CMT scores of 0 and 1 and SCC levels under 200,000/ml were deemed healthy (N=50), whereas milk samples with CMT scores of 2 or 3 and SCC levels above 200,000/ml were deemed to have clinical mastitis (N=50) (Gebrewahid et al., 2012). Table 1. Composition of the concentrate feed mixture (CFM).
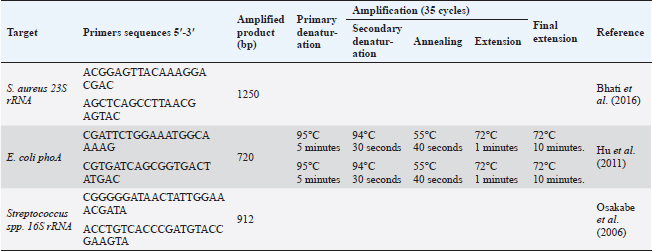
Blood sampleJugular venipuncture was used to get ten ml of blood from each animal. The drawn blood was added to EDTA-containing tubes, heparin-containing tubes, and plain tubes (i.e., anticoagulant-free) to obtain whole blood, heparinized plasma and serum, respectively. All samples were chilled using crushed ice before being brought right away to the laboratory for additional processing. Whole blood tubes were used for both the PCR assay and complete blood count (CBC). Microbiological analysis of milk specimensSamples of milk with a CMT score of +2 or +3 and SCC greater than 200,000/ml were used for the bacteriological test. 10 ml of milk were dispersed using the quadrant streaking method on a 5% sheep blood agar plate. The medium was then examined for typical bacterial colonies after being incubated at 37°C for 24–48 hours. Following their dissemination on brain heart infusion agar, pure colonies on blood agar were incubated for 24 to 48 hours at 37°C in an aerobic environment (Mbindyo et al., 2014). At this stage, the Gram reaction, culture, and shape were used to identify the bacterium (Ewing, 1986). Staphylococcus aureus was located using selective media such as Mannitol salt agar and Baird–Parker agar. As representative colonies, black gram-positive cocci colonies with a clear zone surrounding them were chosen and subjected to a catalase test (Staphylex, Oxoid). Staphylococcus aureus was discovered in colonies that lacked catalase but had hemolysis. To isolate Streptococcus, all isolate clones that seemed to be gram-positive were dispersed on sheep blood agar slants with 1% esculin (Oxoid, UK). Catalase-negative Streptococci isolates were then cultured on kanamycin-esculin-azide agar (Oxoid) after drops of hydrogen peroxide were applied to already-growing cultures. Gram-negative bacilli colonies were then distributed on MacConkey agar plates for isolating Escherichia coli, where the pink color of the bacterial colonies indicates that E. coli is growing. The most probable number (MPN) method, which uses three tubes, was used to confirm the E. coli isolates. The positive MPN tube was disseminated on Eosin methylene blue (EMB) agar and allowed to develop overnight at 35°C. A green metallic sheen dye is produced around the bacterial growth in 24–36 hours. Bacterial identificationStaphylococcus aureus, E. coli, and Strept. spp. standard bacterial reference strains ATCC 25922, 25922, and 15913 were received for comparison from the veterinary quality control on poultry production, Animal Health Research Institute, Cairo, Egypt. The QIAamp DNA Mini kit (Qiagen, Germany, GmbH) was used to extract DNA from milk samples, with certain modifications made in accordance with the manufacturer’s instructions. 200 µl of the sample suspension was incubated with 10 µl of proteinase K and 200 µl of lysis buffer at 56°C for 10 min. After incubation, 200 µl of 100% ethanol was added to the lysate. The sample was then washed and centrifuged following the manufacturer’s recommendations. Elution buffer (100 µl) was used to elute the nucleic acid. The primers utilized were given by Metabion (Germany) and are listed in Table 2. Primers were utilized in a 25 µl reaction containing 12.5 µl of EmeraldAmp Max PCR Master Mix (Takara, Japan), 1 µl of each primer of 20 pmol concentrations, 5.5 µl of water, and 5 µl of DNA template. An applied biosystem 2,720 heat cycler was used to carry out the process. On a 1.5% agarose gel (Applichem, Germany, GmbH) in 1× TBE buffer at room temperature, the PCR products were separated using 5 V/cm gradient electrophoresis. Each gel slot had 20 µl of the m-PCR products put into it for gel analysis. The sizes of the fragments were measured using a gelpilot 100 bp plus ladder (Qiagen, Gmbh, Germany), a generuler 100 bp ladder (Fermentas, Germany), a 100 bp DNA ladder H3 RTU (Genedirex), and a generuler 100 bp ladder. Computer software was used to analyze the data after the gel was photographed by a gel documentation system (Alpha Innotech, Biometra). Table 2. Primers sequences, target genes, amplicon sizes, and cycling conditions.
Table 3. Forward and reverse primer sequence, length of PCR product, and annealing temperature for immune and antioxidant genes used in PCR-DNA sequencing.
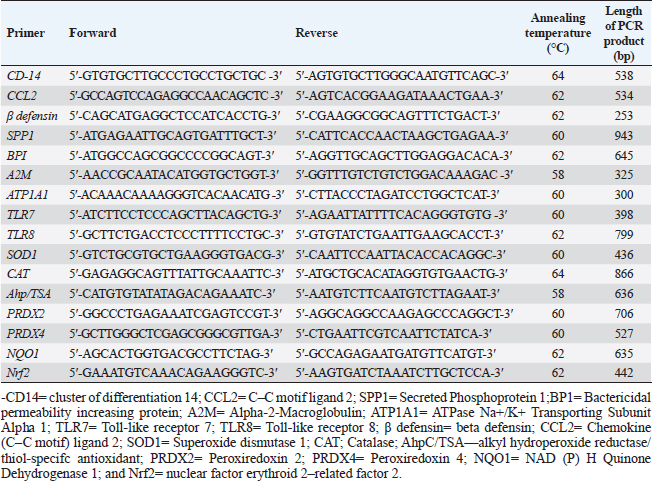
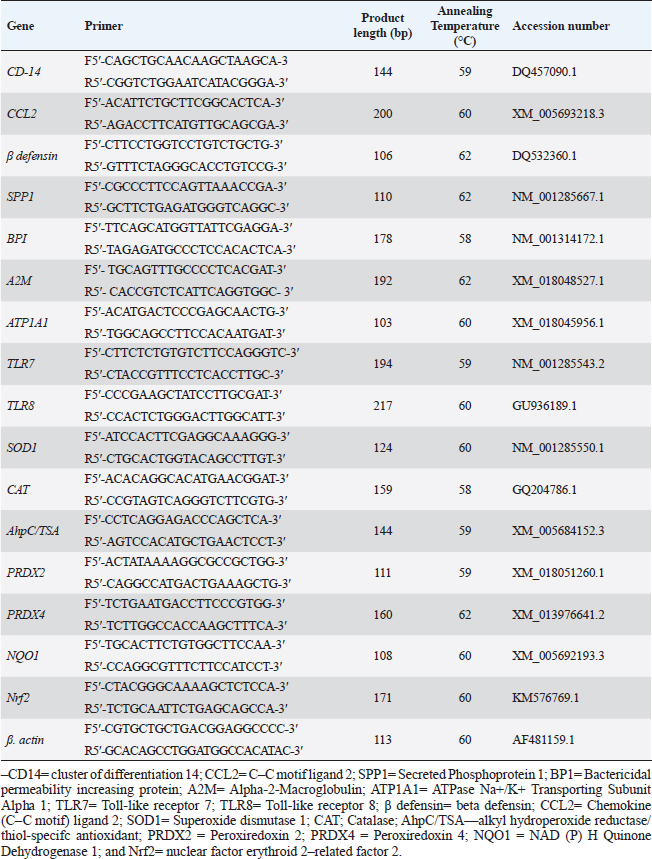
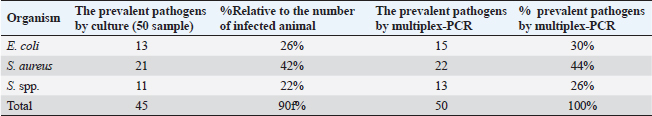
PCR-DNA sequencing of immune and antioxidant genesUtilizing the Gene JET whole blood genomic DNA extraction kit and adhering to the guidelines provided by the company, the genomic DNA was extracted from whole blood (Thermo Scientific, Lithuania). DNA that was high quality, pure, and concentrated was tested by Nanodrop. The amplification of coding elements was done via PCR for immune (CD-14, CCL2, β defensin, SPP1, BP1, A2M, ATP1A1, TLR7, and TLR8) and antioxidant (SOD1, CAT, AhpC/TSA, PRDX2, PRDX4, NQO1, and Nrf2) genes. The sequence of Capra hircus that was published in PubMed was employed in the primer sequence design. Table 3 presents the primers utilized for the amplification. A 100 μl final volume of the polymerase chain reaction mixture was produced in a thermal cycler. 5 μl DNA, 43 μl H2O (d.d water), 50 μl PCR master mix (Jena Bioscience, Germany), and 1 μl of each primer were included in each reaction volume. For six minutes, the reaction mixture was exposed to the starting denaturation temperature of 95°C. The cycling was done in 35 cycles: 45 seconds at 95°C for denaturation, 1 minute for annealing temperatures according to Table 3, 45 seconds at 72°C for extension, and 10 minutes at 72°C for the last extension. The exemplary PCR analysis findings were obtained using agarose gel electrophoresis, and the samples were kept at 4°C for storage. The fragment patterns were then observed using an ultraviolet gel documenting technique. DNA sequencing was preceded by the removal of primer dimmers, nonspecific bands, and other impurities. To purify PCR products with the anticipated size (target bands), a Jena Bioscience #pp-201×s/Germany PCR purification kit was utilized, as stated by (Boom et al., 1990). The manufacturer’s instructions were followed. Using a Nanodrop (Uv-Vis spectrophotometer Q5000/USA), the PCR product was quantified to guarantee sufficient quantities and purity as well as a high product yield (Boesenberg-Smith et al., 2012). Using an enzymatic chain terminator technique developed by Sanger et al. (1977), PCR products with the target band were sent for forward and reverse DNA sequencing using an ABI 3730XL DNA sequencer (Applied Biosystem, USA) to detect SNPs in the genes investigated in the control and mastitis affected does. Utilizing the programs Blast 2.0 and Chromas 1.45, DNA sequencing data analysis was done (Altschul et al., 1990). Genes under investigation were subjected to PCR product comparison with reference sequences from GenBank, and these differences were categorized as single-nucleotide polymorphisms (SNPs). Tamura et al. (2007) evaluated the genes between enrolled does using the MEGA6 software program to do an amino acid sequence variation analysis based on data alignment of DNA sequencing. Quantitative real time PCR for immune and antioxidant genesTrizol reagent was used to extract total RNA from doe blood in accordance with the manufacturer’s instructions (RNeasy Mini Ki, Catalogue no. 74104). Using a NanoDrop® ND-1000 Spectrophotometer, the amount of isolated RNA was measured and certified. The manufacturing protocol (Thermo Fisher, Catalogue no. EP0441) was followed in the synthesis of the cDNA for every sample. Quantitative RT-PCR was used with SYBR Green PCR Master Mix (2× SensiFastTM SYBR, Bioline, CAT No: Bio-98002) to evaluate the gene expression pattern of genes encoding immunity and antioxidants. SYBR Green PCR Master Mix (Quantitect SYBR green PCR kit, Catalogue no. 204141) was used to perform real-time PCR for the relative measurement of the mRNA level. The Capra hircus published sequence in PubMed was followed in the creation of primer sequences, as indicated in (Table 4). For normalization, the housekeeping gene β. actin was employed as a constitutive control. In all, 3 µl of total RNA, 4 µl of 5× Trans Amp buffer, 0.25 µl reverse transcriptase, 0.5 µl of each primer, 12.5 µl 2× Quantitect SYBR green PCR master mix, and 8.25 µl RNase free water made up the 25 µl reaction mixture. Using a thermal cycler, the final reaction mixture was subjected to the following protocol: reverse transcription for 30 minutes at 50°C, initial denaturation for 10 minutes at 94°C, 40 cycles of 94°C for 15 s, annealing temperatures as indicated in Table 4, and 72°C for 30 s. A melting curve analysis was carried out at the conclusion of the amplification stage to verify the PCR product›s specificity. Using the 2−ΔΔCt technique, it was established how each gene’s relative expression in each sample related to the β. actin gene (Pfaffl, 2001). Hematological and biochemical parametersSerum biochemical tests were performed using commercial test kits in accordance with the providers’ established protocols, and hematological parameters (RBCs, Hb, PCV, MCV, MCH, MCHC, WBCs, neutrophils, lymphocytes, monocytes, and eosinophils counts) was manually calculated according to Feldman et al. (2000). GSH, GPx, MDA, SOD, and CAT were detected (Biodiagnostic Egypt, CAT No: MD2529, CA252417, GSH2511, SD 25 21, and GP 2524). Serum haptoglobin (Hp) concentrations were determined by Eagle Biosciences (Columbia) ELISA kits, serum amyloid A (SAA), and plasma fibrinogen (Fb) concentrations were determined by using IBL International Crop (Canada)® ELISA kits. Pro-inflammatory cytokines (IL-1β, IL-6, and TNF-α) levels were determined in serum by using BOSTER BIOLOGICAL TECHNOLOGY PicoKine™ ELISA kits (CAT No: EK0412, EK0393, and EK0526, respectively). Statistical analysisSPSS (version 20, Inc., Chicago, USA) was used to perform the statistical analyses. All parameters were given descriptive statistics, and the means of the two groups were compared by using the Student’s t-test. When p < 0.05, the results were considered significant. ROC curve analysis for the measured APPs and pro-inflammatory cytokines was done using the Graph Pad Prism 8 program in mastitic goats compared to normal ones. Ethical approvalThe processes were carried out in compliance with the regulations set forth by the Ministry of Agriculture and Land Reclamation in Egypt’s Animal and Poultry Health Department, Desert Research Centre, and Ethical Committees. ResultsClinical findingsClinically, none of the healthy Shami producing the typical, regular milk had any mammary anomalies. Does with mastitis had swollen, hot, hard, and extremely painful udders. Milk production was also drastically reduced, and the milk’s appearance was dense and yellow. The breathing and body heat of this group were increased. Prevalence of clinical mastitisOut of 100 goat milk samples analyzed, the results showed that 50 of them tested positive for California mastitis (50%). From 50 samples, 45 had bacteriologically proven bacterial contamination with S. aureus (n=21) (42%), E. coli (n=13) (26%), and Strept. spp. (n=11) (22%), as indicated in (Table 5). When pathogen occurrence used to be decided by the usage of m-PCRPCR on milk samples that had been infected, the effects were greater favorable than those obtained by way of microbiological subculture with 44% S. aureus, 30% E. coli, and 26% Strept. spp. It found two extra E. coli isolates that have been observed in the culture, one greater S. aureus, and two Strept. spp. (Table 5). The presence of combined contamination in 25 animals used to be noted. As indicated in Figure 1, E. coli and S. aureus had been the maximum common combined infections (8/17, 47%), accompanied with the aid of E. coli and Strept. spp. (5/9, 55.5%) and S. aureus, and Strept. spp. (2/14, 14%). Table 4. Oligonucleotide primers sequence, accession number, annealing temperature, and PCR product size of immune and antioxidant genes used in real time PCR.
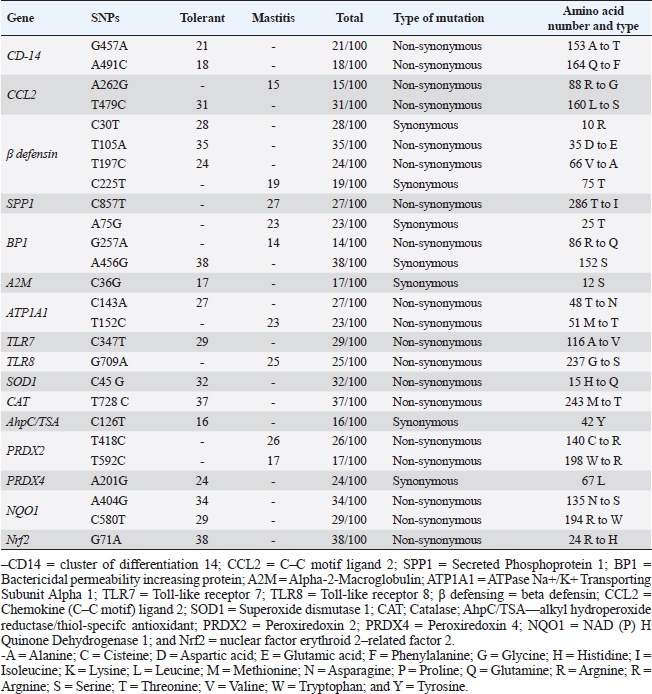
Table 5. Distribution of SNPs, type of mutation in immune, and antioxidant genes for tolerant and affected does to mastitis.
Immunity and antioxidant gene polymorphisms associated with mastitis incidencePCR-DNA sequencing of CD-14 (538-bp), CCL2 (534-bp), β defensin (253-bp), SPP1 (943-bp), BP1 (645-bp), A2M (325-bp), ATP1A1 (300-bp), TLR7 (398-bp), TLR8 (799-bp), SOD1 (436-bp), CAT (866-bp), AhpC/TSA (636-bp), PRDX2 (706-bp), PRDX4 (527-bp), NQO1 (635-bp), and Nrf2 (442-bp) revealed mastitis resistance/susceptibility in the examined does is correlated with nucleotide sequence changes in the formula of SNPs. Nucleotide sequence variation in the genes under study between the affected and resistant does, and between the examined animals and GenBank reference sequences, validated all of the SNPs that were found. Coding mutations between the mastitic did and the healthy doe was caused by the exonic changes that are displayed in Table 6 in all immunological and antioxidant indicators that were investigated.
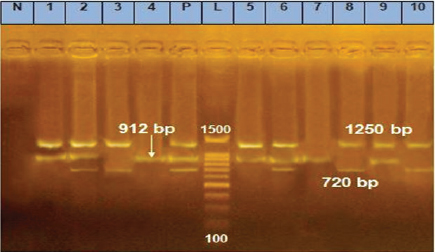
Fig. 1. m-PCR assay, amplifying fragments of 720, 912, and 1250 bp were amplified for E. coli, Strept. spp., and S. aureus, respectively. Lane1: 100–1500 bp marker, Lanes p: positive control of E. coli, Strept. spp., and S. aureus standard strains, Lane N: negative control with no DNA, Lanes 2, 3, 6, 8, and 10 positive for E. coli, Lanes 1, 2, 4, 5, 6, 7, and 9 positive for Strept. spp., Lanes 1, 2, 5, 6, 8, 9, and 10 positive for S. aureus. Table 6. Pathogen existence in milk.
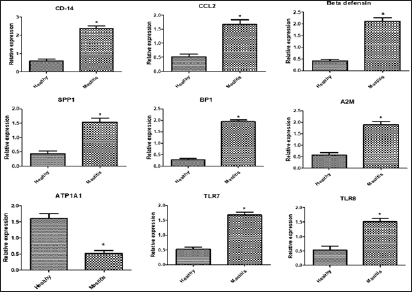
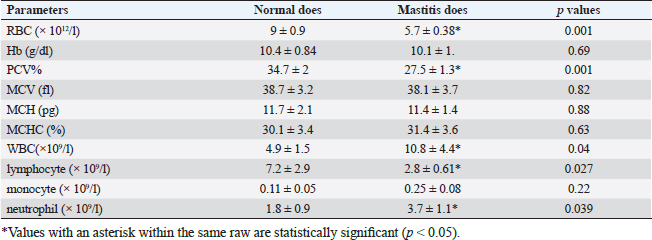
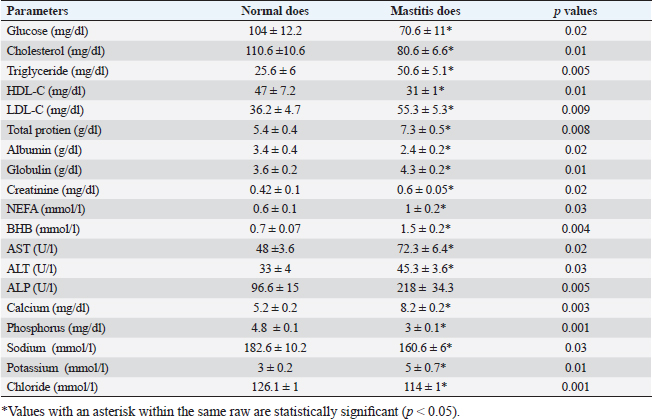
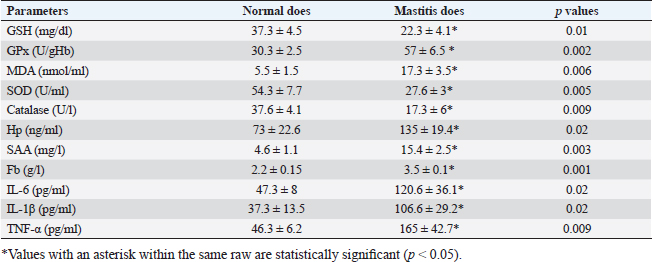
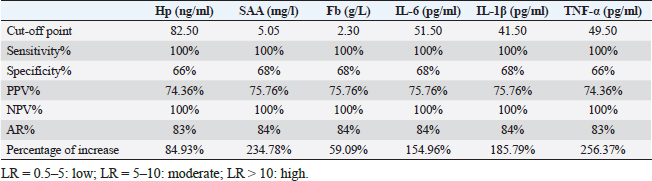
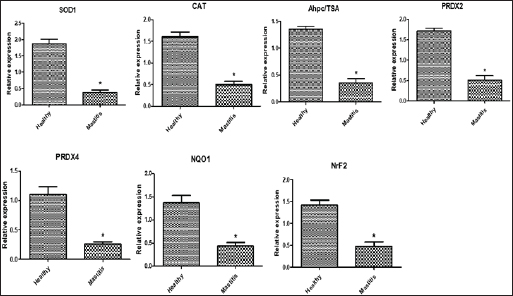
Gene expression configuration of immune and antioxidant markersThe immunological and antioxidant markers’ gene expression profiles are displayed in Figures 2 and 3. Mastitis-affected does have significantly greater expression levels of the genes CD-14, CCL2, β defensin, SPP1, BP1, A2M, TLR7, and TLR8 than resistant does. Mastitis-affected does have significantly lower expression of the genes ATP1A1, SOD1, CAT, AhpC/TSA, PRDX2, PRDX4, NQO1, and Nrf2 than resistant does. Hemato-biochemical and immunological alterations associated with goat mastitisThere were significant (p < 0.05) elevations in WBCs and neutrophils count while significant (p < 0.05) decreases in RBCs, PCV, and lymphocytes count in mastitic does as compared with healthy does (Table 7). However, Hb, MCV, MCH, MCHC, and monocyte count were non-significantly altered (p ≥ 0.05) in mastitic does in relation to the control group (Table 7). There were significant (p < 0.05) augments in serum values of BHB, NEFA, triglycerides, LDL-C, AST, ALT, ALP, creatinine, total protein, globulin, calcium, and potassium with significant (p < 0.05) declines in serum concentrations of glucose, cholesterol, HDL-C, albumin, sodium, chloride, and phosphorus in mastitic does compare to healthy ones (Table 8). There were significant (p < 0.05) rises in serum levels of GPx and MDA with significant (p < 0.05) reductions in serum concentrations of GSH, SOD, and catalase in mastitic does in comparison with healthy ones (Table 9). There was a significant (p < 0.05) increase in APPs and cytokines concentrations in mastitic does in relation to healthy ones (Table 9). ROC curve analysis of the estimated APPs and pro-inflammatory cytokines cleared that they yielded at AUC=1, sensitivity, and NPV as 100%, with high PPVs and ARs (more than 70%) while their specificities and LRs were low. They were almost equal but TNF-α and SAA had unique percentages of increase of 256.37% and 234.78%, respectively (Table 10). DiscussionMastitis, especially in the postpartum period, is the term used to describe the inflammation of one or more of the mammary gland tissues that customarily occur before lactation (Ondieki, 2018). Infections are typically characterized by local inflammatory symptoms, yet depending on how severe the infection is, systemic symptoms may also be present (Mohammed et al., 2016).
Fig. 2. Relative expression patterns of immune genes in healthy and mastitic Shami goats. Results are expressed as means ± SEM. *p < 0.05. Table 7. Hematological parameters of normal (N=50) and mastitic (N=50) Shami goats.
The culture method confirmed the presence of the mastitic pathogens in 95% of milk samples and m-PCR results committed their presence in 100% of milk samples. Whereas, S. aureus was identified in 42% of raw milk samples by culture method and in 44% of raw milk samples by m-PCR method. These results were in agreement with those reported in (43.1%) of dairy does in Italy (Cortimiglia et al., 2015) but lower than found in (50%) of buffalo in Bangladesh (Talukder et al., 2013), (76.9%) of goats in Sardinia (Spanu et al., 2013), and higher than those recorded in (30%) of cow in Egypt (Saad et al., 2019), (38.3%) of sheep and goats in Turkey (Abdallah et al., 2018), (35%) of goat in India (Mishra et al., 2018), (4.5%) of camels at Matrouh Governorates, Egypt (Darwish, 2023), and (13.5%) of goat in Brazil (Rupp et al., 2019). According to Le Loir et al. (2003), inappropriate hygiene procedures, handling of animals during milking, and milk storage could all be contributing factors to the spread of Staphylococcus spp. in milk. In the present data, the isolation rate of E. coli by culture method was 26% and by m-PCR was 30% (focusing on the phoA gene). Our findings were consistent with Sameer et al. (2013) (26.67%) in Egypt, but higher than those reported by Abd El-Tawab et al. (2018) (9.5%) in Sharkia and Dakahlia Governorates, Egypt, Darwish (2023) (12.7%) at Matrouh Governorate, Egypt, Lan et al. (2020) (18.5%) in China, Haftay et al. (2016) (21.3%) in Ethiopia and lower than percentages reported by Abdallah et al. (2018) and Saad et al. (2019) (44% and 36.8%, respectively) in Egypt and Machado et al. (2018) (51.9%) in Brazil. The highest prevalence of E. coli as an environmental pathogen in milk samples from infected goats may be attributed to management errors such as overcrowding, insufficient ventilation, inadequate manure removal, subpar hygienic conditions, and an overall deficiency in farm cleanliness and sanitation (Abdallah et al., 2018). Table 8. Biochemical parameters of healthy (N=50) and mastitic (N=50) Shami goats.
Table 9. Oxidative stress markers, acute phase proteins and pro-inflammatory cytokines in normal does (N=50) and those with clinical mastitis (N=50).
According to our investigation, 22% of Strept. spp. (n=11) were found in milk samples of mastitic Shami goats while m-PCR showed the presence of 26% of Strept. spp. (n=13) (two more isolates than culture). This was almost the same results reported in 22.5% of dairy cows in China (Ding et al., 2016), and 23% of cows in India (Mishra et al., 2018). 25% of goats in Egypt (Mohamed, 2022), but less than other studies reported (31.2%) of cows in Egypt (Saad et al., 2019), 65% of bovine in Nigeria (Amosun et al., 2010), and higher than 8.7% of cattle in China (Han et al., 2022), 9% of cattle in Pakistan (Ali et al., 2021), 12% of cattle in Egypt (Ahmed et al., 2020), 15.4% of camels in Egypt (Darwish, 2023), 17.4% of small ruminant farms in Denmark (Chatzopoulos et al., 2020). Changes in topographical features and management practices may be to blame for the variation in the percentage of isolated Strept. spp. Table 10. ROC curve analysis of the estimated acute phase proteins and cytokines in mastitic does compare to normal does.
Fig. 3. Relative expression patterns of immune genes in healthy and mastitic Shami goats. Results are expressed as means ± SEM. *p < 0.05. Mutations in immune function-related genes are partly responsible for the rumininants’ sensitivity to harmful microorganisms (Michel et al., 2003). Within this context, fragments found by PCR-DNA sequencing of CD-14 (538-bp), CCL2 (534-bp), β defensin (253-bp), SPP1 (943-bp), BP1 (645-bp), A2M (325-bp), ATP1A1 (300-bp), TLR7 (398-bp), TLR8 (799-bp), SOD1 (436-bp), CAT (866-bp), AhpC/TSA (636-bp), PRDX2 (706-bp), PRDX4 (527-bp), NQO1 (635-bp), and Nrf2 (442-bp) genes identified SNPs, or nucleotide sequence changes, in mastitis-resistant and afflicted does. An acceptable explanation for that will be finding SNPs in immune and antioxidant genes is a novel finding in this research that may be relevant to Shami goats’ vulnerability or resistance to mastitis. Upon comparing our findings with the relevant GenBank reference sequence, it was intriguing to observe that the variations discovered in the genes under investigation were initially reported. However (Al-Sharif, 2022), identified DNA polymorphisms in the antioxidant and immunological genes as biomarkers for the tolerance and susceptibility to reproductive diseases in Baladi goats. TLR7 and TLR8 single nucleotide polymorphisms (SNPs) and their relationship to the pro-viral load of small ruminant lentiviruses (SRLVs) in goats were found by (Olech et al., 2021). Relating to mastitis in ruminants and polymorphisms in the immune system and antioxidant genes; TLR4 and SOD SNPs in sheep were linked to postpartum diseases susceptibility in Barki sheep according to Darwish et al. (2023), who assessed the connection between SPP1 and features related to milk yield and composition. A connection between the SPP1SNP and somatic cell score was found by statistical analysis. In addition, discovered a link between mastitis resistance and β-defensin gene polymorphisms (Tolone et al., 2016). Investigated SNPs in immunological (SELL, ABCG2, SLC11A1, and FEZL) and antioxidant (SOD1, CAT, GPX1, and AhpC/TSA) genes linked with mastitis risk in Holstein and Brown Swiss dairy cows were investigated (Ateya et al., 2022). The CD14 gene polymorphism and its link to mastitis was reported (Kumar et al., 2014). The osteopontin transcript (SPP1) was described by (Alain et al., 2009) as hereditary variations that exist in an early innate immunological candidate of E. coli mastitis that may be connected to mastitis resistance. The existence of SNPs in the CCL2 gene and evaluated how they would affect the variation in somatic cell score (SCS) in Holstein bulls was discovered (Leyva-Baca et al., 2007)Schenkel, Sharma, Jansen, & Karrow, 2007. The regulatory changes in the A2M gene contributed to dairy cows’ susceptibility to mastitis (Wang et al., 2014). Mastitis and heat tolerance have both been linked to ATP1A1 gene polymorphism in earlier studies (Elayadeth-Meethal et al., 2021). According to studies on mastitis resistance in buffalo, it was found that variation in the A2M gene was connected to SCC, subclinical mastitis, protein production, fat, milk yield, fat and protein percentages, and mastitis resistance (Mingala et al., 2020). Our findings demonstrated that affected does have a higher pattern of immunological marker expression than resistant does, although antioxidant gene expression showed the reverse tendency. This is the first study to determine the immunological and antioxidant mRNA levels in mastitis-resistant and -susceptible individuals using a real-time PCR approach. We investigated gene polymorphism utilizing SNP genetic markers and gene expression to get beyond the shortcomings of earlier research. We hypothesize that the functional aspects of the genes in caprine mastitis may be impacted by the SNPs found in the examined genes. Consequently, it is well understood how the genes related to immunity and antioxidants that are being studied, both in healthy and mastitis-affected animals, are controlled. Examining the goat mammary gland’s reaction to S. aureus infection was done by Cremonesi et al., (2012) using gene expression profiling in milk somatic and white blood cells. The highest increases in expression were seen in the NFKB1, TNFAIP6, BASP1, IRF1, PLEK, and BATF3 genes after challenge. Goats with S. aureus mastitis compared to healthy goats, Pisoni et al. (2010) showed that pro-inflammatory cytokines, chemokines, and their receptors—IL-1a, lymphotoxin alpha, granulocyte chemotactic protein (CXCL6), and IL-2 receptor gamma were all significantly overexpressed. In previous research examining the gene expression profile of immunological and antioxidant markers linked to ruminant mastitis, Wang et al. (2012) showed that mastitis-infected bovine mammary tissues had considerably greater levels of A2M mRNA and protein expression than did healthy tissues. According to Ateya et al. (2022), compared to tolerant cows, mastitic Holstein and Brown Swiss dairy cows exhibited significantly greater levels of SELL, SLC11A1, and FEZL gene expression. The genes for GPX1, AhpC/TSA, SOD1, CAT, ABCG2, and SOD1 were significantly down-regulated. In addition, Kosciuczuk et al. (2014), found that the expression pattern of β-defensin is significantly higher in bovine mammary glands infected with coagulase-positive or coagulase-negative Staphylococci than in udders free of bacteria. Asadpour et al. (2021) looked into variations in antioxidant gene expression during clinical mastitis in cows brought on by E. coli and S. aureus. They discovered that, in contrast to E. coli, S. aureus-induced mastitis milk had far higher levels of SOD expression. Furthermore, in terms of mRNA levels, GPx was substantially overexpressed in milk from mastitis induced by E. coli as opposed to S. aureus. Ewes with postpartum issues had significantly greater levels of IL-5, IL-6, IL1-ß, TNF-α, TLR4, and Tollip than resistant ewes; conversely, SOD and CAT gene patterns were down-regulated. CD14 is a crucial part of innate immunity. One of the most significant molecules that binds and neutralizes bacterial endotoxins is the anti-bacterial peptide CD14 (Wright et al., 1990). By binding to the CCR2 receptor that is found on the surface of monocytes and neutrophils, respectively, CCL2 functions as an effective chemokine for these cells (Proudfoot, 2002). β-defensins were thought to be the only antibacterial substances made by leucocytes and epithelial cells that acted as the initial barrier against pathogens (Caverly et al., 2003). A cytokine called secreted phosphoprotein 1, also known as SPP1 or osteopontin, is created by activated T cells and macrophages (Chabas et al., 2001). BPI is an endogenous cationic bactericidal/permeability-increasing protein. In addition to eliminating Gram-negative bacteria, endotoxins, and lipopolysaccharide (LPS), which is another name for endotoxin, are neutralized. BPI serves a variety of biological purposes, including enhancing opsonization and complement activation for amplified phagocytosis, inhibiting angiogenesis and the release of inflammatory mediators, and guarding against infection by fungi and protozoan pathogens (Balakrishnan et al., 2013). Along with the complement system’s C3, C4, and C5, alpha-2-macroglobulin (A2M) is a member of the alpha-macroglobulin (aM) family of proteins (Levashina et al., 2001). In addition, it promotes the growth of macrophages and T cells (Bonacci et al., 2007). Genetic variations of TLRs have been discovered to be related to various disease outcomes (Skevaki et al., 2015). Numerous viral infections have been linked to TLR7 and TLR8 polymorphisms (Zhang et al., 2020). A transmembrane enzyme called Na+, K+- ATPase uses ATP to move 3 Na+ out of and 2 K+ into a mammalian cell via the plasma membrane (Rakowski et al., 1989). To regulate cell volume, osmotic balance, and appropriate resting membrane potentials, ionic homeostasis in the cytoplasm is made easier by this (Pavlov and Sokolov, 2000). Antioxidants fight free radicals by scavenging them, detoxifying them, preventing their generation, or sequestering the transition metals that produce them (Masella et al., 2005). The primary inducible defense against oxidative stress is the Keap1-Nrf2 stress response system, which controls the production of cytoprotective genes (Yamamoto et al., 2018). According to Bianchet et al. (2004), the antioxidant NQO1 protects against harmful oxidative by cleansing radicals and peroxides that include sulfur, thiol-specific peroxidase serves as a sensor of hydrogen peroxide-mediated signaling events and supports cellular defense against oxidative stress (Ateya et al., 2021). The significant change in the immune (CD-14, CCL2, β defensin, SPP1, BP1, A2M, TLR7, and TLR8) and antioxidant (SOD1, CAT, AhpC/TSA, PRDX2, PRDX4, NQO1, and Nrf2) expression patterns in mastitic may be ascribed to the severe inflammation that harms the affected tissue and causes the release of cytotoxic radicals and pro-inflammatory cytokines, the excess ROS weakens the immune system in the absence of aentire antioxidant that is appropriate (Sordillo and Aitken, 2009). Our research showed that mastitics do have much lower levels of ATP1A1 mRNA than healthy ones. Mastitis has been proven to have an impact on milk’s potassium and sodium levels (Liu et al., 2012). Milk has a higher concentration of K+ than Na+ because Na+, K+ -ATPase, which is positioned on the basolateral membrane, actively removes Na+ from the secretory cells of the mammary glands (Linzell and Peaker, 1971). However, mastitis causes changes to the K+ and Na+ contents in milk. K+ concentration falls and Na+ concentration rises in cows with subclinical mastitis (Auldist and Hubble, 1998). As a result, milk’s osmolality changes. As a result, since the Na+, K+ -ATPase system is crucial for the osmoregulation of milk, information on its sensitivity to mastitis would be of relevance. When mastitis affects cows, the osmolality of their milk may alter, indicating a decrease in the activity of the Na+, K+ -ATPase enzyme. Our study’s findings unequivocally show that, in comparison to healthy goats, the hematology of mastitic Shami goats was significantly altered. Of particular concern was the marked increase in WBCs, neutrophils count, and lymphopenia, likely due to differential immunomodulatory effects resulting from increased demand for leukocytes in the infected udder (Carvalho-Sombra et al., 2021). Disease-related stress and neutrophilia (subsequent leukocytosis) as well as lymphopenia may develop as a result of the endogenous production of corticosteroids in the mastitic group (Duncan, 1994; Kaneko, 1997). Our finding was similar to that obtained in mastitis in goats (Abba, 2013), in cattle (Saleh et al., 2022), and in camels (Darwish, 2023). In the current study, there were noticeable decreases in RBCs and PCV between mastitic and healthy does. These findings agreed with the findings of Hristov (2018) in goats, Saleh et al. (2022) in cattle, and Darwish (2023) in camels, but away from that reported by Garba et al. (2019) in Red Sokoto goats or Sarvesha et al. (2016) in Indigenous cow who did not observe any significance difference of Hb and RBCs level of mastitic animal as opposed to healthy ones. There were not any noteworthy changes in values of Hb, MCV, MCH, MCHC, and monocytes between the mastitic and control groups. These findings were opposite to those found by Saleh et al. (2022) in cows who confirmed considerably reduced Hb, MCV, MCH, and MCHC values when in contrast to controls, indicating microcytic hypochromic anemia, which is frequent in inflammatory disorders. The authors delivered that, inflammatory prerequisites and the pro-inflammatory cytokines stimulate the mononuclear phagocytic system to elevate iron trapping and storage, which is notion to be a protecting mechanism with the aid of lowering iron availability to microorganisms, ensuing in a reduction in Hb synthesis and the manufacturing of microcytic hypochromic RBCs (Stockham and Scott, 2013). The considerable increase in the pro-inflammatory cytokines IL-6 in the mastitic group in our study lends weight to this opinion. The occurrence of hypoglycemia and the observed increase in serum levels of BHB, NEFA, triglycerides, LDL-C, and total lipids in mastitic goats compared to healthy ones may result from reduced appetite in afflicted does, leading to heightened lipolysis, the onset of acidosis, and a ketotic state (Kaneko, 1997; El-Deeb, 2013). On the other hand, the noted reduction in serum cholesterol and HDL-C in diseased does than in healthy does, may be attributed to the hepatic insufficiency usually observed with the disease (Kaneko, 1997). Our results were consistent with those seen in mastitic does (El-Deeb, 2013) and mastitic she-camels (Darwish, 2023). The current study recorded higher serum total protein levels in mastitic cases compared to healthy ones, which may be explained by the inflammatory process taking place in the tissue of the mammary glands triggering the body’s immunological system. In this study, we also found that mastitic goats’ serum globulin levels were higher than non-mastitic does’. This could happen as a result of the gamma-globulin antibody production that is responsible for blocking the effects of the invasive microorganism (Nicholas, 2005). This finding was in part similar to that given by Garba et al. (2019) in Red Sokoto goats, and Das et al. (2018) in cattle and she-camels Darwish (2023). Contrariwise, the mastitic in our data suffered from obvious hypoalbuminemia, this may be attributed to the albumin nature as a negative APP which decreases in response to inflammation to conserve amino acid for synthesis of positive APPs as haptoglobin (Darwish, 2023). The present research revealed that does afflicted with mastitis exhibited elevated levels of creatinine. This increase was thought to be the result of severe necrosis in the udder tissue, which has an abundance of smooth muscle. This result shared some similarities with those reported for ewes (Çetin et al., 2005) and camels (Darwish, 2023). Similarly, the serum activity of AST, ALT, and ALP markedly increased in the current study, which is indicative of liver dysfunction. This result shared some similarities with those reported in does (Das et al., 2018), cattle (Garba et al., 2019), and camels (Darwish, 2023). Our study’s findings were in contrast to those of Çetin et al. (2005) who studied ewes mastitis, and Sarvesha et al. (2017) and Sarvesha et al. (2016) who studied mastitis in indigenous and crossbred cows, respectively, who found no difference in serum hepatic enzymatic activities between diseased and healthy animals. Our data about mastitis referred to hypercalcemia in the mastitic does, this may be explained by the fact that infected animals produce less milk, which results in less calcium being excreted from the milk (Wegner and Stull, 1978). These consequences had been similar to those proven in preceding reviews by Das et al. (2018) and Sarvesha et al. (2017) in cows. The mastitic does (in relation to healthy does) in the present data also showed a pronounced hyperkalemia. This result was attributed to the above-mentioned ketoacidosis and the subsequent H+ accumulation. The body tries to get rid of accumulated H+ by pumping it intracellularly leading to K+ pump extracellularly (Kaneko et al., 1997). Hyperkalemia was noted in mastitis-affected cow and buffaloes (Das et al., 2018; Sarvesha et al., 2017). On the contrary, prominent hyponatremia, hypochloremia, and hypophosphatemia were depicted between the diseased and healthy ones. Anorexia and renal dysfunctions may be acceptable causes for these deficiencies. Similar opinions were mentioned in mastitis in Sahel goats (Abba, 2013) and in she camels (Darwish, 2023). In the current research, there was a marked higher level of GPx and MDA in mastitic does than healthy ones, along with a significantly lower level of GSH, SOD, and catalase. These alterations could be attributed to the increased demand for antioxidant activities due to elevated oxidative damage resulting from inflammatory responses in mammary gland tissue. Our results were in line with those reported in ewes (Çetin et al., 2005), does (El-Deeb, 2013), and camels (Darwish, 2023). The current records clear that tissue damage and inflammation may be influenced by the raised serum oxidative stress indicators in clinically mastitic does. The present data showed that clinically mastitic does had higher APPs (Hp, SAA, and Fb), and pro-inflammatory cytokines (IL-6, IL-1β, and TNF-α)levels than did healthy does. Gabay and Kushner (1999) mentioned that the release of TNF-α as a result of inflammation may enhance the increased levels of APPs. The elevated APPs and pro-inflammatory cytokines levels are consistent with those found in does with gangrenous mastitis (El-Deeb, 2013), ewes with experimentally induced mastitis with Staphylococcus (Winter et al., 2003). In this work, the signals control neutrophil activity and recruitment to infection places are chemotactic agents produced by infecting bacteria as well as different immune system components. However, there are drawbacks to this neutrophil influx. Tissue damage can result from this, which has the potential to lead to fibrosis and a decline in mammary function. Previous research emphasized the importance of APPs and pro-inflammatory cytokines in diagnosing and predicting various diseases including mastitis (El-Deed, 2013; Darwish, 2023). The ROC curve analysis conducted in the current study further validated this assertion. Despite their relatively low specificities and LRs, these biomarkers exhibited high sensitivity, PPVs, NPVs, and ARs with AUC values. Notably, the percentage increase of TNF-α and SAA singled them out as dependable markers for diagnosing goat mastitis. ConclusionThe current investigation concluded that the m-PCR assay was better than the culture method in the identification of major bacterial species causing mastitis in Shami goats (S. aureus, E. coli, and Strept. spp.). Our results emphasize the importance of SNPs as genetic markers in genes related to immunity and antioxidant defense which would determine the predispose individuals to either resistance or susceptibility to mastitis. Treating caprine mastitis requires a comprehensive approach that considers not only the clinical symptoms but also the associated alterations in immune response and hemato-biochemical parameters. TNF-α and SAA may be reliable biomarkers for the diagnosis of goat mastitis. AcknowledgmentsThe authors acknowledge staff members of the Animal Health and Poultry Department, Desert Research Center, Egypt. Conflict of interestThe authors declare that they have no conflict of interest. FundingThe authors received no financial support for the research, authorship, and/or publication of this article. Authors contributionsAhmed El-Sayed conceived, and designed the experiment, collected blood samples, performed biochemical analysis, and wrote the manuscript. Ahmed Ateya performed real-time PCR and contributed to writing the manuscript. Amani A. Hafez conducted m-PCR and contributed to writing the manuscript Eman Ebissy and Asmaa Darwish analyzed the data and contributed to writing the manuscript. Data availabilityOn reasonable request, the corresponding author will provide the information supporting the study’s conclusions. ReferencesAbd El-Tawab, A., Aggour, G., El-Hofy, F. and El-Mesalami, M. 2018. Antimicrobial resistance and virulence characterization of E. Coli isolated from subclinical mastitic sheep and goats. BVMJ. 34(3), 267‒278. Abba, Y. 2013. Alterations in hematological and serum biochemical parameters of sahel goats with clinical mastitis. J. Agric. Vet. Sci. 4(4), 74‒77. Abdallah, E.S., Eissa, M. and Menaze, A. 2018. The prevalence and etiology of subclinical mastitis in sheep and goats. Zag. Vet. J. 46(2), 96‒104. Ahmed, W., Neubauer, H., Tomaso, H., El Hofy, F.I., Monecke, S., Abdeltawab, A.A. and Hotzel, H. 2020. Characterization of Staphylococci and Streptococci Isolated from milk of bovides with mastitis in Egypt. Pathogens. 9(5), 381. Al-Sharif, M., Marghani, B.H. and Ateya, A. 2022. DNA polymorphisms and expression profile of immune and antioxidant genes as biomarkers for reproductive disorders tolerance/susceptibility in Baladi goat. Anim. Biotechnol. 2022, 1‒12. Alain, K., Karrow, N.A., Thibault, C., St-Pierre, J., Lessard, M. and Bissonnette, N. 2009. Osteopontin: an early innate immune marker of Escherichia coli mastitis harbors genetic polymorphisms with possible links with resistance to mastitis. BMC Genom. 10, 444. Ali, T., Kamran, A.R., Wazir, I., Ullah, R., Shah, P., Ali, M.I., Han, B., and Liu, G. 2021. Prevalence of mastitis pathogens and antimicrobial susceptibility of isolates from cattle and buffaloes in Northwest of Pakistan. Front. Vet. Sci. 8, 746755. Altschul, S.F., Gish, W., Miller, W., Myers, E.W. and Lipman, D.J. 1990. Basic local alignment search tool. J. Mol. Biol. 215(3), 403‒410. Amosun, E., Ajuwape, A. and Adetosoye, A. 2010. Bovine streptococcal mastitis in Southwest and Northern states of Nigeria. African J. Biomed. Res. 13(1), 33‒37. Asadpour, R., Zangiband, P., Nofouzi, K. and Saberivand, A. 2021. Differential expression of antioxidant genes during clinical mastitis of cow caused by Staphylococcus aureus and Escherichia coli. Vet. Arhiv. 91(5), 451‒458. Ateya, A., El-Sayed, A. and Mohamed, R. 2021. Gene expression and serum profile of antioxidant markers discriminate periparturient period time in dromedary camels. Mammal Res. 66(4), 603‒613. Ateya, A.I., Ibrahim, S.S. and Al-Sharif, M.M. 2022. Single nucleotide polymorphisms, gene expression and economic evaluation of parameters associated with mastitis susceptibility in European Cattle Breeds. Vet. Sci. 9(6), 394. Auldist, M. and Hubble, I. 1998. Effects of mastitis on raw milk and dairy products. Aust. J. Dairy Technol. 53(1), 28. Balakrishnan, A., Marathe, S.A., Joglekar, M., and Chakravortty, D. 2013. Bactericidal/permeability increasing protein: a multifaceted protein with functions beyond LPS neutralization. Innate Immun.19(4), 339–347. Barıtçı, I. and Adıgüzel, C. 2017. Aleppo (Damascus) goat breeding. Dicle Univ. J. Inst. Nat. Appl. Sci. 6, 39–42. Bhati, T., Nathawat, P., Sharma, S.K., Yadav, R., Bishnoi, J. and Kataria, A.K. 2016. Polymorphism in spa gene of Staphylococcus aureus from bovine subclinical mastitis. Vet. World. 9(4), 421–424. Bianchet, M.A., Faig, M. and Amzel, L.M. 2004. Structure and mechanism of NAD[P]H:quinone acceptor oxidoreductases (NQO). Methods Enzymol. 382, 144–174. Boesenberg-Smith, K.A., Pessarakli, M.M. and Wolk, D.M. 2012. Assessment of DNA yield and purity: an overlooked detail of PCR troubleshooting. Clin. Microbiol. 34(1), 1–6. Bonacci, G.R., Caceres, L.C., Sanchez, M.C. and Chiabrando, G.A. 2007. Activated alpha(2)-macroglobulin induces cell proliferation and mitogen-activated protein kinase activation by LRP-1 in the J774 macrophage-derived cell line. Arch. Biochem. Biophys. 460(1), 100–106. Boom, R., Sol, C.J., Salimans, M.M., Jansen, C.L., Wertheim-van Dillen, P.M. and van der Noordaa, J. 1990. Rapid and simple method for purification of nucleic acids. J. Clin. Microbiol. 28(3), 495–503. Capurro, A., Artursson, K., Waller, K.P., Bengtsson, B., Ericsson-Unnerstad, H. and Aspan, A. 2009. Comparison of a commercialized phenotyping system, antimicrobial susceptibility testing, and tuf gene sequence-based genotyping for species-level identification of coagulase-negative staphylococci isolated from cases of bovine mastitis. Vet. Microbiol. 134(3–4), 327–333. Carvalho-Sombra, T.C.F., Fernandes, D.D., Bezerra, B.M.O. and Nunes-Pinheiro, D.C.S. 2021. Systemic inflammatory biomarkers and somatic cell count in dairy cows with subclinical mastitis. Vet. Anim. Sci. 2021 11, 100165. Caverly, J.M., Diamond, G., Gallup, J.M., Brogden, K.A., Dixon, R.A. and Ackermann, M.R. 2003. Coordinated expression of tracheal antimicrobial peptide and inflammatory-response elements in the lungs of neonatal calves with acute bacterial pneumonia. Infect. Immun. 71(5), 2950–2955. Çetin, H., Gurgoze, S.Y., Keskin, O., Atli, M.O. and Korkmaz, Ö. 2005. Investigation of antioxidant enzymes and some biochemical parameters in ewes with gangrenous mastitis. Turkish J. Vet. Anim. Sci. 29(2), 301–308. Chabas, D., Baranzini, S.E., Mitchell, D., Bernard, C.C., Rittling, S.R., Denhardt, T., Sobel, R.A., Lock, C., Karpuj, M., Pedotti, R., Heller, R., Oksenberg, J.R. and Steinman, L. 2001. The influence of the proinflammatory cytokine, osteopontin, on autoimmune demyelinating disease. Science. 294(5547), 1731–1735. Chatzopoulos, D.C., Lianou, D.T., Michael, C.K., Gougoulis, D.A., Mavrogianni, V.S., Vasileiou, N.G.C., Katsafadou, A.I., Politis, A.P., Kordalis, N.G., Katsarou, E.I., Ioannidi, K.S., Petinaki, E. and Fthenakis, G.C. 2020. Streptococcus spp. from bulk-tank milk and milking machine teatcups on small ruminant farms, and factors potentially associated with their isolation. J. Dairy Res. 87(3), 277–281. Cortimiglia, C., Bianchini, V., Franco, A., Caprioli, A., Battisti, A., Colombo, L., Stradiotto, K., Vezzoli, F. and Luini, M. 2015. Short communication: prevalence of Staphylococcus aureus and methicillin-resistant S. aureus in bulk tank milk from dairy goat farms in Northern Italy. J. Dairy Sci. 98(4), 2307–2311. Cremonesi, P., Capoferri, R., Pisoni, G., Del Corvo, M., Strozzi, F., Rupp, R., Caillat, H., Modesto, P., Moroni, P., Williams, J.L., Castiglioni, B. and Stella, A. 2012. Response of the goat mammary gland to infection with Staphylococcus aureus revealed by gene expression profiling in milk somatic and white blood cells. BMC Genomics. 13(1), 540. Darwish, A., Ebissy, E., Ateya, A. and El-Sayed, A. 2023. Single nucleotide polymorphisms, gene expression and serum profile of immune and antioxidant markers associated with postpartum disorders susceptibility in Barki sheep. Anim. Biotechnol. 34(2), 327–339. Darwish, A.A. 2023. Clinicopathological study on camel mastitis at Matrouh Governorate. J. Adv. Vet. Anim. Res. 10(2), 284–291. Das, D., Panda, S.K., Kundu, A.K., Jena, B., Das, B.C. and Sahu, R.K. 2018. Haematological and metabolic profile test of mastitis affected bovines in Odisha. J. Entomol. Zool. Stud. 6(2), 3022–3024. Ding, Y., Zhao, J., He, X., Li, M., Guan, H., Zhang, Z. and Li, P. 2016. Antimicrobial resistance and virulence-related genes of Streptococcus obtained from dairy cows with mastitis in Inner Mongolia, China. Pharm. Biol, 54(1), 162–167. Dmitriev, A., Bhide, M. and Mikula, I. 2006. cpn60 gene-based multiplex-PCR assay for simultaneous identification of Streptococcal species. Acta Vet. Brno. 75(2), 235–240. Duncan, J.R., Mahaffey, E.A. and Prasse, K.W. 1994. Veterinary laboratory medicine. Ames, UK: Iowa State University Press, pp: 243. El-Deeb, W.M. 2013. Clinicobiochemical investigations of gangrenous mastitis in does: immunological responses and oxidative stress biomarkers. J. Zhejiang Univ. Sci. B. 14(1), 33–39. Elayadeth-Meethal, M., Thazhathu Veettil, A., Asaf, M., Pramod, S., Maloney, S.K., Martin, G.B., Rivero, M.J., Sejian, V., Naseef, P.P., Kuruniyan, M.S. and Lee, M.R.F. 2021. Comparative expression profiling and sequence characterization of ATP1A1 gene associated with heat tolerance in tropically adapted cattle. Animals 11(8), 2368. Ewing, W.H. 1986. Edwards and ewing’s identification of enterobacteriaceae. 4 the ed. Elsevier, New York, USA. Faraz, A.Y.M., Waheed, A., Yaqoob, M. and Ishaq, K. 2019. Growth performance and hair mineral status of Marecha (Camelus dromedarius) calves reared under different management systems. Pak. J. Zool. 51, 503–509. Feldman BF, Zinkl JC, Jain NC 2000. Schalm’s veterinary hematology, 5th (ed.), Philadelphia, London: Lippincott Williams & Wilkins. Gabay, C. and Kushner, I. 1999. Acute-phase proteins and other systemic responses to inflammation. N. Engl. J. Med. 340(6), 448–454. Garba, B., Habibullah, S.A., Saidu, B. and Suleiman, N. 2019. Effect of mastitis on some hematological and biochemical parameters of Red Sokoto goats. Vet. World. 12(4), 572–577. Gebrewahid, T., Abera, B. and Menghistu, H. 2012. Prevalence and etiology of subclinical mastitis in small ruminants of Tigray Regional State, North Ethiopia. Vet. World. 5(2), 103–109. Haftay, A., Habtamu, T. and Abebe, M. 2016. Bacterial identification and antimicrobial susceptibility of subclinical mastitis causing bacteria from goats in Aba’lla district, Afar, North-Eastern Ethiopia. Rev. Med. Vet. 167(7–8), 170–175. Han, G., Zhang, B., Luo, Z., Lu, B., Luo, Z., Zhang, J., Wang, Y., Luo, Y., Yang, Z., Shen, L., Yu, S., Cao, S., Yao, X. 2022. Molecular typing and prevalence of antibiotic resistance and virulence genes in Streptococcus agalactiae isolated from Chinese dairy cows with clinical mastitis. PLoSOne. 17(5), e0268262. Hogan, J., Gonzalez, R., Harmon, R., Nickerson, S., Oliver, S., Pankey, J. and Smith, K.L. 1999. Laboratory handbook on bovine mastitis. Madison, WI: National Mastitis Council. 78(7), 485–488. Hristov, K., Pepovich, R., Nikolov, B., Stoimenov, G. and Stamberov, P. 2018. Hematological changes associated with subclinical mastitis in goats. Scientific Works. Series C. Vet. Med. LXIV(2), 38-41. Hu, Q., Tu, J., Han, X., Zhu, Y., Ding, C. and Yu, S. 2011. Development of multiplex PCR assay for rapid detection of Riemerella anatipestifer, Escherichia coli, and Salmonella enterica simultaneously from ducks. J. Microbiol. Methods 87(1), 64–69. Jackson, P.G. and Cockcroft, P.D. 2002. Appendix 3: laboratory reference values: biochemistry. Clin. Exam. Farm anim. 150, 303–305. Joint, F. 2011. Food and Agriculture Organization of the United Nations. Caramel Colours. Combined Compendium of Food Additive Specification, Monograph, 11, 1817–7077. Kaneko, J.J. 1997. Clinical biochemistry of domestic animals. 5th (ed.), San Digo, London, Tokyo and Toronto: Academic press. Khazaal, K. 2009. Comparison of the performance of Shami (Damascus) and Saanen goats raised under similar environmental conditions in Lebanon. Paper presented at the Options Mediterranean. Kosciuczuk, E.M., Lisowski, P., Jarczak, J., Krzyzewski, J., Zwierzchowski, L. and Bagnicka, E. 2014. Expression patterns of beta-defensin and cathelicidin genes in parenchyma of bovine mammary gland infected with coagulase-positive or coagulase-negative Staphylococci. BMC Vet. Res. 10, 246. Koskinen, M.T., Holopainen, J., Pyörälä, S., Bredbacka, P., Pitkälä, A., Barkema, H.W., Bexiga, R., Roberson, J., Sølverød, L., Piccinini, R., Kelton, D., Lehmusto, H., Niskala, S. and Salmikivi, L. 2009. Analytical specificity and sensitivity of a real-time polymerase chain reaction assay for identification of bovine mastitis pathogens. J. Dairy Sci. 92(3), 952–959. Kumar, V., Gupta, I.D., Verma, A., Kumar, S.R. and Chaudhari, M.V. 2014. CD14 gene polymorphism using HinfI restriction enzyme and its association with mastitis in Sahiwal cattle. Indian J. Anim. Res. 48(1), 11–13. Lan, T., Liu, H., Meng, L., Xing, M., Dong, L., Gu, M., Wang, J. and Zheng, N. 2020. Antimicrobial susceptibility, phylotypes, and virulence genes of Escherichia coli from clinical bovine mastitis in five provinces of China. Food Agric. Immun. 31(1), 406–423. Le Loir, Y., Baron, F. and Gautier, M. 2003. Staphylococcus aureus and food poisoning. Genet. Mol. Res. 2(1), 63–76. Levashina, E.A., Moita, L.F., Blandin, S., Vriend, G., Lagueux, M. and Kafatos, F.C. 2001. Conserved role of a complement-like protein in phagocytosis revealed by dsRNAknockout in cultured cells of the mosquito, Anopheles gambiae. Cell. 104(5), 709–718. Leyva-Baca, I., Schenkel, F., Sharma, B.S., Jansen, G.B. and Karrow, N.A. 2007. Identification of single nucleotide polymorphismsin the bovine CCL2, IL8, CCR2 and IL8RA genes and their association with health and production in Canadian Holsteins. Anim. Genet. 38(3), 198–202. Linzell, J.L. and Peaker, M. 1971. Intracellular concentrations of sodium, potassium and chloride in the lactating mammary gland and their relation to the secretory mechanism. J. Physiol. 216(3), 683–700. Liu, Y.X., Xu, C.H., Gao, T.Y. and Sun, Y. 2012. Polymorphisms of the ATP1A1 geneassociated with mastitis in dairy cattle. Genet. Mol. Res. 11(1), 651–660. Machado, G.P., Silva, R.C., Guimarães, F.F., Salina, A. and Langoni, H. 2018. Detection of Staphylococcus aureus, Streptococcus agalactiae and Escherichia coli in Brazilian mastitic milk goats by multiplex-PCR. Pesquisa Veterinária Brasileira. 38, 1358–1364.s Masella, R., Di Benedetto, R., Vari, R., Filesi, C. and Giovannini, C. 2005. Novel mechanisms of natural antioxidant compounds in biological systems. J. Nutr. Biochem. 16(10), 577–586. Mavrogenis, A., Antoniades, N. and Hooper, R. 2006. The Damascus (shami) goat of Cyprus. Animal Genetic Resources/Resources génétiques animales/Recursos genéticos animales, 38, 57–65. Mbindyo, C., Gitao, C. and Bebor, L. 2014. A cross-sectional study on the prevalence of subclinical mastitis and antimicrobial susceptibility patterns of the bacterial isolates in milk samples of smallholder dairy goats in Kenya. Am. J. Res. Commun. 2(8), 30–51. Michel, O., LeVan, T.D., Stern, D., Dentener, M., Thorn, J., Gnat, D., Beijer, M.L., Cochaux, P., Holt, P.G. and Martinez, F.D. 2003. Systemic responsiveness to lipopolysaccharide and polymorphisms in the toll-like receptor 4 gene in human beings. J. Allergy Clin. Immunol. 112(5), 923–929. Mingala, C., Wy, A., Cruz, D., Ermitanio, E., Gaban, P. and Castro, D. 2020. Alpha-2-Macroglobulin gene polymorphism in water buffaloes (Bubalus bubalis) with subclinical mastitis. Int. J. Vet. Sci. 9, 116–120. Mishra, A.K., Sharma, N., Singh, D.D., Gururaj, K., Abhishek, Kumar, V. and Sharma, D.K. 2018. Prevalence and bacterial etiology of subclinical mastitis in goats reared in organized farms. Vet. World. 11(1), 20–24. Mohamed, A.A.E., Wahba, A.K.A., Faisal, R.A.S.R. and Yousreya, H.M. 2013. Some bacteriological and biochemical studies on subclinical mastitis in buffaloes. New York Science J. 6, 71–79.Mohamed, H.M.A. and El-Zamkan, M.A. 2022. Detection of Streptococcus thoraltensis in raw milk with special reference to their antibiogram. Adv. Anim. Vet. Sci. 10(3), 630–638. Mohammed, S.A., Razzaque, M.A., Omar, A.E., Albert, S. and Al-Gallaf, W.M. 2016. Biochemical and hematological profile of different breeds of goat maintained under intensive production system. African J. Biotechnol. 15(24), 1253–1257. Mota, R. 2008. Aspectos epidemiológicos, diagnóstico e controle das mastites em caprinos e ovinos. Tecnol. Ciênc. Agropec. 2(3), 57–61. Narenji Sani, R., Mahdavi, A. and Moezifar, M. 2015. Prevalence and etiology of subclinical mastitis in dairy ewes in two seasons in Semnan province, Iran. Trop. Anim. Health Prod. 47(7), 1249–1254. Nicholas, F.W. 2005. Animal breeding and disease. Philos. Trans. R. Soc. Lond. B. Biol. Sci. 360(1459), 1529–1536. Olech, M., Ropka-Molik, K., Szmatoła, T., Piórkowska, K. and Kuźmak, J. 2021. Single nucleotide polymorphisms in genes encoding toll-like receptors 7 and 8 and their Association with proviral load of SRLVs in goats of polish carpathian breed. Animals. 11(7), 1908. Ondiek, J.O., Ogore, P.B. and Kemboi F. 2018. Clinical mastitis gives off-flavor and reduces quality of milk in smallholder goat farms. Int. J. Curr. Microbiol. App. Sci. 7(1): 2387–2396. Osakabe, Y., Yaguchi, C., Miyai, T., Miyata, K., Mineo, S., Nakamura, M. and Amano, S. 2006. Detection of Streptococcus species by polymerase chain reaction in infectious crystalline keratopathy. Cornea, 25(10), 1227–1230. Pavlov, K.V. and Sokolov, V.S. 2000. Electrogenic ion transport by Na+,K+-ATPase. Membr. Cell Biol. 13(6), 745–788. Pfaffl, M. 2001. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 29, e45. Pisoni, G., Moroni, P., Genini, S., Stella, A., Boettcher, P.J., Cremonesi, P., Scaccabarozzi, L., Giuffra, E. and Castiglioni, B. 2010. Differentially expressed genes associated with Staphylococcus aureus mastitis in dairy goats. Vet. Immunol. Immunopathol. 135(3–4), 208–217. Proudfoot, A.E. 2002. Chemokine receptors: multifaceted therapeutic targets. Nat. Rev. Immunol. 2(2), 106–115. Pugh, D.G. and Baird, N. (Ed.) 2012. sheep and goat medicine, 2nd ed. Philadelphia, London: W.B. Saunders Company. Rakowski, R.F., Gadsby, D.C. and De Weer, P. 1989. Stoichiometry and voltage dependence of the sodium pump in voltage-clamped, internally dialyzed squid giant axon. J. Gen. Physiol. 93(5), 903–941. Rupp, R., Huau, C., Caillat, H., Fassier, T., Bouvier, F., Pampouille, E., Clément, V., Palhière, I., Larroque, H., Tosser-Klopp, G., Jacquiet, P. andTosser-Klopp, G. (2019). Divergent selection on milk somatic cell count in goats improves udder health and milk quality with no effect on nematode resistance. J. Dairy Sci. 102(6), 5242–5253. Saad, W., El-Tawab, A. and Nabih, A. 2019. Bacteriological and molecular diagnosis of most common bacteria causing subclinical mastitis in cow. Benha Vet. Med. J. 37(2), 28–32. Saleh, N., Allam, T.S., Omran, A. and Abdelfattah, A.M. 2022. Evaluation of changes in hemato-biochemical, inflammatory, and oxidative stress indices as reliable diagnostic biomarkers for subclinical mastitis in Cows. Alexandria J. Vet. Sci. 72(2), 23–34. Sameer, M.M., El-Kabbany, M., Abeer, M. and Eid, S. 2013. Inflammatory markers as laboratory tool for early diagnosis of subclinical mastitis in goats with special reference to their effect on the milk quality. Zag. Vet. J. 41(4), 171–178. Sanger, F., Nicklen, S. and Coulson, A.R. 1977. DNA sequencing with chain-terminating inhibitors. Proc. Natl. Acad. Sci. USA. 74(12), 5463–5467. Sarvesha, K., Satyanarayana, M., Narayanaswamy, H., Rao, S., Yathiraj, S., Isloor, S., Mukartal, S.Y., Singh, S.V. and Anuradha, M.E. 2017. Haemato-biochemical profile and milk leukocyte count in subclinical and clinical mastitis affected crossbred cattle. J. Exp. Biol. Agric. Sci. 5(1), 1–6. Sarvesha, K., Satyanarayana, M., Narayanaswamy, H., Rao, S., Yathiraj, S., Isloor, S., Mukartal, S.Y., Srikanth, M., Anuradha, M.E. and Kamal, H. 2016. Effect of subclinical and clinical mastitis on haematobiochemical profile and milk leukocyte count in indigenous cows. J. Cell Tissue Res. 16(3), 5829–5834. Schukken, Y.H., González, R.N., Tikofsky, L.L., Schulte, H.F., Santisteban, C.G., Welcome, F.L., Bennett, G.J., Zurakowski, M.J. and Zadoks, R.N. 2009. CNS mastitis: nothing to worry about? Vet. Microbiol. 134(1–2), 9–14. Shome, B.R., Das Mitra, S., Bhuvana, M., Krithiga, N., Velu, D., Shome, R., sloor, S., Barbuddhe, S.B. and Rahman, H. 2011. Multiplex PCR assay for species identification of bovine mastitis pathogens. J. Appl. Microbiol. 111(6), 1349–1356. Singh, S. 2000. Udder health profiles with special reference to acute phase proteins and supplementation therapy. MV Sc thesis submitted to the GB Pant University of Agriculture and Technology, Pantnagar, India. Skevaki, C., Pararas, M., Kostelidou, K., Tsakris, A. and Routsias, J.G. 2015. Single nucleotide polymorphisms of toll-like receptors and susceptibility to infectious diseases. Clin. Exp. Immunol. 180(2), 165–177. Sordillo, L.M. and Aitken, S.L. (2009). Impact of oxidative stress on the health and immune function of dairy cattle. Vet. Immunol. Immunopathol. 128(1–3), 104–109. Spanu, V., Scarano, C., Virdis, S., Melito, S., Spanu, C. and De Santis, E.P. 2013. Population structure of Staphylococcus aureus isolated from bulk tank goat’s milk. Foodborne Pathog. Dis. 10(4), 310–315. Stockham, S.L. and Scott, M.A. 2013. Fundamentals of veterinary clinical pathology. Hoboken, NJ: John Wiley & Sons. Talukder, A.A., Rahman, H.H., Mahmud, S.J., Alam, F. and Dey, S.K. 2013. Isolation, identification andresistance pattern of microorganisms associated with mastitis in buffalo. Bangladesh J. Microbiol. 30(1–2), 1–5.Tamura, K., Dudley, J., Nei, M. and Kumar, S. 2007. MEGA4: molecular evolutionary genetics analysis (MEGA) software version 4.0. Mol. Biol. Evol. 24(8), 1596–1599. Tolone, M., Mastrangelo, S., Gerlando, R., Sutera, A., Monteleone, G., Sardina, M. and Portolano, B. 2016. Association study between β-defensin gene polymorphisms and mastitis resistance in Valle del Belice dairy sheep breed. Small Ruminant Res. 136, 18–21. Wang, X., Huang, J., Feng, M., Ju, Z., Wang, C., Yang, G., Yuan, J.D. and Zhong, J. 2014. Regulatory mutations in the a 2M gene are involved in the mastitis susceptibility in dairy cows. Anim. Genet. 45(1), 28–37. Wang, X., Huang, J., Zhao, L., Wang, C., Ju, Z., Li, Q., Qi, C., Zhang, Y., Zhang, Z., Zhang, W., Hou, M., Yuan, J. and Zhang, W. 2012. The exon 29 c. 3535A> T in the alpha-2-macroglobulin gene causing aberrant splice variants is associated with mastitis in dairycattle. Immunogenetics. 64, 807–816. Wegner, T. and Stull, J. 1978. Relation between mastitis test score, mineral composition of milk, and blood electrolyte profiles in Holstein cows. J. Dairy Sci. 61(12), 1755–1759. Wilson, R. 1987. Livestock production in central Mali: environmental factors affecting weight in traditionally managed goats and sheep. Anim. Sci. 45(2), 223–232. Winter, P., Fuchs, K., Walshe, K. and Colditz, I.G. 2003. Serum amyloid A in the serum and milk of ewes with mastitis induced experimentally with Staphylococcus epidermidis. Vet. Rec. 152(18), 558–562. Wright, S.D., Ramos, R.A., Tobias, P.S., Ulevitch, R.J. and Mathison, J.C. 1990. CD14, a receptor for complexes of lipopolysaccharide (LPS) and LPS binding protein. Sci. 249(4975), 1431–1433. Yamamoto, M., Kensler, T.W. and Motohashi, H. 2018. The KEAP1-NRF2 System: a Thiol-based sensor-effector apparatus for maintaining redox homeostasis. Physiol. Rev. 98(3), 1169–1203. Zenebe, N., Habtamu, T. and Endale, B. 2014. Study on bovine mastitis and associated risk factors in Adigrat, Northern Ethiopia. Afr. J. Microbiol. Res. 8(4), 327–331. Zhang, T., Zhu, J., Su, B., Cao, L., Li, Z., Wei, H., Huang, X., Zheng, K., Li, A., Chen, N., Liu, L., Xia, W., Wu, H. and He, Q. 2020. Effects of TLR7 polymorphisms on the susceptibility and progression of HIV-1 infection in Chinese MSM population. Front. Immunol. 11,s 589010. | ||
| How to Cite this Article |
| Pubmed Style Ebissy E, Darwish A, Hafez AA, Ateya A, El-Sayed A. Individual genomic loci, transcript level and biochemical profile of immune and antioxidant markers associated with genetically identified bacterial mastitis in Shami goats in Egypt. Open Vet J. 2024; 14((1) (Zagazig Veterinary Conference)): 370-388. doi:10.5455/OVJ.2024.v14.i1.34 Web Style Ebissy E, Darwish A, Hafez AA, Ateya A, El-Sayed A. Individual genomic loci, transcript level and biochemical profile of immune and antioxidant markers associated with genetically identified bacterial mastitis in Shami goats in Egypt. https://www.openveterinaryjournal.com/?mno=175632 [Access: May 08, 2024]. doi:10.5455/OVJ.2024.v14.i1.34 AMA (American Medical Association) Style Ebissy E, Darwish A, Hafez AA, Ateya A, El-Sayed A. Individual genomic loci, transcript level and biochemical profile of immune and antioxidant markers associated with genetically identified bacterial mastitis in Shami goats in Egypt. Open Vet J. 2024; 14((1) (Zagazig Veterinary Conference)): 370-388. doi:10.5455/OVJ.2024.v14.i1.34 Vancouver/ICMJE Style Ebissy E, Darwish A, Hafez AA, Ateya A, El-Sayed A. Individual genomic loci, transcript level and biochemical profile of immune and antioxidant markers associated with genetically identified bacterial mastitis in Shami goats in Egypt. Open Vet J. (2024), [cited May 08, 2024]; 14((1) (Zagazig Veterinary Conference)): 370-388. doi:10.5455/OVJ.2024.v14.i1.34 Harvard Style Ebissy, E., Darwish, . A., Hafez, . A. A., Ateya, . A. & El-Sayed, . A. (2024) Individual genomic loci, transcript level and biochemical profile of immune and antioxidant markers associated with genetically identified bacterial mastitis in Shami goats in Egypt. Open Vet J, 14 ((1) (Zagazig Veterinary Conference)), 370-388. doi:10.5455/OVJ.2024.v14.i1.34 Turabian Style Ebissy, Eman, Asmaa Darwish, Amani A. Hafez, Ahmed Ateya, and Ahmed El-Sayed. 2024. Individual genomic loci, transcript level and biochemical profile of immune and antioxidant markers associated with genetically identified bacterial mastitis in Shami goats in Egypt. Open Veterinary Journal, 14 ((1) (Zagazig Veterinary Conference)), 370-388. doi:10.5455/OVJ.2024.v14.i1.34 Chicago Style Ebissy, Eman, Asmaa Darwish, Amani A. Hafez, Ahmed Ateya, and Ahmed El-Sayed. "Individual genomic loci, transcript level and biochemical profile of immune and antioxidant markers associated with genetically identified bacterial mastitis in Shami goats in Egypt." Open Veterinary Journal 14 (2024), 370-388. doi:10.5455/OVJ.2024.v14.i1.34 MLA (The Modern Language Association) Style Ebissy, Eman, Asmaa Darwish, Amani A. Hafez, Ahmed Ateya, and Ahmed El-Sayed. "Individual genomic loci, transcript level and biochemical profile of immune and antioxidant markers associated with genetically identified bacterial mastitis in Shami goats in Egypt." Open Veterinary Journal 14.(1) (Zagazig Veterinary Conference) (2024), 370-388. Print. doi:10.5455/OVJ.2024.v14.i1.34 APA (American Psychological Association) Style Ebissy, E., Darwish, . A., Hafez, . A. A., Ateya, . A. & El-Sayed, . A. (2024) Individual genomic loci, transcript level and biochemical profile of immune and antioxidant markers associated with genetically identified bacterial mastitis in Shami goats in Egypt. Open Veterinary Journal, 14 ((1) (Zagazig Veterinary Conference)), 370-388. doi:10.5455/OVJ.2024.v14.i1.34 |