| Research Article | ||
Open Veterinary Journal, (2024), Vol. 14(1): 176-185 Original Research Screening of antibiogram, virulence factors, and biofilm production of Staphylococcus aureus and the bio-control role of some probiotics as alternative antibioticsAya R. Mohammed, Esmat I. El-Said, Salah F. Abd ElAal and Rania M. Kamal*Department of Food Control, Zagazig University, Zagazig, Egypt *Corresponding Author: Rania M. Kamal. Department of Food Control, Zagazig University, Zagazig, Egypt. Email: raniakamal79 [at] gmail.com Submitted: 01/10/2023 Accepted: 15/12/2023 Published: 31/01/2024 © 2024 Open Veterinary Journal
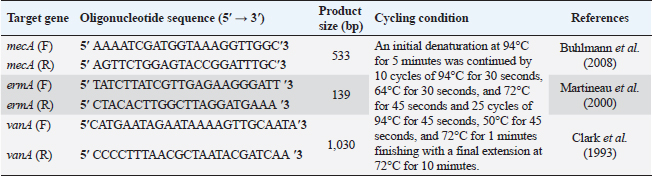
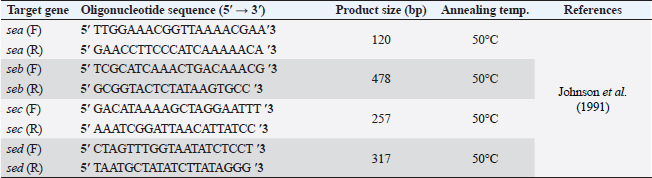
AbstractBackground: Food safety is a serious challenge in the face of increasing population and diminishing resources. Staphylococcus aureus is a critical foodborne pathogen characterized by its capability to secret a diverse range of heat-resistant enterotoxins. Antibiotic usage in dairy herds resulted in the occurrence of antimicrobial resistance (AMR) patterns among bacterial species, which were consequently transmitted to humans via dairy products. Lactic acid bacteria (LAB) produce bacteriocins, which provide an excellent source of natural antimicrobials with the further advantage of being environmentally friendly and safe. Aim: Detection of multidrug resistance (MDR) S. aureus isolates in concerned samples, molecular characteristics, biofilm production, and the inhibitory role of LAB against it. Methods: Random samples of raw milk and other dairy products were analyzed for S. aureus isolation. Phenotypic and genotypic assessment of AMR was performed, in addition to detection of classical enterotoxin genes of S. aureus. Finally, evaluation of the antimicrobial action of some Lactobacillus strains against S. aureus. Results: Incidence rates of presumptive S. aureus in raw milk, Kariesh cheese, and yogurt samples were 50%, 40%, and 60%, respectively. The highest resistance of S. aureus was to Kanamycin (100%) and Nalidixic acid (89.3%), respectively. (78.66%) of S. aureus were MDR. 11.1% of S. aureus carried mecA gene. In concern with enterotoxins genes, PCR showed that examined isolates harbored sea with a percentage of (22.2%), while sed was found in (11.1%) of isolates. Regarding biofilm production, (88.88%) of S. aureus were biofilm producers. Finally, agar well diffusion showed that Lactobacillus acidophilus had the strongest antimicrobial action against S. aureus with inhibition zone diameter ranging from 18 to 22 mm. Conclusion: There is a widespread prevalence of MDR S. aureus in raw milk and dairy products. Production of staphylococcal enterotoxins, as well as biofilm production are responsible for public health risks. Therefore, installing proper hygienic routines and harsh food safety policies at food chain levels is substantial. Keywords: MecA gene, Staphylococcal enterotoxins, Biofilm formation, Lactic acid bacteria. IntroductionMilk is an essential nutrient for humans of all ages, especially children and teenagers, to promote their physiological functions and growth (Abunna et al., 2019). Milk, in addition, is a great medium for pathogenic bacteria growth and development, chemical transfer, and the dissemination of other contaminants (Nirwal et al., 2013). Ingestion of unheated treated milk and dairy products has been associated with the incidence of foodborne illness outbreaks, owing to the development of pathogens such as Salmonella spp., Campylobacter jejuni, Escherichia coli, Listeria monocytogenes, Staphylococcus aureus, and Bacillus cereus (Biernbaum et al., 2021). Staphylococcus aureus is globally the third most common cause of foodborne illness and usually contaminates milk or its products via multiple pathways (Şanlıbaba, 2022). It serves as one of the most prevalent contributing factors of mastitis in dairy animals, with diseased animals usually releasing S. aureus into milk (Li et al., 2017). Antimicrobial resistance (AMR), biofilm formation, enterotoxin production, and other virulence characteristics, including nucleases, proteases, hyaluronidase, lipases, and collagenase production, are all associated with S. aureus pathogenicity (Shettigar and Murali, 2020). Staphylococcal enterotoxins (SEs) are exotoxins that cause food poisoning in human beings (Balaban and Rasooly, 2000). Several classical SEs were incriminated in food poisoning as SEA, SEB, SEC, SED, and SEE (Bergdoll, 1983 and 1989). Recently, other types were identified as potential agents of food poisoning including SEG, SEH, SEI, SER, SES, and SET (Omoe et al., 2004). Nowadays, living organisms are facing an urgent threat from AMR due to increasing the distribution of antibiotic resistance genes (ARGs) among different populations (Van Boeckel et al., 2015; Zhang et al., 2022). Raw milk can be contaminated and transmit several types of these ARGs (T´oth et al., 2020). On the other hand, the emergence of several ARGs within clinically relevant bacterial pathogens is a global issue that causes human death and provides massive economic costs (Mestrovic et al., 2022). Biofilms are structures of extracellular matrices of microbial communities characterized by heterogeneity that can easily colonize various substrates such as soil, water, and organic matter (Donlan and Costerton, 2002). Microorganisms with biofilms provide numerous advantages: improved metabolic cooperation between species (Shapiro, 1998), higher resistance to host immunological reactions, demanding larger dosages of antibiotics (Ceri et al., 1999), and enhanced bacterial association capability (Hennequin et al., 2012). The cycle of formation, maturation, and propagation is the primary reason for surface-to-food cross-contamination (Kumar et al., 2017). Lactobacilli are popularly recognized for their ability to protect food and prevent illnesses caused by food contamination (Adams, 1999). In the food industry, multiple Lactobacillus species and their metabolic products were granted a “generally regarded as safe” certification (Wells, 2011). In recent years, Lactobacilli strains have been used to replace synthetic preservatives in the food industry as natural antimicrobials due to the rising attention to healthy food, which has sparked curiosity among scientific organizations (Arena et al., 2016). The present study aims to investigate S. aureus prevalence in raw milk and some dairy product samples obtained from various markets in Zagazig city as well as to evaluate its phenotypic and genotypic antimicrobial susceptibility patterns, also to detect several virulence genes related to its pathogenicity. Furthermore, the ability for biofilm production and the antimicrobial effect of some Lactobacillus strains against this food-borne pathogen. Materials and MethodsSamples collection150 samples of raw milk and other dairy products (Kariesh cheese and plain yogurt), (50 of each) were obtained from various shops in Zagazig city, under optimum conditions of hygiene and delivered without delay for a microbiological assessment. Isolation and identification of S. aureusIsolation was performed following ISO 6888–1:1999 +A1:2003 guidelines protocol (ISO, 2003) on Baired Parker agar plates after enrichment, Incubation of cultures was performed at 37°C and examined for two successive days, and further biochemical identification was done. Antibiotic sensitivity testingAll bacterial isolates were evaluated for their antibiotics’ susceptibility in vitro, according to the guidelines of the Clinical and Laboratory Standards Institute, by Kirby–Bauer disk diffusion method CLSI (2020). Against the discs of Ampicillin (AM), Oxacillin (OX), Azithromycin (AZ), Erythromycin (E), Tetracycline (T), Cefotaxime (CF), Kanamycin (K), Gentamicin (G), Nalidixic acid (NA), Ciprofloxacin (CP), Imipenem (IPM), Clindamycin (CL), and Sulphamethoxazol (SXT). The diameters of inhibition zones were recorded in millimeters, and the measurements were classified as sensitive, moderate, or resistant based on the interpretative manual of CLSI (2020). In addition, S. aureus isolates were tested using the broth microdilution method to determine the minimum inhibitory concentrations (MICs) of Vancomycin. Then, according to CLSI (2020) interpretation, Vancomycin susceptible S. aureus isolates had MICs ≤2 mg/ml, VISA isolates had MICs of 4–8 mg/ml, and Vancomycin resistant S. aureus (VRSA) isolates had MICs > 16 mg/ml. The formula of Singh et al. (2010) was used to measure the multiple antibiotic resistance (MAR) index. Moreover, the multidrug resistance (MDR) isolates were that showed resistance to ≥3 classes of antimicrobial tested (Magiorakos et al., 2012). Multiplex polymerase chain reaction techniqueDetection of S. aureus enterotoxins and resistance genesIdentification of ARGs represented by methicillin-resistant “MRSA” (mecA), E (ermA), and vancomycin (vanA) of S. aureus was applied using primers in Table 1. Specific primers of SEs (sea, seb, sec, and sed) genes were listed in Table 2. DNA extractionDNA extraction was done for isolates by Qiagene DNA extraction kits (QIAGEN, GmbH, Hilden, Germany) according to manufacturers’ recommendations. PCR amplification and analysis of PCR productsBy using purified bacterial DNA, the resistance genes were identified by multiplex PCR according to Perez-Roth et al. (2001) (Table 1). Also, PCR procedures were based on Rall et al. (2008) to identify the classical enterotoxins. Biofilm formation by microtiter plate assay (MTP)The method MTP was performed on a 96-well, flat-bottomed, sterile MTP to examine the biofilm formation ability of S. aureus isolates. 200 µl of brain heart infusion broth were placed into wells of polystyrene microplate and 20 µl of each strain culture were dispersed in triplicate into the wells. Then, the microplate was incubated overnight at 37°C in aerobic conditions. The plate was drained and properly washed with phosphate buffer saline three times. Afterward, 200 µl of methanol was transferred to each well to fix those attached cells. The adhering bacterial cells were stained with 200 µl of 0.5% (w/v) crystal violet for 10 minutes. Subsequent to staining, phosphate buffer saline (PBS) was used to wash the wells appropriately then dried aerobically before being resolved with 250 µl 33% glacial acetic acid. The optical density of the stained adherent bacteria was measured at 570 nm using a microplate reader. Following the formulas developed by Stepanovic´ et al. (2004), the bacterial strains were classified as non-producers, weak, moderate, and strong biofilm producers. Table 1. Primer sequences of mecA gene and other resistance genes of S. aureus.
Table 2. Primer sequences of SE genes.
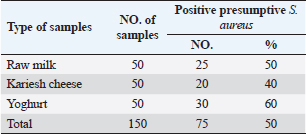
Table 3. Prevalence of S. aureus in raw milk and dairy products.
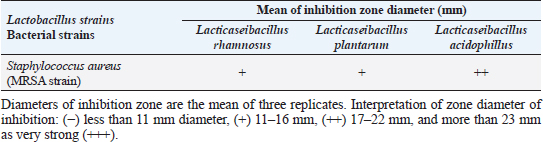
Agar well diffusion assayCell-free supernatant (CFS) preparation of Lactobacillus strainsLacticaseibacillus rhamnosus LMG23522, Lacticaseibacillus plantarum MK 806485, and Lacticaseibacillus acidophillus MK850930 strains were used to measure the inhibitory effect against S. aureus. Inoculation of probiotic strains in MRS broth was performed and then incubated at 37°C in a CO2 incubator for 2 days. After that, the bacterial suspension was centrifuged at 4,000 rpm for 10 minutes and CFS was extracted aseptically then sterilized with a 0.20-µm pore size filter and utilized freshly. Antimicrobial activityThe agar well diffusion method was applied by loading the wells of 6 mm diameter with CFS from Lactobacillus strains (100 µl/well). After one night of incubation at 37°C, a digital caliper was used to record the inhibition zone diameters. Then, Lactobacillus strains were categorized into 4 groups as follows: inhibition zone of <11 mm diameter was regarded as negative (−), 11–16 mm as moderate (+), 17–22 mm as strong (++), and >23 mm as extremely strong (+++), respectively (Rammelsberg and Radler, 1990). ResultsIsolation and identification of S. aureusData presented in Table 3 showed that the highest incidence of presumptive S. aureus was in yogurt samples, followed by raw milk samples, then Kariesh cheese samples at 60%, 50%, and 40%, respectively. Table 4. Antimicrobial susceptibility of S. aureus isolates (n=75).
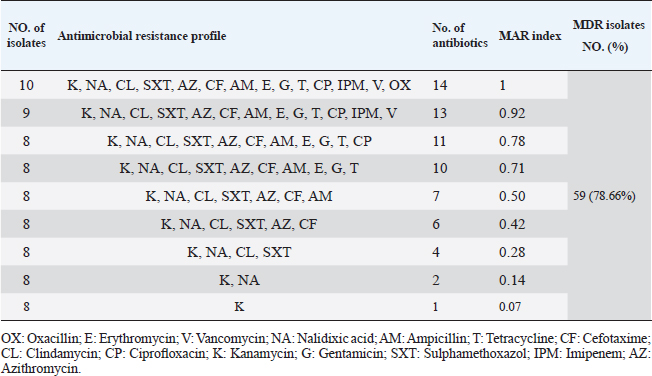
Table 5. Multiple antibiotic resistance phenotypes of S. aureus strains (n=75).
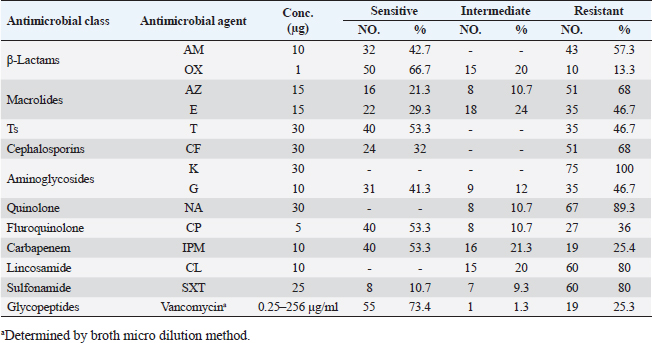
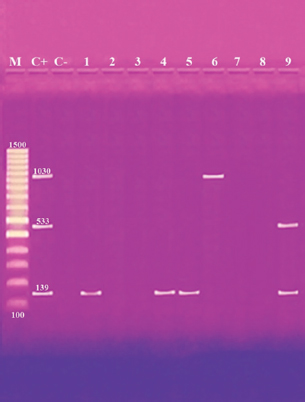
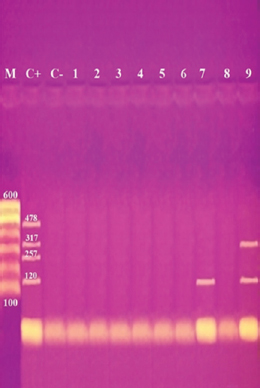
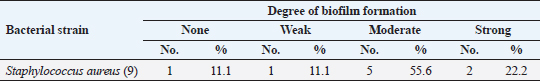
Antibiotic sensitivity testingAntimicrobial susceptibility testing of S. aureus isolates showed that the highest resistance was to K and NA by percentages being (100%) and (89.3%), respectively, as mentioned in Table 4. MDR and MAR indexData in Table 5 declared that 59 out of total 75 S. aureus isolates showed MDR by a percentage of (78.66%). MAR ranged from 0.07 to 1 and (78.66%) of total isolates had a MAR index ≥0.2. Multiplex PCR assayARGs of S. aureusFigure 1 and Table 6 revealed the incidence of mecA, ermA, and vanA genes in nine strains of S. aureus where vanA and mecA genes presented a percentage of (11.1%) of each. While ermA gene appeared in four out of nine strains (44.4%). The incidence rate of MRSA was (11.1%). Staphylococcal enterotoxins Figure 2 and Table 6 showed that sea was detected in 2 out of 9 (22.2%) of S. aureus strains while sed in 1/9 (11.1%). However, seb and sec failed to be detected in any isolate. Ability to form biofilmTable 7 represents the ability of S. aureus strains to produce biofilm. 8 out of 9 (88.88%) of S. aureus were biofilm producers as follows: 5 out of 9 S. aureus isolates (55.6%) were moderate biofilm producers, 2/9 (22.2%) were strong biofilm producers, while only one isolate was weak biofilm producer and so only one isolate was non-biofilm producer. Antibacterial activity of Lactobacillus strains against S. aureus Table 8 assessed the antimicrobial activity of three Lactobacillus strains against the most virulent strain of S. aureus (MRSA strain). Lactobacillus acidophilus MK850930 had the strongest antimicrobial action against S. aureus with inhibition zone diameter ranging from 18 to 22 mm. DiscussionImproper personal hygiene practices, environmental contamination with wastes of infected animals, cross-contamination, and faulty handling during shipping to milk collecting centers may all contribute to the high occurrence of S. aureus in raw milk and investigated dairy products (Addis et al., 2011). The predominant way of Staphylococcus pathogen into milk is through shedding from diseased mammary tissues (Rahimi, 2013). It is worth noting that, if some strains of this organism multiply heavily in foods, they can secrete food-poisoning enterotoxins (Pereira et al., 2009). The findings revealed that the highest prevalence of presumptive S. aureus was in yogurt, then raw milk and Kariesh cheese with percentages of 60%, 50%, and 40%, respectively (Table 3). These results agreed with Ahmed et al. (2019) results, who isolated S. aureus by a percentage of (42%) from raw milk samples, while greater percentages were detected by both Ibrahim et al. (2015) and Kandil et al. (2018), where they isolated S. aureus from (100%) and (80%) raw milk samples, respectively. Zeinhom and Abed (2020) obtained a lower incidence rate in raw milk (13%). Regarding Kariesh cheese, Ibrahim et al. (2015) agreed with our results as S. aureus reported in (66%) of Kareish cheese samples. However, extremely higher results were detected by Kandil et al. (2018) where (92%) of Kariesh cheese samples were contaminated with S. aureus. Our results were higher than those obtained by Ahmed et al. (2019) and Zeinhom and Abed (2020) who identified (38%) and (18%) prevalence of S. aureus in Kariesh cheese samples, respectively. Yogurt had the highest incidence rate (60%). Several studies detected lower incidence rates of S. aureus in yogurt as El-Ansary (2014), Ahmed et al. (2019), and Kandil et al. (2018) found S. aureus in (42%), (40%), and (36%) of yogurt samples, respectively. Ingestion of inadequate or non-heat-treated dairy products leads to infection with antibiotic-resistant bacteria, which develops a major public health threat. Inappropriate utilization of antibiotics in the treatment of mastitis can end up in the propagation of resistant strains serving as crucial public health issues (De Jong et al., 2018). Table 4 demonstrated the resistance rates of S. aureus against examined discs of antimicrobials and showed that the highest resistance was to K, NA, CL, and SXT by percentages being (100%), (89.3%), (80%), and (80%), respectively. Moderate resistance to AM (57.3%) followed by E, T, and G by a percentage of (46.7%) of each. Low resistance to Vancomycin (25.3%) where 20 out of 75 S. aureus isolates had MIC values between 16 and 256 mg/ml. Similarly, Alnakip et al. (2023) declared that examined S. aureus isolates resisted AM by a percentage of (46.66%), T (44.45%), and E (40%), respectively. Nearly similar results reported by Thaker et al. (2013) showed low resistance to AM (40%) of S. aureus isolates. Higher results were detected by Gundogan and Avci (2014) who revealed that isolates of S. aureus showed resistance to AM (92.6%) and T (54.3%), respectively. Contrary to our findings, Feyissa et al. (2023) demonstrated that (100%) of S. aureus isolates were T resistant. While Bissong and Ateba (2020) reported high resistance to Vancomycin (83.1%) among S. aureus isolates.
Fig. 1. Multiplex PCR of ermA (139 bp), mecA (533 bp) and vanA (1,030 bp) ARGs of S. aureus. Lane M: 100 bp ladder. Lane C+: Control positive for ermA, mecA, and vanA genes. Lane C-: Control negative. Lanes 1, 4 & 5: Positive S. aureus strains for ermA gene. Lanes 2, 3, 7 & 8: Negative strains for ermA, mecA and vanA genes. Lane 6: Positive S. aureus strain for vanA genes. Lane 9: Positive S. aureus strain for ermA and mecA genes. Table 6. Distribution of SEs and some ARGs.
Fig. 2. Multiplex PCR of sea (120 bp), seb (478 bp), sec (257 bp), and sed (317 bp) as enterotoxin genes. Lane M: 100 bp ladder as molecular size DNA marker. Lane C+: Control positive for sea, seb, sec, and sed genes. Lane C-: Control negative. Lane 7: Positive S. aureus strain for sea gene. Lane 9: Positive S. aureus strain for sea & sed genes. Lanes from 1 to 6 and Lane 8: Negative S. aureus for sea, seb, sec, and sed genes Table 5 recorded that (78.66%) 59 out of 75 S. aureus isolates were MDR to at least 3 different classes of antibiotics. Furthermore, Abd El Halem (2019) detected that the MDR in S. aureus isolates had a percentage of (39.5%), while (95.5%) of isolates were multidrug resistant as mentioned by Samaha et al. (2012). To achieve reliable, sensitive, and specific determination of MDR S. aureus strains, molecular diagnostics like PCR must be applied. Data in Figure 1 and Table 6 revealed that both mecA and vanA were detected in 1/9 (11.1%) of S. aureus isolates while ermA in 4/9 (44.4%). Therefore, MRSA was detected by a percentage of (11.1%). Our results agreed with results obtained by Saka and Terzi Gulel (2018) who demonstrated that 9% of S. aureus isolates were methicillin-resistant (mecA gene positive). On the contrary, Nam et al. (2011) declared that 17/402 of S. aureus isolates were identified genotypically as MRSA (carrying mecA gene) by a percentage of (4.2%). However, Zhao et al. (2021) found that the prevalence of MRSA was (0.7%) which was mecA positive. In concern with ermA and vanA genes, Ning et al. (2023) revealed that none of S. aureus isolates harbored vanA or ermA. Kou et al. (2021) identified that 2 out of 62 (3.22%) isolates carried ermA gene while 11/62 (17.7%) carried vanA gene in S. aureus isolates. Furthermore, Pajohesh et al. (2019) determined that the prevalence of mecA and ermA was (22.22%) and (13.33%), respectively, in S. aureus isolates. SEs are extremely firm and withstand against high temperatures and proteases such as pepsin and trypsin (Clarisse et al., 2013). Universally, staphylococcal food poisoning is commonly caused by SEA, followed by SED and SEB (Argudin et al., 2010). As shown in Figure 2 and Table 6, 2 out of 9 strains (22.2%) harbored sea gene and 1/9 (11.1%) harbored sed gene. Unfortunately, the detection of seb and sec was failed. In line with our results, Saka and Terzi Gulel (2018) mentioned that 12 out of 99 S. aureus isolates were positive for SEs (harbored one or two genes) by a percentage of (12%). Among them, 5/12 (41.6%) were positive for sea, 1/12 (8.3%) carried sed and no strain carried seb. In contrast to our study, Zhao et al. (2021) detected the presence of sed gene in 16/121, followed by sec in 10/121 and seb in 8/121 while, no isolate harbored sea gene. Pajohesh et al. (2019) declared that sed was reported in 15 isolates (33.3%), sea in 8 isolates (17.7%), sec in 7 isolates (15.5%), and seb in 4 isolates (8.8%), respectively. Table 7. Ability of S. aureus strains to form biofilm.
Table 8. Antimicrobial activity of some Lactobacillus strains against S. aureus.
The existence of bacteria capable of creating a biofilm is critically important in the food industry. A microbiological biofilm developed on the equipment and machinery in food sector plants not only threatens the hygienic quality of the processed food items but also additionally represents a risk to consumer health since the structure of the biofilm could incorporate several food-borne pathogens (Van Houdt and Michiels, 2010). Data in Table 7 represents the biofilm production ability of S. aureus strains. Moderate biofilm production was detected in five out of nine with the highest percentage (55.6%), followed by strong producers in two out of nine (22.2%), then weak and non-producers in one out of nine (11.1%) of each, respectively. Several investigations evaluated the biofilm production ability in S. aureus. Ballah et al. (2022) examined isolates of S. aureus and found that (97%) of isolates were able to form biofilm. In addition, 20 S. aureus isolates were identified as strong biofilm formers. Pajohesh et al. (2019) revealed that biofilm formation was shown in 35 strains (77.77%) of S. aureus, 20 out of 35 showed strong biofilm formation (44.44%), 15 out of 35 were weak producers (33.33%) and 10 out of 35 (22.22%) had no ability to produce biofilms. Bissong and Ateba (2020) illustrated that (90%) of S. aureus isolates were biofilm producers, out of which the majority 66 out of 70 (94.3%) were detected as strong producers. Organic acids, acetoin, diacetyl, bacteriocins, and hydrogen peroxide are among the antimicrobial metabolites synthesized by probiotics. These compounds help to minimize microbiological risk by suppressing pathogenic bacteria and restricting the growth of other microorganisms (Pyar and Peh, 2014). Table 8 assessed the antimicrobial activity of three Lactobacillus strains against the most virulent strain of S. aureus (MRSA strain). Lactobacillus acidophillus MK850930 had the strongest antimicrobial action against S. aureus with inhibition zone diameter ranged from 18 to 22 mm. While L. plantarum MK806485 and L. rhamnosus LMG23522 showed moderate action against S. aureus (14–16 mm) and (12–16 mm), respectively. Anas et al. (2008) stated that L. plantarum strain gives an inhibition diameter of 20 mm for S. aureus. While the antagonistic activity of different Lactobacillus (CFS) assessed by Koohestani et al. (2018) mentioned that the highest inhibition zone diameter (16 mm) against S. aureus was recorded with CFS of L. acidophilus LA5. ConclusionThe findings of this investigation revealed that S. aureus was excessively dispersed in the investigated specimens and contributed to life-threatening situations for consumers. The elevated incidence of S. aureus showing MDR highlights the serious issue of AMR in the dairy sector that threatens public health by traveling through the food chain. Moreover, the formation of biofilms is a worrisome issue due to the high survival rate in the environment. Therefore, it is crucial to apply firm measures that achieve the hygienic quality of milk and its products at all levels of production to minimize their cross-contamination hazards. In addition, this study provides evidence that the screened Lactobacillus strains possess a significant ability to suppress the growth of S. aureus under in vitro conditions. However, in-vivo trials are additionally required to assess whether they operate as probiotics in real-life conditions for human health benefits. ReferencesAbd El Halem, S.G. 2019. Prevalence and antibiotic resistance of Staphylococcus aureus isolated from raw milk and dairy products collected from Alexandria, Egypt. Alex. J. Food. Sci. Technol. 16, 25–33. Abunna, F., Tasew, N., Ragassa, F., Ayana, D. and Amenu, K. 2019. Handling practices, quality and safety of milk along the dairy value chains in selected sub cites of Addis Ababa, Ethiopia. Biomed. J. Sci. Tech. Res. 13, 9652–9665. Adams, M.R. 1999. Safety of industrial lactic acid bacteria. J. Biotechnol. 68, 171–178. Addis, M., Pal, M. and Kyule, M.N. 2011. Isolation and identification of Staphylococcus species from raw bovine milk in Debre Zeit, Ethiopia. Vet. Res. 4, 45–49. Ahmed, A.A.H., Maharik, N.M.S., Valero, A. and Kamal, S.M. 2019. Incidence of enterotoxigenic Staphylococcus aureus in milk and Egyptian artisanal dairy products. Food. Control. 104, 20–27. Alnakip, M.E., Youssef, M.Z., Abd-Elaal, S.F. and Bayoumi, M.A. 2023. Screening of food-borne Staphylococcus aureus and E. coli pathogens in Artisanal white soft Cheese in Delta Region, Egypt. J. Adv. Vet. Res. 13, 1203–1209. Anas, M., Eddine, H.J. and Mebrouk, K. 2008. Antimicrobial activity of Lactobacillus species isolated from Algerian raw goat’s milk against Staphylococcus aureus. World. J. Dairy. Food. Sci. 3, 39–49. Arena, M.P., Silvain, A., Normanno, G., Grieco, F., Drider, D. and Spano, G. 2016. Use of Lactobacillus plantarum strains as a bio-control strategy against food-borne pathogenic microorganisms. Front. Microbiol. 7, 464. Argudin, M.Á., Mendoza, M.C. and Rodicio, M.R. 2010. Food poisoning and Staphylococcus aureus enterotoxins. Toxins 2, 1751–1773. Balaban, N. and Rasooly, A. 2000. Staphylococcal enterotoxins. Int. J. Food. Microbiol. 61, 1–10. Ballah, F.M., Islam, M.S., Rana, M.L., Ferdous, F.B., Ahmed, R., Pramanik, P.K. and Rahman, M.T. 2022. Phenotypic and genotypic detection of biofilm-forming Staphylococcus aureus from different food sources in Bangladesh. Biology 11, 949. Bergdoll, M.S. 1983. Enterotoxins. In Staphylococci and staphylococcal infections. Eds., Easton, C.S.F. and Adlam, C. London, UK: Academic Press, pp: 559–598. Bergdoll, M.S. 1989. Staphylococcus aureus. In Foodborne bacterial pathogens. Ed., Doyle, M.P. New York, NY: Marcel Dekker, Inc., pp: 463–523. Biernbaum, E.K., Gnezda, A., Akbar, S., Franklin, R., Venturelli, P.A. and McKillip, J.L. 2021. Lactoferrin as an antimicrobial against Salmonella enterica and Escherichia coli O157:H7 in raw milk. Int. J. Dairy. Sci. 2, 92–97. Bissong, M.E.A. and Ateba, C.N. 2020. Genotypic and phenotypic evaluation of biofilm production and antimicrobial resistance in Staphylococcus aureus isolated from milk, North West Province, South Africa. Antibiotics 9, 156. Buhlmann, M.K., Bögli-Stuber, K., Droz, S. and Mühlemann, K. 2008. Rapid screening for carriage of methicillin-resistant Staphylococcus aureus by PCR and associated costs. J. Clin. Microbiol. 46, 2151–2154. Ceri, H., Olson, M.E., Stremick, C., Read, R.R., Morck, D. and Buret, A. 1999. The calgary biofilm device: new technology for rapid determination of antibiotic susceptibilities of bacterial biofilms. J. Clin. Microbiol. 37, 1771–1776. Clarisse, T., Michèle, S., Olivier, T., Valérie, E., Jacques-Antoine, H., Michel, G. and Florence, V. 2013. Detection and quantification of staphylococcal enterotoxin A in foods with specific and sensitive polyclonal antibodies. Food. Control. 32, 255–261. Clark, N.C., Cooksey, R.C., Hill, B.C., Swenson, J.M. and Tenover, F.C. 1993. Characterization of glycopeptide-resistant enterococci from US hospitals. Antimicrob. Agents. Chemother. 37, 2311–2317. CLSI. 2020. CLSI M100 performance standards for antimicrobial susceptibility testing, 30th ed. CLSI, Wayne, PA, USA. De Jong, A., El Garch, F., Simjee, S., Moyaert, H., Rose, M., Youala, M. and Vet Path Study Group. 2018. Monitoring of antimicrobial susceptibility of udder pathogens recovered from cases of clinical mastitis in dairy cows across Europe: vet path results. Vet. Microbiol. 213, 73–81. Donlan, R.M. and Costerton, J.W. 2002. Biofilms: survival mechanisms of clinically relevant microorganisms. Clin. Microbiol. Rev. 15, 167–193. Feyissa, N., Alemu, T., Birri, D.J. and Dessalegn, A. 2023. Isolation, identification, and determination of antibiogram characteristics of Staphylococcus aureus in cow milk and milk products (yoghurt and cheese) in West Showa Zone, Ethiopia. Int. Dairy. J. 137, 105503. Gundogan, N. and Avci, E. 2014. Occurrence and antibiotic resistance of Escherichia coli, Staphylococcus aureus and Bacillus cereus in raw milk and dairy products in Turkey. Int. J. Dairy. Technol. 67, 562–569. Hennequin, C., Aumeran, C., Robin, F., Traore, O. and Forestier, C. 2012. Antibiotic resistance and plasmid transfer capacity in biofilm formed with a CTX-M-15-producing Klebsiella pneumoniae isolate. J. Antimicrob. Chemother. 67, 2123–2130. Ibrahim, G.A., Sharaf, O.M. and El-Khalek, A.B.A. 2015. Microbiological quality of commercial raw milk, domiati cheese and kareish cheese. Middle. East. J. Appl. Sci. 5, 171–176. ISO. 2003. Part 3: microbiology of food and animal feeding stuffs horizontal method for the detection and identification of Staphylococci. Geneva, Switzerland: ISO. Johnson, W.M., Tyler, S.D., Ewan, F.E., Ashton, F.R., Pollard, D.R. and Rozee, K.R. 1991. Detection of genes for enterotoxins, exfoliative toxins, and toxic shock syndrome toxin 1 in Staphylococcus aureus by the polymerase chain reaction. J. Clin. Microbiol. 29, 426–430. Kandil, A.A., Elhadidy, M., El-Gamal, A. and Al-Ashmawy, M.A. 2018. Identification of S. aureus and E. coli from dairy products intended for human con-sumption. Adv. Anim. Vet. Sci. 6, 509–513. Koohestani, M., Moradi, M., Tajik, H. and Badali, A. 2018. Effects of cell-free supernatant of Lactobacillus acidophilus LA5 and Lactobacillus casei 431 against planktonic form and biofilm of Staphylococcus aureus. Vet. Res. Forum. 9(4), 301. Kou, X., Cai, H., Huang, S., Ni, Y., Luo, B., Qian, H. and Wang, X. 2021. Prevalence and characteristics of Staphylococcus aureus isolated from retail raw milk in Northern Xinjiang, China. Front. Microbiol. 12, 705947. Kumar, A., Alam, A., Rani, M., Ehtesham, N.Z. and Hasnain, S.E. 2017. Biofilms: survival and defense strategy for pathogens. Int. J. Med. Microbiol. 307, 481–489. Li, T., Lu, H., Wang, X., Gao, Q., Dai, Y., Shang, J. and Li, M. 2017. Molecular characteristics of Staphylococcus aureus causing bovine mastitis between 2014 and 2015. Front. Cell. Infect. Microbiol. 7, 127. Magiorakos, A.P., Srinivasan, A., Carey, R.B., Carmeli, Y., Falagas, M.E., Giske, C.G. and Monnet, D.L. 2012. Multidrug-resistant, extensively drug-resistant and pandrug-resistant bacteria: an international expert proposal for interim standard definitions for acquired resistance. Clin. Microbiol. Infect. 18, 268–281. Martineau, F., Picard, F.J., Lansac, N., Ménard, C., Roy, P.H., Ouellette, M. and Bergeron, M.G. 2000. Correlation between the resistance genotype determined by multiplex PCR assays and the antibiotic susceptibility patterns of Staphylococcus aureus and Staphylococcus epidermidis. Antimicrob. Agents. Chemother. 44, 231–238. Mestrovic, T., Aguilar, G.R., Swetschinski, L.R., Ikuta, K.S., Gray, A.P., Weaver, N.D. and Naghavi, M. 2022. The burden of bacterial antimicrobial resistance in the WHO European region in 2019: a cross-country systematic analysis. Lancet. Public. Health. 7, 897–913. Nam, H.M., Lee, A.L., Jung, S.C., Kim, M.N., Jang, G.C., Wee, S.H. and Lim, S.K. 2011. Antimicrobial susceptibility of Staphylococcus aureus and characterization of methicillin-resistant Staphylococcus aureus isolated from bovine mastitis in Korea. Foodborne. Pathog. Dis. 8, 231–238. Ning, K., Zhou, R. and Li, M. 2023. Antimicrobial resistance and molecular typing of Staphylococcus aureus isolates from raw milk in Hunan Province. Peer. J. 11, 15847. Nirwal, S., Pant, R. and Rai, N. 2013. Analysis of milk quality, adulteration and mastitis in milk samples collected from different regions of Dehradun. Int. J. Pharm. Tech. Res. 5, 359–364. Omoe, K., Imanishi, K.I., Hu, D.L., Kato, H., Takahashi-Omoe, H., Nakane, A. and Shinagawa, K. 2004. Biological properties of staphylococcal enterotoxin-like toxin type R. Infect. Immun. 72, 3664–3667. Pajohesh, R., Tajbakhsh, E., Momtaz, H. and Rahimi, E. 2019. Genotyping and distribution of putative virulence factors of Staphylococcus aureus isolated from dairy products in Shahrekord, Iran. Arch. Pharm. Pract. 10, 63–75. Pereira, V., Lopes, C., Castro, A., Silva, J., Gibbs, P. and Teixeira, P. 2009. Characterization for enterotoxin production, virulence factors, and antibiotic susceptibility of Staphylococcus aureus isolates from various foods in Portugal. Food. Microbiol. 26, 278–282. Perez-Roth, E., Claverie-Martın, F., Villar, J. and Mendez-Alvarez, S. 2001. Multiplex PCR for simultaneous identification of Staphylococcus aureus and detection of methicillin and mupirocin resistance. J. Clin. Microbiol. 39, 4037–4041. Pyar, H. and Peh, K.K. 2014. Characterization and identification of Lactobacillus acidophilus using biolog rapid identification system. Int. J. Pharm. Sci. 6, 189–193. Rahimi, E. 2013. Enterotoxigenicity of Staphylococcus aureus isolated from traditional and commercial dairy products marketed in Iran. Braz. J. Microbiol. 44, 393–399. Rall, V., Vieira, F., Rall, R., Vieitis, R., Fernandes, A., Candeias, J., Cardoso, K. and Araujo, J. 2008. PCR detection of staphylococcal enterotoxin genes in Staphylococcus aureus strains isolated from raw and pasteurized milk. Vet. Microbiol. 132, 408–413. Rammelsberg, M. and Radler, F. 1990. Antibacterial polypeptides of Lactobacillus spp. J. Appl. Bacteriol. 69,177–184. Saka, E. and Terzi Gulel, G. 2018. Detection of enterotoxin genes and methicillin-resistance in Staphylococcus aureus isolated from water buffalo milk and dairy products. J. Food. Sci. 83, 1716–1722. Samaha, H.A., Haggag, Y.N., Nossair, M.A. and Mohammad, H.S. 2012. Using some recent techniques in diagnosis of some zoonotic bacterial diseases transmitted through milk. Alex. J. Vet. Sci. 35, 11–21. Şanlıbaba, P. 2022. Prevalence, antibiotic resistance, and enterotoxin production of Staphylococcus aureus isolated from retail raw beef, sheep, and lamb meat in Turkey. Int. J. Food. Microbiol. 361, 109461. Shapiro, J.A. 1998. Thinking about bacterial populations as multicellular organisms. Annu. Rev. Microbiol. 52, 81–104. Shettigar, K. and Murali, T.S. 2020. Virulence factors and clonal diversity of Staphylococcus aureus in colonization and wound infection with emphasis on diabetic foot infection. Eur. J. Clin. Microbiol. Infect. Dis. 39, 2235–2246. Singh, S., Yadav, A.S., Singh, S.M. and Bharti, P. 2010. Prevalence of Salmonella in chicken eggs collected from poultry farms and marketing channels and their antimicrobial resistance. Food. Res. Int. 43, 2027–2030. Stepanovic´, S., Cirkovic´, I., Ranin, L. and Svabic´-Vlahovic´, M. 2004. Biofilm formation by Salmonella spp. and Listeria monocytogenes on plastic surface. Lett. Appl. Microbiol. 38, 428–432. T´oth, A.G., Csabai, I., Krik´o, E., T˝ozs´er, D., Mar´oti, G., Patai, ´A.V., Makrai, L., Szita, G. and Solymosi, N. 2020. Antimicrobial resistance genes in raw milk for human consumption. Sci. Rep. 10, 7464. Thaker, H.C., Brahmbhatt, M.N., Nayak, J.B., and Thaker, H.C. 2013. Isolation and identification of Staphylococcus aureus from milk and milk products and their drug resistance patterns in Anand, Gujarat. Vet. World. 6, 10–13. Van Boeckel, T.P., Brower, C., Gilbert, M., Grenfell, B.T., Levin, S.A., Robinson, T.P., Teillant, A. and Laxminarayan, R. 2015. Global trends in antimicrobial use in food animals. Proc. Natl. Acad. Sci. U. S. A. 112, 5649–5654. Van Houdt, R. and Michiels, C.W. 2010. Biofilm formation and the food industry, a focus on the bacterial outer surface. J. Appl. Microbiol. 109, 1117–1131. Wells, J.M. 2011. Immunomodulatory mechanisms of lactobacilli. Microb. Cell. Fact. 10, 1–15. Zeinhom, M. and Abed, A. 2020. Prevalence, characterization, and control of Staphylococcus aureus isolated from raw milk and Egyptian soft cheese. J. Vet. Med. Res. 27, 152–160. Zhang, J., Wang, J., Jin, J., Li, X., Zhang, H., Shi, X. and Zhao, C. 2022. Prevalence, antibiotic resistance, and enterotoxin genes of Staphylococcus aureus isolated from milk and dairy products worldwide: a systematic review and meta-analysis. Food. Res. Int. 162, 111969. Zhao, X., Yuan, X., Hu, M., Zhang, Y., Li, L., Zhang, Q. and Liu, Y. 2021. Prevalence and characterization of Staphylococcus aureus and methicillin-resistant Staphylococcus aureus isolated from bulk tank milk in Shandong dairy farms. Food. Control. 125, 107836. | ||
| How to Cite this Article |
| Pubmed Style Mohammed AR, El-Said EI, ElAal SFA, Kamal RM, . Screening of antibiogram, virulence factors and biofilm production of Staphylococcus aureus and the bio-control role of some probiotics as alternative antibiotics. Open Vet J. 2024; 14((1) (Zagazig Veterinary Conference)): 176 -185. doi:10.5455/OVJ.2024.v14.i1.16 Web Style Mohammed AR, El-Said EI, ElAal SFA, Kamal RM, . Screening of antibiogram, virulence factors and biofilm production of Staphylococcus aureus and the bio-control role of some probiotics as alternative antibiotics. https://www.openveterinaryjournal.com/?mno=175633 [Access: May 08, 2024]. doi:10.5455/OVJ.2024.v14.i1.16 AMA (American Medical Association) Style Mohammed AR, El-Said EI, ElAal SFA, Kamal RM, . Screening of antibiogram, virulence factors and biofilm production of Staphylococcus aureus and the bio-control role of some probiotics as alternative antibiotics. Open Vet J. 2024; 14((1) (Zagazig Veterinary Conference)): 176 -185. doi:10.5455/OVJ.2024.v14.i1.16 Vancouver/ICMJE Style Mohammed AR, El-Said EI, ElAal SFA, Kamal RM, . Screening of antibiogram, virulence factors and biofilm production of Staphylococcus aureus and the bio-control role of some probiotics as alternative antibiotics. Open Vet J. (2024), [cited May 08, 2024]; 14((1) (Zagazig Veterinary Conference)): 176 -185. doi:10.5455/OVJ.2024.v14.i1.16 Harvard Style Mohammed, A. R., El-Said, E. I., ElAal, S. F. A., Kamal, R. M. & (2024) Screening of antibiogram, virulence factors and biofilm production of Staphylococcus aureus and the bio-control role of some probiotics as alternative antibiotics. Open Vet J, 14 ((1) (Zagazig Veterinary Conference)), 176 -185. doi:10.5455/OVJ.2024.v14.i1.16 Turabian Style Mohammed, Aya R., Esmat I. El-Said, Salah F. Abd ElAal, Rania M. Kamal, and . 2024. Screening of antibiogram, virulence factors and biofilm production of Staphylococcus aureus and the bio-control role of some probiotics as alternative antibiotics. Open Veterinary Journal, 14 ((1) (Zagazig Veterinary Conference)), 176 -185. doi:10.5455/OVJ.2024.v14.i1.16 Chicago Style Mohammed, Aya R., Esmat I. El-Said, Salah F. Abd ElAal, Rania M. Kamal, and . "Screening of antibiogram, virulence factors and biofilm production of Staphylococcus aureus and the bio-control role of some probiotics as alternative antibiotics." Open Veterinary Journal 14 (2024), 176 -185. doi:10.5455/OVJ.2024.v14.i1.16 MLA (The Modern Language Association) Style Mohammed, Aya R., Esmat I. El-Said, Salah F. Abd ElAal, Rania M. Kamal, and . "Screening of antibiogram, virulence factors and biofilm production of Staphylococcus aureus and the bio-control role of some probiotics as alternative antibiotics." Open Veterinary Journal 14.(1) (Zagazig Veterinary Conference) (2024), 176 -185. Print. doi:10.5455/OVJ.2024.v14.i1.16 APA (American Psychological Association) Style Mohammed, A. R., El-Said, E. I., ElAal, S. F. A., Kamal, R. M. & (2024) Screening of antibiogram, virulence factors and biofilm production of Staphylococcus aureus and the bio-control role of some probiotics as alternative antibiotics. Open Veterinary Journal, 14 ((1) (Zagazig Veterinary Conference)), 176 -185. doi:10.5455/OVJ.2024.v14.i1.16 |