| Research Article | ||
Open Veterinary Journal, (2024), Vol. 14(1): 214-224 Original Research Escherichia coli and Salmonella enterica isolated from Egyptian dairy cattle herds: The prevalence and molecular characteristicsMahmoud S.A. Zaki, Amr M.M. Abd-El-All, Amira S.A. Attia*, Hesham Dahshan, Manal A. Al-Ashery and Ayman MegahedDepartment of Veterinary Public Health, Faculty of Veterinary Medicine, Zagazig University, Zagazig, Egypt *Corresponding Author: Amira S.A. Attia. Department of Veterinary Public Health, Faculty of Veterinary Medicine, Zagazig University, Zagazig, Egypt. Email: dr.attiamirasamir [at] gmail.com Submitted: 01/10/2023 Accepted: 15/12/2023 Published: 31/01/2024 © 2024 Open Veterinary Journal
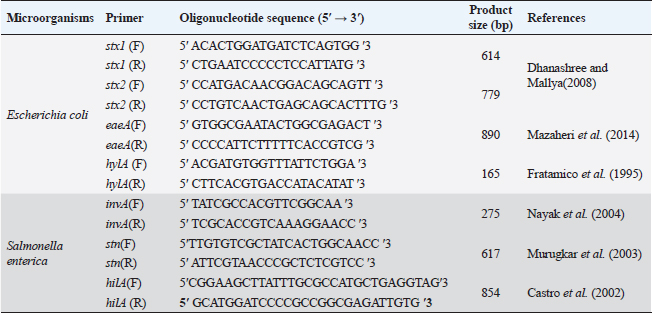
ABSTRACTBackground: The pathogens Escherichia coli and Salmonella enterica that caused substantial health problems and financial losses were believed to have originated primarily from Egypt’s dairy farms. Aim: The purpose of this study was to ascertain the occurrence of E. coli and S. enterica in three large dairy farms located in the Egyptian governorate of Sharkia. Furthermore, biochemical and serological characteristics of the isolated isolates were described. Further analysis revealed that several E. coli serovars had the genes stx1, stx2, eaeA, and hylA, while invA, stn, and hilA genes were found in several S. enterica serotypes using a multi-plex PCR. Methods: A total of 540 samples of fresh raw cow milk, water, feedstuffs, feces, (108 each), as well as swabs from feeders, milker hands and cattle crushes (36 each ), were gathered and analyzed. Results: The recovery of E. coli from various sampling sources was shown to have an overall prevalence of 62.2% (336/540) in the results. Fecal samples had isolated S. enterica, with a frequency of 0.74% (4/540). The existence of various groups of serovars, such as O26, O44, O55, O78 and O111 for E. coli and Salmonella enteritidis, Salmonella typhimurium and Salmonella inganda for S. enterica was revealed by serological identification of the two species. However, it was discovered that a number of E. coli serovars had much higher percentages of the eaeA and hylA genes as well as shiga-toxin types 1 and 2 (stx1 and stx2). The presence of the invA gene, a diagnostic marker for S. enterica was 100% across all serovars. Salmonella enteritidis possessed both the enterotoxin gene (stn) and the hyper-invasive locus gene (hilA). Salmonella typhimurium had the hilA gene, whereas S. inganda had the stn gene. Conclusion: Escherichia coli and S. enterica recovered in this study have significant genetic risk factors for high pathogenicity and virulence, posing a real threat to dairy population productivity and health, which could spread to the general public through milk. Keywords: Dairy herds, E. coli, S. enterica, Multiplex PCR, Shiga toxins. IntroductionAmong all African nations, Egypt has one of the biggest populations of dairy products. Due to their significant significance in income generation and job creation, dairy farms make up a significant portion of Egypt’s overall economy. With a national plan that extends until 2030 to meet the growing local market demand for dairy products and meat, Egypt’s national sector and the special investment sector are collaborating to increase the country’s dairy population. Animal health, farm environment, production and storage conditions, farm management, season, and geographic location are some of the variables that influence the quantity of harmful bacteria in milk (Santorum et al., 2012). The production, distribution, or manufacturing procedures are all potential sources of bacterial contamination in milk (Garedew et al., 2012). The consumption of milk and dairy products has been related to about 5% of human foodborne diseases (Holt et al., 2011). Enteropathogenic food-born Escherichia coli bacteria have been linked to severe, occasionally deadly diarrhea in children, and cases have been documented in developing nations (Mora et al., 2011). The most prevalent strains of shiga toxin-producing E. coli (STEC) are those that belong to serogroups O26, O91, O103, O111, O118, O145, and O166. This is because these strains are more likely to be found in the environment since they have adapted to live in the colons of healthy humans and animals. Furthermore, according to Paton and Paton (1998), “STEC” describes strains of E. coli that express the stx1 and stx2 genes, respectively, and are able to produce either type 1 (stx1), type 2 (stx2), or both. These strains are believed to be responsible for the vascular endothelial damage seen in patients with hemorrhagic colitis (HC), hemolyticuraemic syndrome and acute or chronic renal failure (Mayer et al., 2012; Bitzan and Lapeyraque, 2016). Mousa and Shama (2020) carried out additional investigation on sixteen E. coli strains that were obtained from dairy farms in the Menoufiya area of Egypt. They used PCR to look at six virulence genes (iss, fimH, tsh, iutA, stx2, and eaeA). Nonetheless, Hailu et al. (2021) discovered that aadA, aphA1 and mphA were the most often discovered resistance genes. It was found that E. coli O157 isolates from small-scale agricultural settings in Northeastern Ohio treated with dairy cattle manure were prevalent, with a percentage of 1.8. A zoonotic disease known as salmonellosis can infect adult cattle as well as calves and result in serious sickness. Anaemia, diarrhea, dehydration, fever, abortion, and endotoxemia can all result from bovine salmonellosis. Since invasion A (invA) contains sequences unique to the genus Salmonella, it is one of the greatest pathogenicity factors and is employed as a biomarker for the identification of Salmonella species (Nikiema et al., 2021). In addition to the AvrA gene, which increases the virulence of Salmonella spp. by inhibiting TNF-α and IL-8 and restricting the host’s inflammatory responses by inducing cell death, especially in macrophages, the plasmid’s spvC virulence-related gene is also required for Salmonella survival within the host cell. Diarrhea occurs on by an accelerated loss of intestinal fluids, which is caused by the Salmonella enterotoxin gene (Ben-Barak et al., 2006). However, Salmonella enteritidis, Salmonella typhimurium, Salmonella heidelberg, Salmonella infants, Salmonella tsevie, and Salmonella haifa were found to be present in 24% of the row farm milk in Mansoura, Egypt, according to El-Baz et al. (2017). These serovars had percentages of 33.3%, 25.9%, 14.8%, 11.11%, 11.11%, and 3.7%, respectively. Wang et al. (2023) found 3% of positive samples for Salmonella in Henan, China. The samples came from 35 lactating cows that were resistant to tetracycline and florfenicol. Preventive measures include sufficient animal feeding supplies, excellent hygiene procedures with consumer safety knowledge, and the application of control points across the dairy chain must be done in order to limit the risks connected with milk intake (Owusu-Kwarteng et al., 2020). Finding out how common E. coli and Salmonella enterica are right now in three sizable dairy farms is the aim of this investigation. Seasonal fluctuation and the source of recovery were taken into consideration while characterizing these infections. Using multiplex PCR, the organisms were identified serologically and they harbor for certain genes associated with patogenicity and virulence, such as stx1, stx2, eaeA and hylA for E. coli and invA, stn and hilA for S. enterica. Materials and MethodsStudy farms and populationThree sizable dairy farms in Egypt’s El-Sharkia area served as the study’s sites: Farm I: The Sami Asaad farm, with roughly 1,000 Holstein dairy cows, is situated in the village of Hana Merham. Farm II: The Al-Salheya dairy farm is situated in the city of Al-Salheya and has roughly 2,000 Holstein dairy cows. Farm III: A dairy farm with approximately 1,250 Holstein dairy cows, situated in the village of Al-Talline. Earthen flooring provided shelter for all the animals, who were kept in open yards with some cover from direct sunlight and rain. The milk parlor automatically milked every animal twice a day. Teat dipping with iodine 2,500 ppm by 1:6 ratio was the only pre-milking procedure allowed. Post-milking teat dipping used the same ratio of iodine 2,500 ppm by 3:1. During the period of July 2022 to June 2023, each farm was visited on a monthly basis for the four seasons. Transportation and sample collectionThroughout the experiment, 540 samples were collected from the farms that were being studied. The following samples were taken at random: 108 samples each from raw milk, water, feedstuffs and fecal matter; 36 samples each from milker’s hands, cattle crushes, and feeders swabs. To avoid any unanticipated alterations, the obtained samples were stored in an ice tank before being sent to the laboratory for quick investigation. Test tubes that had been sanitized, polyethylene plastic bags, cotton-tipped swabs that had been moistened with buffered peptone water (BPW) and sterile plastic bottles with a 150 ml capacity were all used to gather samples. All sample and handling techniques, such as the use of sterile materials, flaming and freezing were carried out according to aseptic technique. Sampling technique and preparationRaw milkFrom dairy animals on the farms under investigation, raw milk samples were randomly taken and placed in aseptic plastic bottles that had been cleaned, dried, and sterilized. Drinking water and feedstuffsFrom the individual drinking troughs of the animals, water samples were collected into single-use, sterile, dry and clean test tubes. While feed samples were being taken from each individual cow feeding bucket and placed in a sterile polyethylene plastic bag. Faecal matterDirect collection of feces from the rectum was done via back racking. A sterile polyethylene plastic bag held about 50 g of excrement. Nonetheless, animal feces samples that had diarrhea were placed individually in sterile plastic vials. Milker hands, feeders and cattle crushesA sterile wooden cotton swab was used to collect swabs from milker hands, feeders, and cow crushes. The swab was wiped on the material’s surface and then placed into a 10 ml test tube along with 5 ml of sterile BPW. Every sample was correctly coded according to the date of collection, the source of the sample, and the type of sample. Following that, it was sent in an ice box to be analyzed at the Microbiological Laboratory of the College of Veterinary Medicine at Zagazig University. Bacterial isolation and identificationEscherichia coliAfter adding 5 ml of the obtained samples to sterilized tubes containing 45 ml of BPW, the tubes were incubated at 37°C for a full 24 hours. The enrichment was achieved by adding one loopfull of the incubated broth to 10 ml of MacConkey broth, which was then incubated aerobically for 24 hours at 37°C. A loopful of incubated MacConkey broth was spread out and incubated for 18 to 24 hours at 37°C on the surface of Eosin Methylene Blue agar. Methyl red (MR) and voges-proskaure (VP) tests were performed, along with biochemical testing (urease production, lysine decarboxylase production, citrate utilization, H2S production, and indole production) on small green fluorescence colonies that had been selected and stained with gram stain. On the other hand, the nutrient agar slopes were streaked with the purified colony and left to incubate at 37°C for 18 to 24 hours. Once it was ready for PCR and serological identification, it was then frozen (Quinn et al., 1994). Salmonella entericaFive ml of the samples that were obtained were placed into sterilized tubes with 45 ml of BPW, and the tubes were then incubated for 24 hours at 37°C. Ten ml of Rappaport Vassiliadis Soya broth were mixed with 1 ml of the pre-enriched culture and the mixture was incubated for 24 hours at 42°C. Finally, a single loop of broth culture was streaked over Xylose Lysine Deoxycholate, and the mixture was incubated for 24 hours at 37°C. The suspected colonies displayed a somewhat translucent reddish-colored zone with a black center (Humphrey et al., 1989). Biochemical tests (urease production, MR and VP testing, lysine decarboxylase production, citrate utilization, H2S production and indole production) were carried out on these colonies. It was then put onto a nutrient agar slant and cultured for 24 hours at 37°C to perform additional tests (Macfaddin, 2000). Serological identificationEscherichia coli isolates were serologically identified using fast diagnostic E. coli antisera sets (DENKA SEIKEN Co., Japan) for Enteropathogenic type diagnosis (Kok et al., 1996). On the other hand, Salmonella was detected using Salmonella antiserum (DENKA SEIKEN Co., Japan) in compliance with the Kauffman–White technique (Kauffman, 1974) for the identification of Somatic (O) and Flagellar (H) antigens. Multiplex PCRDifferent E. coli serotypes were screened for Stx1, Stx2, eaeA and hlyA. To find out which Salmonella serovars possessed the stn, hilA and invA genes, tests were conducted. Bacterial DNA was extracted using the Gene JET Genomic DNA Purification Kit (Fermentas) (Sambrook et al., 1989). A total volume of 25 ml [6 ml DNA template, 20 promol of each primer, and 12.5 ml of Emerald Amp GT PCR mastermix (2x premix)] were used in the reaction. The PCR mixture was then heated in a thermal cycler for the following cycling conditions: 95°C (3 minutes), 95°C (20 seconds), 58°C (20 seconds), 72°C (1.5 minutes), and 72°C (5 minutes). For E. coli and Salmonella molecular identification, eight primers were utilized in addition to six primers, respectively (Table 1). Statistical analysisThe correlation or difference between groups (farms, season, and sources) with the prevalence of Salmonella and E. coli was tested using the chi-square test and Fisher’s exact test. A statistically significant result is defined as p < 0.05. IBM Corp., Armonk, NY’s SPSS version 24.0 was used for all analyses (McHugh, 2013). Table 1. PCR primers of virulence gene in E. coli and S. enterica.
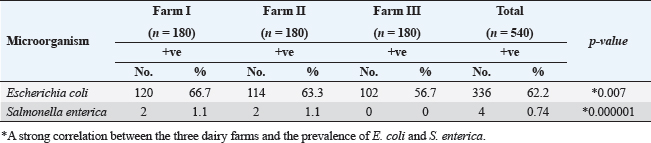
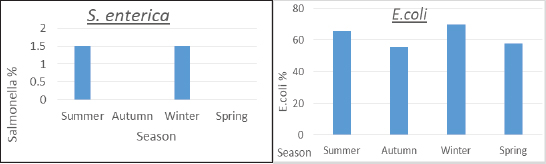
ResultsTable 2 showed that farm III had a lower prevalence of E. coli isolates (56.7%), while farms I and II had the least variation in prevalence (66.7% and 63.3%, respectively). About the frequency of S. enterica on different farms, Table 2 also reveals that samples from farm III did not include any of the bacteria, while it was isolated at equal percentages from farms I and II (1.1%). Between the three farms under investigation, there was a stronger association between E. coli and S. enterica, according to statistical analysis ( p=0.007 and 0.000001). In the dairy farms that were examined, the prevalence rate of E. coli was much higher in the winter (69.6%) than it was in the summer (65.9%), and it was higher in the spring (57.8%) than it was in the autumn (55.6%). Salmonella enterica was also detected with a similar prevalence (1.5%) in the summer and winter, but was absent in the other seasons, as shown in Figure 1. Table 2. Prevalence of E. coli and S. enterica in the dairy farms under investigation.
Fig. 1. Seasonal recovery of E. coli and S. enterica from dairy cattle under examination. Table 3. Escherichia coli and S. enterica relative recovery percentages from various sampling sites in the dairy populations under investigation.
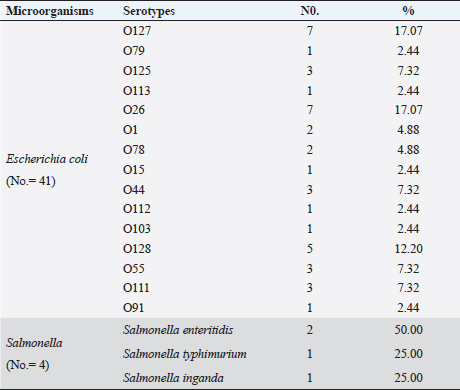
Table 4. Serological identification of the isolated E. coli and Salmonella species.
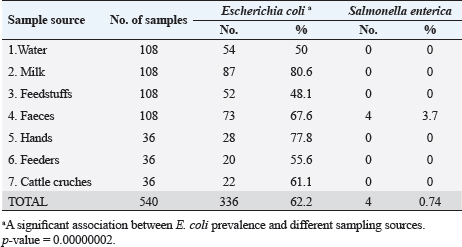
Escherichia coli was found in 336 of the 540 samples, representing a rate of 62.2% overall (Table 3). A total of 50%, 48.1%, 55.6% and 61.1% of E. coli was recovered from water, feedstuffs, feeders and cattle crushes swaps, respectively. The highest recovery percentage of E. coli came from milk (80.6%), hand swabs (77.8%) and feces (67.6%). A significant association was observed between E. coli prevalence and different sampling sources ( p-value=0.00000002). However, only 3.7% of the cow fecal samples (4/108) across all sampling locations had S. enterica isolated, with an overall recovery percentage of 0.74% (4/540). There was no significant correlation between S. enterica and different sampling sources. Out of 336 E. coli isolates that were recovered, 41 isolates were identified using serology and were chosen at random based on factors such as farms, season, and sampling site. The most prevalent serovars of E. coli were O127 and O26 (17.07%), which were followed by O128 (12.20%), O125, O44, O55, and O111 (7.32% each), O78, and O1 (4.88% each). Among the additional E. coli strains identified (2.44% each) were serovars O79, O113, O15, O112, O103, and O91, as shown in Table 4. The recovered S. enterica isolates were identified as S. typhimurium and Salmonella inganda, with a percentage of 25% each and a proportion of 50% (2/4) for S. enteritidis (Table 4).
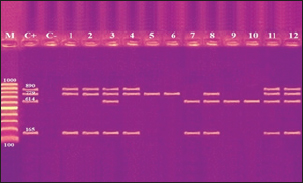
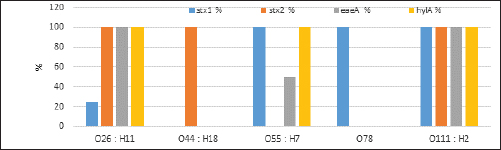
Fig. 2. Agarose gel electrophoresis product of multiplex PCR for presence of stx1 (614 bp), stx2 (799 bp), eaeA (890 bp) and hlyA (165 bp) genes in different E. coli strains. Lane M: 100 bp ladder as molecular size DNA marker;Lane C+: Control +ve E. coli for stx1, stx2,eaeA & hlyA genes; Lane C-: -ve control E. coli for stx1, stx2, eaeA & hlyA genes. Lanes 1, 2, 4 (O26) & 8 (O55): +ve strains for stx2, eaeA and hlyA genes.Lanes 3 (O26), 11 & 12 (O111): +ve strains for stx1, stx2, eaeA and hlyA genes.Lanes 5 & 6 (O44): +ve strain for stx2 gene. Lane 7 (O55): +ve strain for stx1and hlyA gene. Lanes 9 & 10 (O78): +ve strain for stx1 gene. The relative frequency of virulent genes in various E. coli strains obtained from Egyptian dairy cattle farms is displayed in Figures 2 and 3. Shiga-toxin 1 gene (stx1) was found to be absent in O44 E. coli strains, but present in 100% of the tested O55, O78 and O111 strains as well as 25% of O26 strains. Out of the three strains of E. coli tested, O26, O44, and O111, all had 100% of the shiga-toxin 2 gene (stx2), while the other two (O55 and O78) did not have any detectable results. Furthermore, intimin gene (eaeA) appeared in all three tested strains of E. coli (O26, O111 and O55), but not in the other two (O44 and O78). The haemolysin gene (hylA) was found in all studied serovars of E. coli (O26, O55 and O111), but not in O44 or O78.
Fig. 3. Relative estimates for the frequency of virulence genes in enteropathogenic E. coli isolated from dairy cattle farms.
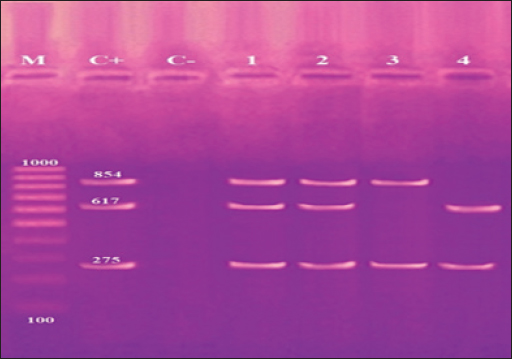
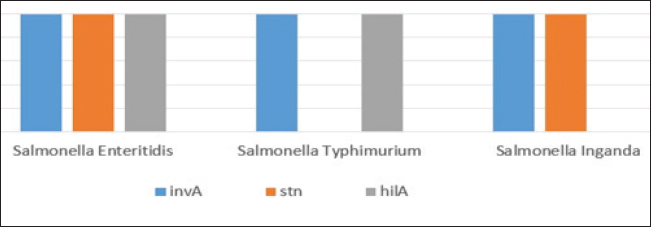
Fig. 4. Agarose gel electrophoresis product of multiplex PCR for presence of invA (275 bp), stn (617 bp) and hilA (854 bp) genes virulence genes in different S. enterica serovars. Lane M: 100 bp ladder as molecular size DNA marker. Lane C+: Control +ve strain for invA, stn and hilA genes.Lane C-: Control -ve.Lanes1 & 2 (S. enteritidis): +ve for invA, stn & hilAgenes. Lane 3 (S. typhimurium): +ve for invA and hilA genes. Lane 4 (S. inganda): +ve for invA and stn genes. All three Salmonella serovars (S. enteritidis, S. typhimurium, and S. inganda) had the invasion gene (invA), according to a pattern of distribution of virulence and diagnostic genes in distinct S. enterica seovars. Salmonella enteritidis possessed both the enterotoxin gene (stn) and the hyper-invasive locus gene (hilA). Salmonella typhimurium had the hilA gene, whereas S. inganda had the stn gene (Figs. 4 and 5). DiscussionOne of the most significant sources of nutrients for human health is milk and its by products. To prevent any biological, physical, or chemical hazards, it must be manufactured in a fully sterile environment. In addition to the degree of hygiene standards followed in the dairy farms, the detection of foodborne pathogens in milk mostly relates to both direct and indirect contact between dairy animals and their milk (Owusu-Kwarteng et al., 2020).
Fig. 5. Distribution pattern of virulence genes in different S. enterica serovars isolated from dairy cattle farms. There was a high overall prevalence (62.2%) of E. coli recovered in this investigation, regardless of the farm, season, or source of recovery. Ameen et al. (2019) recorded a lower frequency (17%) on dairy farms in Egypt. However, Makkia et al. (2022) discovered that 43.64% of dairy farms in Egypt’s Dakahlya area had E. coli infections. Conversely, data from Tanzania showed a very high (90.67%) prevalence of E. coli (Lubote et al., 2014). In this case, the main cause of significant difference may be the variations in sample locations and procedures, as our observation does not pose a conflicting issue. Seasonally, E. coli was recovered by 65.9% in the summer and 69.6% in the winter. Approximately 1.5% of cases were found in the summer and winter, with a reduced overall prevalence of 0.74%. Comparing this study to earlier literature, the prevalence rate of Salmonella recovery is significantly lower (Blau et al., 2005; Sobur et al., 2019; Abrar et al., 2020; Fesseha et al., 2020; Moustafa et al., 2020; Geletu et al., 2022). Changes in host and microbial density, as well as seasonal variations in the occurrence of enteric infections, can all contribute to this phenomenon in dairy farms. In Egypt, the winter months were the times when pathogenic bacteria were most prevalent because to increased calving rates and cold stress. These bacteria may have returned over the summer because of the optimal humidity and temperature, which encouraged the development and abundance of environmental bacteria in bedding material (Zeinhom et al., 2016). Furthermore, summer heat stress affects cows’ susceptibility to infection by reducing their resistance to harmful bacteria or increasing their exposure to them because the dairy environment is suitable to their growth and multiplication. In contrast to our results, Moustafa et al. (2020) found that the spring season in Egypt had the highest seasonal rate of salmonellosis in cow and buffalo calves, followed by winter and summer seasons. Moreover, Lambertini et al. (2015) examined seasonal variation on three US dairy farms and discovered that for two of the farms, there was no apparent seasonal influence. Furthermore, Heuvelink et al. (1998) discovered that O157 VTEC thrives and survives better in the Dutch dairy environment throughout the summer because to the warmer and more humid weather. The most significant biological hygiene indicator in animal farms is thought to be E. coli. Because of this, initial information regarding the hygiene score of the farms under investigation can be obtained from the location and recovery rates of this bacterium. Different levels of E. coli were isolated from each sampling site in the current investigation. With an 80.6% recovery rate, milk had the greatest percentage, followed by hand swabs (77.8%), fecal matter (67.6%), cattle crush swabs (61.1%), feeder swabs (55.6%), water (50%) and feed (48.1%). The present study’s heightened E. coli recovery percentage from milk is greater than the 63% reported by Ali and Abdelgadir (2011) from El-Khartoum. Conversely, Lubote et al. (2014) showed better results in row milk that was taken from Arsha, Tanzania (90.67%). Additionally, lower values (23.7%) were reported by Abebe et al. (2014) in Tigray, Ethiopia, and lower results (9.6%) were reported by Geletu et al. (2022) in Central Ethiopia. Additionally, in dairy cattle fecal testing, the prevalence rates of O157 (0.2% to 48.8%) and non-O157 STEC (0.4% to 74.0%) differed significantly (Hussein and Sakuma, 2005). It is important to draw attention to the greater percentage of E. coli recovered from milk at the dairy farms under investigation in this study. It is therefore very concerning to raise the hygiene standards in these farms in order to reduce the high milk contamination along the milk chain (during the milking, storage, handling, and transportation stages) (Ahmedsham et al., 2018). High levels of E. coli contamination on workers’ hands, as demonstrated by this study, should be given careful consideration because they pose a risk to other animals and environments (feed stores, water, etc.) as occupational carriers of infection. In addition, the study’s widely dispersed, elevated recovery percentage of E. coli from various sampling sites (milk, feed, water, feces, swabs from hands, cattle crush and feeders) could encourage farm owners and managers to implement strict biosecurity procedures in order to ensure a decreased risk of disease and clean milk production (Singh et al., 2020). It is evident from this study that all Salmonellae were recovered from feces at the S. enterica recovery sites, with a rate of 3.7% (4/108). The extremely low prevalence rate (0.74%) of S. enterica may be the reason for its absence in other sample sites. The use of anti-Salmonella feed additives may help to partially explain the study’s lower overall prevalence rate of Salmonella (0.74%) (Adetoye et al., 2018). A similar discovery was made by Galal et al. (2013) in Egyptian dairy farms at Kafr El Shiek governorate, and our serological results confirm this. They found O25, O26, O55, O78, O86, O111, O119, O127, and O158. In French raw milk, however, O26, O103, O111, O145, and O157 were found by Madic et al. (2011). Also found in Scottish cattle in the Scottish State of the United Kingdom were O26, O103, and O145 (Pearce et al., 2006). Through the presence of virulence genes, the recovered E. coli serotypes from dairy cattle farms in this investigation demonstrated a strong capacity for pathogenicity and virulence. Higher pathogenicity is caused by the EaeA gene, which increases attachment to epithelial cells in vitro and in the body of an animal (Yang et al., 2020). Out of all the O26:H11 and O111:H2 samples that failed multi-plex PCR testing, the later gene was found in 50% of them. Additionally, it was established through analysis of the sample from stx1 and stx2 that the recovered E. coli serotypes were schigo toxigenic. Due to their ability to inactivate eukaryotic ribosomes enzymatically, these genes are important virulence factors for E. coli strains (STEC), allowing them to cause hemolytic uremic syndrome and severe HC (Pacheco and Sperandio, 2012). Of the serovars tested in our study, stx1 was found in 100% of O55:H7, O78 and O111:H2 serovars and in 25% of O26:H11 serovars; stx2 was found in 100% of O26:H11, O44:H18 and O111:H2 serovars. Unfortunately, significant but fatal pathological alterations can result from the hypershigatoxigenicity observed at a very high percentage in different E. coli serotypes from Egyptian dairy cattle. Lastly, 100% of O26: H11; O55:H7, and O111:H2 serovars were discovered to harbor the hylA gene. According to Karam et al. (2019), this gene is thought to be a significant virulence factor and may be a major cytotoxin that causes severe urinary tract infections and peritonitis in humans and animals. While the majority of the E. coli serotypes under investigation exhibited noticeable pathogenicity and virulence characteristics, O26:H11 and O111:H2 were the most affected, posing a significant risk to cattle and consequently human health. Despite the study’s lower prevalence (0.74%) of S. enterica recovered from dairy cattle farms, the virulence parameters associated with the detected Salomonella serovars indicated their considerable threat to cattle populations in Egypt. The invA gene, which is thought to be a biomarker for Salmonella identification, regulates the invasion of intestinal epithelial cells, which determines the intracellular pathogenicity of Salmonella (Mohammed, 2022). Undoubtedly, the invA gene was present in all Salmonella serovars that this investigation detected, including S. enteritidis, S. typhimurium, and S. inganda. Furthermore, the stn gene has been found in S. enteritidis and S. inganda. According to Nakano et al. (2012), this gene maintains bacterial hemostasis and improves membrane integrity. On the other hand, hilA gene was recovered from both S. enteritidis and S. typhimurium. The hilA gene is responsible for encoding an activator for invasion gene expression (Durant et al., 2000). The presence of the genes stn, hilA and invA in the S. enteritidis recovered in this investigation is noteworthy and suggests the epidemiological significance of these genes in Egyptian dairy farms. ConclusionThe study concludes that the high pathogenicity and virulence of the recovered E. coli and S. enterica were caused by important genetic risk factors, posing a serious threat to dairy population productivity and health, which could potentially spread to the general public through consumption of milk. Preventive steps include maintaining adequate supplies of animal feed, maintaining high standards of hygiene and providing consumer safety information, as well as putting control points in place across the dairy chain, are required to lower the hazards connected with consuming milk. AcknowledgmentNone. Conflict of interestThe authors declare that there is no conflict of interest. FundingNone. Data availabilityAll data are provided in the manuscript. ReferencesAbebe, M., Hailelule, A., Abrha, B., Nigus, A., Birhanu, M., Adane, H. and Haftay, A. 2014. Antibiogram of Escherichia coli strains isolated from food of bovine origin in selected Woredas of Tigray, Ethiopia. J. Bacteriol. Res. 6, 17–22. Abrar, A., Beyene, T. and Furgasa, W. 2020. Isolation, identification and antimicrobial resistance profiles of Salmonella from dairy farms in Adama and Modjo Towns, Central Ethiopia. Eur. J. Med. Health. Sci. 2, 1–11. Adetoye, A., Pinloche, E., Adeniyi, B.A. and Ayeni, F.A. 2018. Characterization and anti-Salmonella activities of lactic acid bacteria isolated from cattle faeces. BMC. Microbiol. 18, 1–11. Ahmedsham, M., Amza, N. and Tamiru, M. 2018. Review on milk and milk product safety, quality assurance and control. Int. J. Livest. Prod. 9, 67–78. Ali, A.A. and Abdelgadir, W.S. 2011. Incidence of Escherichia coli in raw cow’s milk in Khartoum state. Br. J. Dairy. Sci. 2, 23–26. Ameen, F., Reda, S.A., El-Shatoury, S.A., Riad, E.M., Enany, M.E. and Alarfaj, A.A. 2019. Prevalence of antibiotic resistant mastitis pathogens in dairy cows in Egypt and potential biological control agents produced from plant endophytic actinobacteria. Saudi. J. Biol. Sci. 26, 1492–1498. Ben-Barak, Z., Streckel, W., Yaron, S., Cohen, S., Prager, R. and Tschäpe, H. 2006. The expression of the virulence-associated effector protein gene avrA is dependent on a Salmonella enterica specific regulatory function. In. J. Med. Microbiol. 296, 25–38. Bitzan, M. and Lapeyraque, A.L. 2016. Postinfectious hemolytic uremic syndrome. In Pediatric kidney disease. Eds., Geary, D.F. and Schaefer, F. Berlin, Germany: Springer, pp: 653–731. Blau, D.M., McCluskey, B.J., Ladely, S.R., Dargatz, D.A., Fedorka-Cray, P.J., Ferris, K.E. and Headrick, M.L. 2005. Salmonella in dairy operations in the United States: prevalence and antimicrobial drug susceptibility. J. Food. Prot. 68, 696–702. Castro, N., Pineda, E. and Ochoa, M. 2002. Detection of hilA gene sequences in serovars of Salmonella enterica subspecies enterica. Mem. Inst. Oswaldo. Cruz. 97, 1153–1156. Dhanashree, B. and Mallya, S. 2008. Detection of shiga-toxigenic Escherichia coli (STEC) in diarrhoeagenic stool and meat samples in Mangalore, India. Indian J. Med. Res. 128, 271–277. Durant, J.A., Corrier, D.E., Stanker, L.H. and Ricke, S.C. 2000. Expression of the hil A Salmonella typhimurium gene in a poultry Salm. enteritidis isolate in response to lactate and nutrients. J. Appl. Microbiol. 89, 63–69. El-Baz, A.H., El-Sherbini, M., Abdelkhalek, A. and Al-Ashmawy, M.A. 2017. Prevalence and molecular characterization of Salmonella serovars in milk and cheese in Mansoura city, Egypt. J. Adv. Vet. Anim. Res. 4, 45–51. Fesseha, H., Aliye, S., Kifle, T. and Mathewos, M. 2020. Isolation and multiple drug resistance patterns of Salmonella isolates from selected dairy farms in Hawassa town, Ethiopia. J. Vet. Sci. Med. 8, 7–14. Fratamico, P., Sackitey, S., Wiedmann, M. and Deng, M. 1995. Detection of Escherichia coli O157:H7 by multiplex PCR. J. Clin. Microbiol. 33, 2188–2191. Galal, H.M., Hakim, A.S. and Dorgham, S.M. 2013. Phenotypic and virulence genes screening of Escherichia coli strains isolated from different sources in delta Egypt. Life. Sci. J. 10, 352–361. Garedew, L., Berhanu, A., Mengesha, D. and Tsegay, G. 2012. Identification of gram-negative bacteria from critical control points of raw and pasteurized cow milk consumed at Gondar town and its suburbs, Ethiopia. BMC. Public. Health. 12, 1–7. Geletu, U.S., Usmael, M.A. and Ibrahim, A.M. 2022. Isolation, identification, and susceptibility profile of E. coli, Salmonella, and S. aureus in dairy farm and their public health implication in Central Ethiopia. Vet. Med. Int. 2022, 1–13. Hailu, W., Helmy, Y.A., Carney-Knisely, G., Kauffman, M., Fraga, D. and Rajashekara, G. 2021. Prevalence and antimicrobial resistance profiles of foodborne pathogens isolated from dairy cattle and poultry manure amended farms in northeastern Ohio, the United States. Antibiotics 10, 1450–1472. Heuvelink, A.E., Van Den Biggelaar, F.L.A.M., Zwartkruis-Nahuis, J.T.M., Herbes, R.G., Huyben, R., Nagelkerke, N., Melchers, W.J., Monnens, L.A. and De Boer, E. 1998. Occurrence of verocytotoxin-producing Escherichia coli O157 on Dutch dairy farms. J. Clin. Microbiol. 36, 3480–3487. Holt, H.R., Eltholth, M.M., Hegazy, Y.M., El-Tras, W.F., Tayel, A.A. and Guitian, J. 2011. Brucella spp. infection in large ruminants in an endemic area of Egypt: cross-sectional study investigating seroprevalence, risk factors and livestock owner’s knowledge, attitudes and practices (KAPs). BMC. Public. Health. 11, 1–10. Humphrey, T.J., Baskerville, A., Mawer, S., Rowe, B. and Hopper, S. 1989. Salmonella enteritidis phage type 4 from the contents of intact eggs: a study involving naturally infected hens. Epidemiol. Infect. J. 103, 415–423. Hussein, H.S. and Sakuma, T. 2005. Invited review: prevalence of Shiga toxin-producing Escherichia coli in dairy cattle and their products. J. Dairy. Sci. 88, 450–465. Karam, M.R.A., Habibi, M. and Bouzari, S. 2019. Urinary tract infection: pathogenicity, antibiotic resistance and development of effective vaccines against uropathogenic Escherichia coli. Mol. Immunol. 108, 56–67. Kauffman, G. 1974. Kauffmann white scheme. J. Acta. Path. Microbiol. Sci. 61, 385. Kok, T., Worswich, D. and Gowans, E. 1996. Some serological techniques for microbial and viral infections. In Practical medical microbiology, 14th ed. Eds., Collee, J., Fraser, A., Marmion, B. and Simmons, A. Edinburgh, UK: Churchill Livingstone. Lambertini, E., Karns, J.S., Van Kessel, J.A.S., Cao, H., Schukken, Y.H., Wolfgang, D.R. and Pradhan, A.K. 2015. Dynamics of Escherichia coli virulence factors in dairy herds and farm environments in a longitudinal study in the United States. Appl. Environ. Microbiol. 81, 4477–4488. Lubote, R., Shahada, F. and Matemu, A. 2014. Prevalence of Salmonella spp. and Escherichia coli in raw milk value chain in Arusha, Tanzania. Am. J. Res. Commun. 2, 1–13. Macfaddin, J.F., 2000. Biochemical testes for identification medical bacteria. Los Anglos, CA: Warery Press Inc. Madic, J., Vingadassalon, N., de Garam, C.P., Marault, M., Scheutz, F., Brugere, H. and Auvray, F. 2011. Detection of Shiga toxin-producing Escherichia coli serotypes O26: H11, O103: H2, O111: H8, O145: H28, and O157: H7 in raw-milk cheeses by using multiplex real-time PCR. Appl. Environ. Microbiol. 77, 2035–2041. Makkia, D.I., Mahmoud, A.H., Bahout, A.A., Bayoumi, M.A. and Alnakip, M.E. 2022. Molecular studies on some emerging pathogens in dairy products retailed in Dakahlia Governorate, Egypt. J. Adv. Vet. Res. 12, 392–398. Mayer, C.L., Leibowitz, C.S., Kurosawa, S. and Stearns-Kurosawa, D.J. 2012. Shiga toxins and the pathophysiology of hemolytic uremic syndrome in humans and animals. Toxins 4, 1261–1287. Mazaheri, S., Ahrabi, S. and Aslani, M. 2014. Shiga toxin-producing Escherichia coli isolated from lettuce samples in Tehran, Iran. Jundishapur. J. Microbiol. 7, 1–6. McHugh, M.L. 2013. The chi-square test of independence. Biochem. Med. (Zagreb). 23, 143–149. Mohammed, B.T. 2022. Identification and bioinformatic analysis of invA gene of Salmonella in free range chicken. Braz. J. Biol. 84, e263363. Mora, A., Herrrera, A., López, C., Dahbi, G., Mamani, R., Pita, J.M. and Blanco, J. 2011. Characteristics of the Shiga-toxin-producing enteroaggregative Escherichia coli O104: H4 German outbreak strain and of STEC strains isolated in Spain. Int. Microbiol. 14, 121–141. Mousa, W.S. and Shama, U.H.A. 2020. Prevalence, antimicrobial resistance and substantial virulence-associated genes of Escherichia coli isolated from colibacillosis in neonatal calves in Egypt. J. Microbiol. Biotechnol. Food. Sci. 9, 1145–1450. Moustafa, A.M., Elsayed, S., Abo-Sakaya, R. and Ali, A. 2020. Prevalence and molecular characterization of Salmonella serovars isolated from diarrheic cattle and buffalo-calves. Zagazig. Vet. J. 48, 273–283. Murugkar, H., Rahman, H. and Dutta, P. 2003. Distribution of virulence genes in Salmonella serovars isolated from man and animals. Indian. J. Med. Res. 117, 66–70. Nakano, M., Yamasaki, E., Ichinose, A., Shimohata, T., Takahashi, A., Akada, J.K. and Kurazono, H. 2012. Salmonella enterotoxin (Stn) regulates membrane composition and integrity. Dis. Model. Mech. 5, 515–521. Nayak, R., Stewart, T., Wang, R., Lin, J., Cerniglia, C. and Kenney, P. 2004. Genetic diversity and virulence gene determinants of antibiotic-resistance Salmonella isolated from preharvest turkey production sources. Int. J. Food. Microbiol. 91, 51–62. Nikiema, M.E., Kakou-Ngazoa, S., Ky/Ba, A., Sylla, A., Bako, E., Addablah, A.Y.A., Ouoba, J.B., Sampo, E., Gnada, K., Zongo, O. and Sangaré, L. 2021. Characterization of virulence factors of Salmonella isolated from human stools and street food in urban areas of Burkina Faso. BMC. Microbiol. 21, 338–350. Owusu-Kwarteng, J., Akabanda, F., Agyei, D. and Jespersen, L. 2020. Microbial safety of milk production and fermented dairy products in Africa. Microorganisms 8, 752–776. Pacheco, A.R. and Sperandio, V. 2012. Shiga toxin in enterohemorrhagic E. coli: regulation and novel anti-virulence strategies. Front. Cell. Infect. Microbiol. 2, 81–93. Paton, A.W. and Paton, J.C. 1998. Detection and characterization of shiga toxigenic Escherichia coli by using multiplex PCR assays for stx1, stx2 eaeA, enterohemorrhagic E. coli HlyA, rfbO157. J. Clin. Microbiol. 36, 598–602. Pearce, M.C., Evans, J., McKendrick, I.J., Smith, A.W., Knight, H.I., Mellor, D.J. and Low, J.C. 2006. Prevalence and virulence factors of Escherichia coli serogroups O26, O103, O111, and O145 shed by cattle in Scotland. Appl. Environ. Microbiol. 72, 653–659. Quinn, P.J., Carter, M.E., Markey, B. and Carter, G.R., 1994. Clinical veterinary microbiology. Madrid, Spain: Wolfe Publishing, pp: 220–226. Sambrook, J., Fritsch, E.F. and Maniatis, T. 1989. Molecular cloning: laboratory manual, 2nd ed. New York, NY: Cold Spring, Harbor. Santorum, P., Garcia, R., Lopez, V. and Martínez-Suárez, J.V. 2012. Dairy farm management and production practices associated with the presence of Listeria monocytogenes in raw milk and beef. Span. J. Agric. 2, 360–371. Singh, J., Singh, B.B., Tiwari, H.K., Josan, H.S., Jaswal, N., Kaur, M. and Dhand, N.K. 2020. Using dairy value chains to identify production constraints and biosecurity risks. Animals 10, 2332–2354. Sobur, M.A., Sabuj, A.A.M., Sarker, R., Rahman, A.T., Kabir, S.L. and Rahman, M.T., 2019. Antibiotic-resistant Escherichia coli and Salmonella spp. associated with dairy cattle and farm environment having public health significance. Vet. World. 12, 984–993. Wang, J., Zhu, X., Zhao, Y., Liu, H., Zhang, Z., Yan, L. and Aleri, J.W. 2023. Prevalence and antimicrobial resistance of Salmonella and ESBL E. coli isolated from dairy cattle in Henan Province, China. Prev. Vet. Med. 213, 105856. Yang, X., Sun, H., Fan, R., Fu, S., Zhang, J., Matussek, A. and Bai, X. 2020. Genetic diversity of the intimin gene (eae) in non-O157 Shiga toxin-producing Escherichia coli strains in China. Sci. Rep. 10, 3275–3287. Zeinhom, M.M., Aziz, R.L.A., Mohammed, A.N. and Bernabucci, U. 2016. Impact of seasonal conditions on quality and pathogens content of milk in Friesian cows. Asian-australas. J. Anim. Sci. 29, 1207–1213. | ||
| How to Cite this Article |
| Pubmed Style Zaki MS, Abd-El-All AM, Attia AS, Dahshan H, Al-Ashery MA, Megahed A, . Escherichia coli and Salmonella enterica isolated from Egyptian dairy cattle herds: The prevalence and molecular characteristics. Open Vet J. 2024; 14((1) (Zagazig Veterinary Conference)): 214-224. doi:10.5455/OVJ.2024.v14.i1.19 Web Style Zaki MS, Abd-El-All AM, Attia AS, Dahshan H, Al-Ashery MA, Megahed A, . Escherichia coli and Salmonella enterica isolated from Egyptian dairy cattle herds: The prevalence and molecular characteristics. https://www.openveterinaryjournal.com/?mno=175952 [Access: May 09, 2024]. doi:10.5455/OVJ.2024.v14.i1.19 AMA (American Medical Association) Style Zaki MS, Abd-El-All AM, Attia AS, Dahshan H, Al-Ashery MA, Megahed A, . Escherichia coli and Salmonella enterica isolated from Egyptian dairy cattle herds: The prevalence and molecular characteristics. Open Vet J. 2024; 14((1) (Zagazig Veterinary Conference)): 214-224. doi:10.5455/OVJ.2024.v14.i1.19 Vancouver/ICMJE Style Zaki MS, Abd-El-All AM, Attia AS, Dahshan H, Al-Ashery MA, Megahed A, . Escherichia coli and Salmonella enterica isolated from Egyptian dairy cattle herds: The prevalence and molecular characteristics. Open Vet J. (2024), [cited May 09, 2024]; 14((1) (Zagazig Veterinary Conference)): 214-224. doi:10.5455/OVJ.2024.v14.i1.19 Harvard Style Zaki, M. S., Abd-El-All, A. M., Attia, A. S., Dahshan, H., Al-Ashery, M. A., Megahed, A. & (2024) Escherichia coli and Salmonella enterica isolated from Egyptian dairy cattle herds: The prevalence and molecular characteristics. Open Vet J, 14 ((1) (Zagazig Veterinary Conference)), 214-224. doi:10.5455/OVJ.2024.v14.i1.19 Turabian Style Zaki, Mahmoud S.A., Amr M.M. Abd-El-All, Amira S.A. Attia, Hesham Dahshan, Manal A. Al-Ashery, Ayman Megahed, and . 2024. Escherichia coli and Salmonella enterica isolated from Egyptian dairy cattle herds: The prevalence and molecular characteristics. Open Veterinary Journal, 14 ((1) (Zagazig Veterinary Conference)), 214-224. doi:10.5455/OVJ.2024.v14.i1.19 Chicago Style Zaki, Mahmoud S.A., Amr M.M. Abd-El-All, Amira S.A. Attia, Hesham Dahshan, Manal A. Al-Ashery, Ayman Megahed, and . "Escherichia coli and Salmonella enterica isolated from Egyptian dairy cattle herds: The prevalence and molecular characteristics." Open Veterinary Journal 14 (2024), 214-224. doi:10.5455/OVJ.2024.v14.i1.19 MLA (The Modern Language Association) Style Zaki, Mahmoud S.A., Amr M.M. Abd-El-All, Amira S.A. Attia, Hesham Dahshan, Manal A. Al-Ashery, Ayman Megahed, and . "Escherichia coli and Salmonella enterica isolated from Egyptian dairy cattle herds: The prevalence and molecular characteristics." Open Veterinary Journal 14.(1) (Zagazig Veterinary Conference) (2024), 214-224. Print. doi:10.5455/OVJ.2024.v14.i1.19 APA (American Psychological Association) Style Zaki, M. S., Abd-El-All, A. M., Attia, A. S., Dahshan, H., Al-Ashery, M. A., Megahed, A. & (2024) Escherichia coli and Salmonella enterica isolated from Egyptian dairy cattle herds: The prevalence and molecular characteristics. Open Veterinary Journal, 14 ((1) (Zagazig Veterinary Conference)), 214-224. doi:10.5455/OVJ.2024.v14.i1.19 |