| Research Article | ||
Open Vet J. 2024; 14(2): 699-706 Open Veterinary Journal, (2024), Vol. 14(2): 699-706 Original Research Molecular detection of extended-spectrum β-lactamase-producing Escherichia coli from bat caves on Lombok IslandYolla Rona Mustika1,2, Kurnia Nisa Kinasih1,2, Mustofa Helmi Effendi3*, Yulianna Puspitasari4, Shendy Canadya Kurniawan5, Aswin Rafif Khairullah6, Muhammad Esa Erlang Samodra7, Abdullah Hasib8, Alfiana Laili Dwi Agustin9, Ikechukwu Benjamin Moses10 and Otto Sahat Martua Silaen111Profession Program in Veterinary Medicine, Faculty of Veterinary Medicine, Universitas Airlangga, Surabaya, Indonesia 2Master Program in Veterinary Science and Public Health, Faculty of Veterinary Medicine, Universitas Airlangga, Surabaya, East Java, Indonesia 3Division of Veterinary Public Health, Faculty of Veterinary Medicine, Universitas Airlangga, Surabaya, East Java, Indonesia 4Division of Veterinary Microbiology, Faculty of Veterinary Medicine, Universitas Airlangga, Surabaya, East Java, Indonesia 5Master Program of Animal Sciences, Department of Animal Sciences, Specialisation in Molecule, Cell and Organ Functioning, Wageningen University and Research, Wageningen, The Netherlands 6Division of Animal Husbandry, Faculty of Veterinary Medicine, Universitas Airlangga, Surabaya, East Java, Indonesia 7Bachelor Program in Veterinary Medicine, Faculty of Veterinary Medicine, Universitas Airlangga, Surabaya, East Java, Indonesia 8School of Agriculture and Food Sustainability, The University of Queensland, Gatton, QLD, Queensland 9Doctoral Program in Veterinary Science, Faculty of Veterinary Medicine, Universitas Airlangga, Surabaya, East Java, Indonesia 10Department of Applied Microbiology, Faculty of Science, Ebonyi State University, Abakaliki, Nigeria 11Doctoral Program in Biomedical Science, Faculty of Medicine, Universitas Indonesia, Senen, Jakarta, Indonesia *Corresponding Author: Mustofa Helmi Effendi. Division of Veterinary Public Health, Faculty of Veterinary Medicine, Universitas Airlangga, Surabaya, East Java, Indonesia. Email: mhelmieffendi [at] gmail.com Submitted: 23/11/2023 Accepted: 27/01/2024 Published: 29/02/2024 © 2024 Open Veterinary Journal
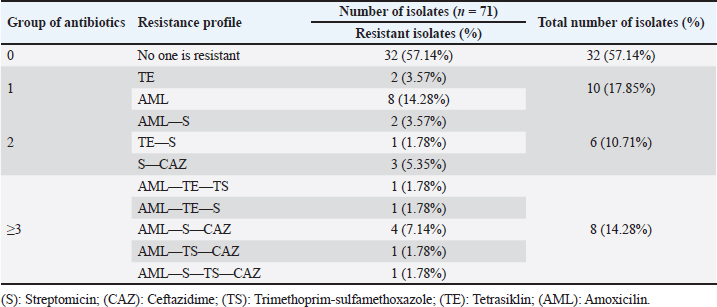
ABSTRACTBackground: The discovery of antibiotic-resistant Enterobacteriaceae bacteria in wild animals is an indication of their potential for wildlife as a reservoir. Bats are natural reservoir hosts and a source of infection for several microorganisms and have the potential to become vectors for the spread of zoonotic diseases. Aim: A study was conducted based on these characteristics to identify and detect the blaTEM gene in Eschericia coli isolated from bat excrements in Tanjung Ringgit Cave, East Lombok. Methods: Bat fecal samples were firstly inoculated onto eosin methylene blue agar media. Recovered bacterial isolates were further characterized using standard microbiological techniques. Antimicrobial susceptibility testing was done using the Kirby-Bauer disc diffusion method. blaTEM gene detection was carried out using polymerase chain reaction (PCR). Results: Out of the 150 bat fecal samples obtained from Tanjung Ringgit cave, Lombok Island, Indonesia, 56 (37%) were positive for E. coli. Eight (8) out of the 56 E. coli isolates that underwent antimicrobial susceptibility testing using the disc diffusion method were confirmed to be multidrug-resistant as they exhibited resistance to at least three different classes of antibiotics. Out of the eight (8) multidrug resistance E. coli isolates recovered from fecal samples of bats, 2 (two) harbored the blaTEM gene. Conclusion: The discovery of the blaTEM gene in bat fecal samples indicates the potential for wild animals, especially bats, to spread ESBL resistance genes to the environment and to humans. Keywords: Escherichia coli, Multidrug resistance, Bats, blaTEM, Human health. IntroductionEscherichia coli is a typical intestinal flora found in both humans and animals (Widodo et al., 2022). Some strains of E. coli are pathogenic, which can cause gastroenteritis, cystitis, pneumonia, and septicemia (Dutta et al., 2017). Escherichia coli is generally used as an indicator of antibiotic resistance because it can easily transfer antibiotic-resistance genes to other bacterial strains in the Enterobacteriaceae family, especially through plasmid mediation (Radhouani et al., 2012). Feces have been identified as a major reservoir of E. coli that harbor extended-spectrum beta-lactamase (ESBL) genes such as blaCTX-M, blaTEM, and blaSHV (ElBaradei et al., 2020). Numerous antibiotic-resistant Enterobacteriaceae bacteria have been isolated from wild animals in recent years (Benavides et al., 2021; Benavides et al., 2022). The discovery of antibiotic-resistant Enterobacteriaceae bacteria in wild animals is a sign of the potential of wild animals as reservoirs (Dolejska and Literak, 2018). Activities such as human encroachment into wild animal habitats, transportation of wild animals, development of captive industries, and management of abused wild animals could be possible causes of antibiotic-resistant E. coli transmission from wild animals to humans (Rhyan and Spraker, 2010; Cunningham et al., 2017). ESBLs display an extended substrate spectrum with the ability to hydrolyze a broader spectrum of antimicrobials belonging to the beta-lactam class that contains the oxyimino-group such as oxyimino-cephalosporins (e.g., ceftazidime and cefotaxime) as well as oxyimino-monobactam (aztreonam) (Guenther et al., 2011). Among microbes belonging to the Enterobacteriaceae family, the most common ESBLs are divided into four major families; TEM (Temoneira)-type beta-lactamases, CTX (cefotaximase)-M-type beta-lactamases, SHV (Sulfhydryl variable)-type beta-lactamases, and OXA (oxacillinase)-type beta-lactamases (Guenther et al., 2011). TEM-type beta-lactamases are well-known derivatives of TEM-1 and TEM-2. While the majority of TEM beta-lactamases are ESBLs, TEM-1, TEM-2, and TEM-13 are only able to hydrolyze penicillin derivates and thus are not regarded as ESBLs (Guenther et al., 2011). The blaTEM resistance gene to β-Lactam antibiotics in wild animals is an emerging phenomenon that is increasingly reported (Guenther et al., 2011). Since most wild animals are not given third-generation cephalosporin treatments, it is believed that contaminated food scraps, wastewater, domestic animal feces, and human contact are the main sources of ESBL-producing E. coli detected in wild animals (Homeier-Bachmann et al., 2022). Although there has not been much research on ESBL-producing E. coli in wildlife up to this point, it is concerning to think that wildlife may eventually serve as a reservoir for bacterial infections with the same pathogen that infects humans or other animals (Lagerstrom and Hadly, 2021). According to the latest data, ESBL-producing E. coli in wild animals is found to be widespread in several wild animal populations such as mammals, waterfowl, birds of prey, and rodents even though they are not exposed to antibiotics continuously (Atterby et al., 2017). It is estimated that there are 230 bat species in Indonesia or around 21% of the bat species in the world (Fajri et al., 2014). Of these species, 77 species are grouped into the Megachiroptera suborder while 153 species are grouped into the Microchiroptera suborder (Fajri et al., 2014; Allocati et al., 2016). Lombok Island is an island that has quite a high diversity of bat species (Fajri et al., 2014). Bats are natural reservoir hosts and a source of infection for several microorganisms and have the potential to become vectors for the spread of zoonotic diseases (Letko et al., 2020). Several bacteria such as Salmonella spp., Pasturella spp., E. coli, Leptospira sp., and Bartonella spp. have been isolated from bats in various countries around the world (Allocati et al., 2016). Caves are well-known roosting places for several types of bats; however, they have also been found in residential areas (Maulany et al., 2019). Based on a research survey conducted by Fajri et al. (2014), there is a cave in Lombok that has various bat species, namely the Tanjung Ringgit Giant Cave. Based on the above background, it is necessary to conduct a study on the detection and identification of ESBL-producing E. coli in bats residing in caves on Lombok Island. The results are expected to illustrate the potential of bats as reservoirs for the spread of ESBL-producing E. coli to the community. Materials and MethodsSample collectionThe research was carried out from March 2023 to June 2023. A total of 150 bat fecal swab samples were aseptically collected in Tanjung Ringgit cave, Lombok Island, Indonesia, and immediately transported within 1–2 hours in ice packs to the laboratory for bacteriological analysis. Isolation and identification of E. coliAll bat fecal swab samples were streaked onto eosin methylene blue agar (EMBA) medium and incubated for 18–24 hours at 37ºC. After incubation, colonies typical of E. coli (green metallic sheen) were further subjected to other physiological and biochemical characterization such as Gram staining, indole, methyl red, and Voges-Proskauer tests. Antibiotic sensitivity testAntimicrobial susceptibility testing was done by disc diffusion method on Mueller-Hinton agar according to the guidelines of the Clinical and Laboratory Standards Institute (CLSI, 2017). A suspension was prepared from a 24-hour growth of the test organisms in sterile water to match the 0.5 McFarland turbidity standard. This was seeded on the entire surface of the solidified Mueller-Hinton (MH) agar plate. The following antimicrobials (Oxoid, UK) were tested against the isolates: amoxicillin (10 μg), trimethoprim-sulfamethoxazole (23.75 μg), ceftazidime (30 μg), streptomycin (10 μg), and tetracycline (30 μg). The inoculated MH agar plates were then incubated at 37°C for 18–24 hours. Inhibition zone diameters were measured, recorded, and interpreted as resistant or susceptible according to the CLSI guidelines (CLSI, 2017). Isolates with intermediate resistance were classified as “resistant” in this study. blaTEM gene detectionThe primer sequences used in this study were primers blaTEM-Forward 5’-ATGAGTATTCAACATTTCCG-3’ and blaTEM-Reverse 5’-CTGACAGTTACCAATGCTTA-3’ (Ballhausen et al., 2014). The marker used in this PCR method is Invitrogen 867 bp. The initial stage of PCR amplification begins with making a reaction mixture which is carried out in cold conditions. The reaction mixture contained 12.5 μl of Go tag green master mix, 1 μl of blaTEM gene forward and reverse primers each, 0.5 μl of DNase-free water and 5 μl of DNA template (Ballhausen et al., 2014). A total of 20 μl of reaction mixture was put into Eppendorf PCR tubes, then amplified using a thermal cycler machine according to the protocol carried out by Fouladi et al. (2011) with slight modifications, namely predenaturation at 95°C for 1 minute followed by 40 denaturation cycles at 95°C for 1 minute, annealing at 55°C for 1 minute, and extension at 72°C for 1 minute. The amplification stage ended with a final extension at 72°C for 2 minutes. Electrophoresis was run in a 2% agarose gel with SYBR® safe DNA gel stain at 100 V and 250 mA for 35 minutes. The amplified PCR products were visualized in a UV transilluminator. Ethical approvalAnimal ethics approval was obtained via the ethical clearance committee of the Faculty of Veterinary Medicine, Universitas Airlangga, Indonesia (Ethics no: 1.KEH.046.03.2023). ResultIsolation of E. coli bacteriaPure colonies of recovered isolates had a green metallic sheen appearance on the EMBA medium (Fig. 1). Physiological and biochemical test results indicated that the isolates were Gram-negative rods (coccobacilli), and were positive for motility, indole, and methyl red tests but negative for Voges-Proskauer and citrate utilization tests. Out of the 150 bat fecal samples obtained from Tanjung Ringgit cave, Lombok Island, Indonesia, 56 (37%) were positive for E. coli. Antimicrobial susceptibility testing resultsThe E. coli isolates recovered in this study showed resistance to amoxicillin, tetracycline, trimethoprim-sulfamethoxazole, streptomycin, and ceftazidime (Fig. 2, Tables 1 and 2). Eight (14.28%) isolates were observed to be multidrug-resistant as they exhibited resistance to at least 3 (≥3) different classes of antibiotics (Tables 1 and 2).
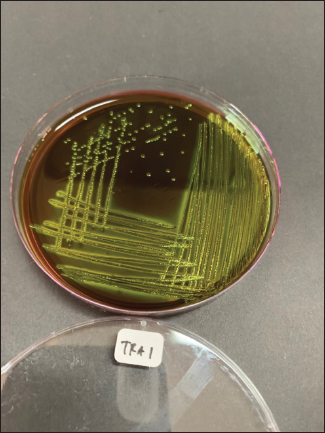
Fig. 1. Metallic green colonies of E. coli isolates on EMBA medium.
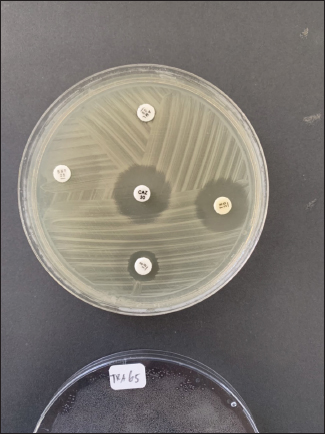
Fig. 2. Antibiotic sensitivity test results showing an antibioticresistant E. coli isolate. Table 1. Antimicrobial susceptibility profiles of the recovered E. coli isolates.
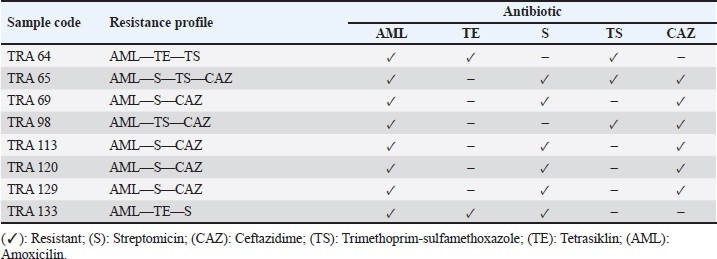
Table 2. Antibiotypes of the recovered multidrug-resistant E. coli isolates.
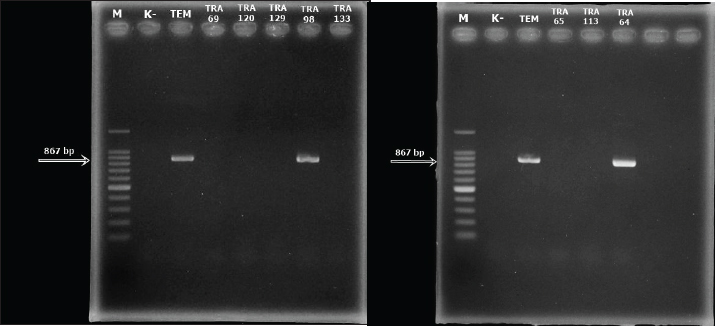
Detection of the E. coli encoding blaTEM ESBL geneAll 8 multidrug resistance (MDR) E. coli isolates recovered in this study were screened for the presence of blaTEM ESBL gene by PCR. Out of the 8 MDR E. coli screened, two (TRA 64 and TRA 98) harbored blaTEM ESBL gene (Fig. 3). DiscussionIn this investigation, we identified antibiotic-resistant E. coli in bat fecal samples from Tanjung Ringgit cave, Lombok Island, Indonesia. The recovered E. coli isolates showed resistance to amoxicillin, tetracycline, trimethoprim-sulfamethoxazole, streptomycin, and ceftazidime. Eight (14.28%) isolates were noted to exhibit multidrug-resistant traits (resistance to at least three classes of antibiotics) with two of them harboring the blaTEM gene. The resistance profiles found in Salmonella spp., Staphylococcus spp., and E. coli in bat feces which contaminate water have been found to increase the transmission of resistant and pathogenic bacteria between wildlife, livestock, and humans (Uddin et al., 2020). Escherichia coli is a bacterium that has the potential to harbor resistance genes to various antibiotics (Ansharieta et al., 2021; Yanestria et al., 2022). The transmission of resistance genes to other bacteria in the vicinity is facilitated by bacterial plasmids, which are identified as the source of antibiotic resistance (Promite et al., 2017). E. coli can act as a reservoir for the spread of multidrug resistance to humans and the environment (Promite et al., 2017). Wibisono et al. (2020) reported a relatively high resistance frequency to aztreonam (86.36%), trimethoprim-sulfamethoxazole (77.27%), gentamicin (72.73%), and ciprofloxacin (68.18%) among ESBL-producing bacterial pathogens.
Fig. 3. The results of the detection of the blaTEM ESBL-encoding gene in MDR E. coli recovered from bat feces taken from Tanjung Ringgit Cave, East Lombok. There have been reports of multidrug-resistant Enterobacteriaceae in wildlife all over the world (Kabali et al., 2021; Lagerstrom and Hadly, 2021; Homeier-Bachmann et al., 2022), but it is still unclear how wildlife contributes to the spread of antibiotic resistance. ESBL-producing E. coli isolated from bats was found to have a fairly high prevalence of antimicrobial resistance (Benavides et al., 2022). MDR E. coli has been reported to be found in fruit bats in Africa, Portugal, and Nigeria. MDR E. coli was found to be resistant to beta-lactamase, carbapenem, and fluroquinolone antibiotics (McDougall et al., 2022). Resistance found in wild animals may be acquired through direct contact with livestock, humans, or the environment (Kabali et al., 2021). The level of tetracycline resistance is quite high due to its common use in the veterinary field, followed by the use of other classes of antibiotics used in this study, namely aminoglycosides such as streptomycin, and sulfonamides such as trimetroprim-sulfamethoxazole (Hunter et al., 2010). The tetracyclines, aminoglycosides, and the combination drug trimetroprim-sulfamethoxozole have a significant impact on microbial activity in the digestive tract; so, they are often used to treat gastrointestinal diseases (Ramandinianto et al., 2020; Kakoullis et al., 2021). To the best of our knowledge, and for the first time in Indonesia, we isolated E. coli harboring blaTEM ESBL gene in bat feces from Lombok Island Cave. Research conducted by Effendi et al. (2022) found that MDR E. coli bacteria isolated from pigs harbored 15.6% of the blaTEM gene. A beta-lactamase enzyme known as blaTEM is frequently found in ESBL-producing bacterial isolates, especially E. coli strains. Beta-lactamase family members of TEM, SHV, and CTX-M are also frequently found in other Enterobacteriaceae bacteria (Guenther et al., 2011). Research conducted on E. coli isolates that were positive for the DDST test found the blaTEM gene and the blaCTX-M gene using multiplex PCR (Ansharieta et al., 2021). This is proven by research conducted by Benavides et al. (2022) detecting eight β-lactamase genes including the blaTEM, blaOXA, and blaCTX-M genes from bat feces samples. In another previous study, Benavides et al. (2018) also found the same ESBL-encoding gene, blaCTX-M-15, in the plasmid of E. coli bacteria isolated from pig and bat feces, indicating cross-contamination of ESBL-producing bacteria between bats and livestock. The spread of ESBL-producing Enterobacteriaceae bacteria is one of the public health problems in the world today (Effendi et al., 2021) as it could complicate treatment options and further result in serious economic burden and losses. Enzymes generated by bacteria harboring ESBL genes can hydrolyze beta-lactam antibiotics, including third and fourth-generation cephalosporins (Riwu et al., 2020; Faridah et al., 2023). Several ESBL variants, such as blaTEM, blaSHV, and blaCTX-M, are currently found on bacterial plasmids (Benavides et al., 2021; Wibisono et al., 2021). In general, wild animals do not come into direct contact with antibiotics; wild animals may be exposed to antibiotic resistance through food and drink consumed in contaminated environments (Savin et al., 2020). International organizations such as the Food and Agriculture Organization are also starting to pay attention to the interconnectedness of wildlife, humans, and livestock regarding antibiotic resistance (Wall et al., 2016). The increasing interaction between humans, livestock, and wild animals can increase the potential for transmission of ESBL-producing E. coli (Kabali et al., 2015). Thus, the detection of antibiotic resistance, especially ESBL-producing E. coli in nature and wild animals, is a concern and requires further research. ConclusionIn total, from 150 bat feces samples isolated, 8 samples were positive for MDR E. coli. Eight (14.28%) E. coli isolates were noted to exhibit multidrug-resistant traits with two of them harboring the blaTEM gene. This indicates the potential for wild animals, especially bats, to spread ESBL resistance genes to the environment and humans. AcknowledgmentThe authors thank the Faculty of Veterinary Medicine, Universitas Airlangga. Conflict of interestThe authors declare that there is no conflict of interest. FundingThis study was supported in part by the Penelitian Unggulan Airlangga (PUA) Universitas Airlangga, Indonesia, in the fiscal year 2023, with grant number: 1710/UN3.LPPM/PT.01.03/2023. Data availabilityAll data supporting the findings of this study are available within the manuscript and no additional data sources are required. Author’s contributionsConceptualization and design: YRM and KNK; acquisition of data: AH and IBM; formal analysis and interpretation of data: OSMS and MEES; writing-original draft preparation: ARK and SCK; writing-review and editing: ALDA, YP, and MHE. All authors have read and agreed to the published version of the manuscript. ReferencesAllocati, N., Petrucci, A.G., Di Giovanni, P., Masulli, M., Di Ilio, C. and De Laurenzi, V. 2016. Bat-man disease transmission: zoonotic pathogens from wildlife reservoirs to human populations. Cell. Death Discov. 2(1), 16048. Ansharieta, R., Ramandinianto, S.C., Effendi, M.H. and Plumeriastuti, H. 2021. Molecular identification of blaCTX-M and blaTEM genes encoding extended-spectrum β-lactamase (ESBL) producing Escherichia coli isolated from raw cow’s milk in East Java, Indonesia. Biodiversitas 22(4), 1600–1605. Atterby, C., Börjesson, S., Ny, S., Järhult, J.D., Byfors, S. and Bonnedahl, J. 2017. ESBL-producing Escherichia coli in Swedish gulls-A case of environmental pollution from humans? PLoS One 12(12), e0190380. Ballhausen, B., Kriegeskorte, A., Schleimer, N., Peters, G. and Becker, K. 2014. The mecA homolog mecC confers resistance against β-lactams in Staphylococcus aureus irrespective of the genetic strain background. Antimicrob. Agents Chemother. 58(7), 3791–3798. Benavides, J.A., Godreuil, S., Opazo-Capurro, A., Mahamat, O.O., Falcon, N., Oravcova, K., Streicker, D.G. and Shiva, C. 2022. Long-term maintenance of multidrug-resistant Escherichia coli carried by vampire bats and shared with livestock in Peru. Sci. Total Environ. 810(1), 152045. Benavides, J.A., Salgado-Caxito, M., Opazo-Capurro, A., González Muñoz, P., Piñeiro, A., Medina, M.O., Rivas, L., Munita, J. and Millán, J. 2021. ESBL-Producing Escherichia coli carrying CTX-M genes circulating among livestock, dogs, and wild mammals in small-scale farms of central chile. Antibiotics (Basel) 10(5), 510. Benavides, J.A., Shiva, C., Virhuez, M., Tello, C., Appelgren, A., Vendrell, J., Solassol, J., Godreuil, S. and Streicker, D.G. 2018. Extended-spectrum beta-lactamase-producing Escherichia coli in common vampire bats Desmodus rotundus and livestock in Peru. Zoonoses Public Health 65(4), 454–458. Cunningham, A.A., Daszak, P. and Wood, J.L.N. 2017. One Health, emerging infectious diseases and wildlife: two decades of progress? Philos. Trans. R. Soc. Lond. B. Biol. Sci. 372(1725), 20160167. Clinical Laboratory Standards Institute (CLSI). 2017. Performance Standards for Antimicrobial Susceptibility Testing. 27th Edition, CLSI supplements M100Wayne: Clinical and Laboratory Standards Institute. Dolejska, M. and Literak, I. 2019. Wildlife is overlooked in the epidemiology of medically important antibiotic-resistant bacteria. Antimicrob. Agents Chemother. 63(8), e01167–19. Dutta, A., Jalal, M.S., Nath, S.K., Dhar, P.K., Das, A. and Uddin, M.M. 2017. Multidrug resistance pattern of Escherichia coli isolated from hospital effluent and determination of tetracycline resistance gene. J. Inf. Mol. Biol. 4(3), 49–53. Effendi, M.H., Hartadi, E.B., Witaningrum, A.M., Permatasari, D.A. and Ugbo, E.N. 2022. Molecular identification of blaTEM gene of extended-spectrum beta-lactamase-producing Escherichia coli from healthy pigs in Malang district, East Java, Indonesia. J. Adv. Vet. Anim. Res. 9(3), 447–452. Effendi, M.H., Tyasningsih, W., Yurianti, Y.A., Rahmahani, J., Harijani, N. and Plumeriastuti, H. 2021. Presence of multidrug resistance (MDR) and extended-spectrum beta-lactamase (ESBL) of Escherichia coli isolated from cloacal swab of broilers in several wet markets in Surabaya, Indonesia. Biodiversitas 22(1), 304–310. ElBaradei, A., Maharem, D.A., Kader, O., Ghareeb, M.K. and Naga, I.S. 2020. Fecal carriage of ESBL-producing Escherichia coli in Egyptian patients admitted to the Medical Research Institute Hospital, Alexandria University. AIMS Microbiol. 6(4), 422–433. Fajri, S.R., Al Idrus, A. and Hadiprayitno, G. 2014. the richness of bat species order chiiroptera in the southern caves of Lombok Island West Nusa Tenggara. Bioedukasi 7(2), 5–9. Faridah, H.D., Wibisono, F.M., Wibisono, F.J., Nisa, N., Fatimah, F., Effendi, M.H., Ugbo, E.N., Khairullah, A.R., Kurniawan, S.C. and Silaen, O.S.M. 2023. Prevalence of the blaCTX-M and blaTEM genes among extended-spectrum beta lactamase–producing Escherichia coli isolated from broiler chickens in Indonesia. J Vet Res. 67 (2023): 179–186. Fouladi, I., Choopani, A. and Mehrabadi, F. 2011. Study of prevalence of enterotoxin type B gene in Meticillin Resistant Staphylococcus aureus (MRSA) isolated from wound. Kowsar Med. J. 16(1), 21–25. Guenther, S., Ewers, C. and Wieler, L.H. 2011. Extended-spectrum beta-lactamases producing E. coli in wildlife, yet another form of environmental pollution? Front. Microbiol. 2, 246. Homeier-Bachmann, T., Schütz, A.K., Dreyer, S., Glanz, J., Schaufler, K. and Conraths, F.J. 2022. Genomic analysis of ESBL-Producing E. coli in wildlife from North-Eastern Germany. Antibiotics (Basel) 11(2), 123. Hunter, P.A., Dawson, S., French, G.L., Goossens, H., Hawkey, P.M., Kuijper, E.J., Nathwani, D., Taylor, D.J., Teale, C.J., Warren, R.E., Wilcox, M.H., Woodford, N., Wulf, M.W. and Piddock, L.J. 2010. Antimicrobial-resistant pathogens in animals and man: prescribing, practices and policies. J. Antimicrob. Chemother. 65(Suppl 1), i3–17. Kabali, E., Pandey, G.S., Maboshe, M. and Munyeme, M. 2015. The wildlife-livestock-human interface, legislation and its impact on communities around Mosi-Oa-Tunya National Park in Zambia. J. Sustain. Dev. Africa 17(6), 125–145. Kabali, E., Pandey, G.S., Munyeme, M., Kapila, P., Mukubesa, A.N., Ndebe, J., Muma, J.B., Mubita, C., Muleya, W., Muonga, E.M., Mitoma, S., Hang’ombe, B.M., Wiratsudakul, A., Ngan, M.T., Elhanafy, E., Daous, H.E., Huyen, N.T., Yamazaki, W., Okabayashi, T., Abe, M., Norimine, J. and Sekiguchi, S. 2021. Identification of Escherichia coli and related enterobacteriaceae and examination of their phenotypic antimicrobial resistance patterns: a pilot study at a wildlife-livestock interface in Lusaka, Zambia. Antibiotics (Basel) 10(3), 238. Kakoullis, L., Papachristodoulou, E., Chra, P. and Panos, G. 2021. Mechanisms of antibiotic resistance in important gram-positive and gram-negative pathogens and novel antibiotic solutions. Antibiotics 10(4), 415. Lagerstrom, K.M. and Hadly, E.A. 2021. The under-investigated wild side of Escherichia coli: genetic diversity, pathogenicity and antimicrobial resistance in wild animals. Proc. Biol. Sci. 288(1948), 20210399. Letko, M., Seifert, S.N., Olival, K.J., Plowright, R.K. and Munster, V.J. Bat-borne virus diversity, spillover and emergence. Nat. Rev. Microbiol. 18(8), 461–471. Maulany, R.I., Wolor, F.S., Nasri, N. and Achmad, A. 2019. Habitat characteristics and population of cave-dwelling bats in Mara Kallang Cave of Maros-Pangkep Karst Area of South Sulawesi. IOP Conf. Ser.: Earth Environ. Sci. 270(1), 012030. McDougall, F., Boardman, W. and Power, M. 2022. High prevalence of beta-lactam-resistant Escherichia coli in South Australian grey-headed flying fox pups (Pteropus poliocephalus). Microorganisms 10(8), 1589. Promite, S., Saha, S.K. and Roy, P.K. 2017. Emerging antibiotic resistance and plasmid DNA in Escherichia coli isolated from respiratory tract infections in Bangladesh. Open Access J. Trans. Med. Res. 1(2), 47–51. Radhouani, H., Poeta, P., Gonçalves, A., Pacheco, R., Sargo, R. and Igrejas, G. 2012. Wild birds as biological indicators of environmental pollution: antimicrobial resistance patterns of Escherichia coli and enterococci isolated from common buzzards (Buteo buteo). J. Med. Microbiol. 61(Pt6), 837–843. Ramandinianto, S.C., Khairullah, A.R., Effendi, M.H. and Hestiana, E.P. 2020. Profile of multidrug resistance (MDR) and methicillin resistant Staphylococcus aureus (MRSA) on dairy farms in East Java province, Indonesia. Indian J. Forensic Med. Toxicol. 14(4), 3439–3445. Rhyan, J.C. and Spraker, T.R. 2010. Emergence of diseases from wildlife reservoirs. Vet. Pathol. 47(1), 34–39. Riwu, K.H.P., Effendi, M.H. and Rantam, F.A. (2020) A review of extended-spectrum β-lactamase (ESBL) producing Klebsiella pneumoniae and multidrug-resistant (MDR) on companion animals. Syst. Rev. Pharm. 11(7), 270–277. Savin, M., Bierbaum, G., Hammerl, J.A., Heinemann, C., Parcina, M., Sib, E., Voigt, A. and Kreyenschmidt, J. 2020. Antibiotic-resistant bacteria and antimicrobial residues in wastewater and process water from German pig slaughterhouses and their receiving municipal wastewater treatment plants. Sci. Total Environ. 727(1), 138788. Uddin, M., Samad, M.A., Rahman, M.K., Islam, S., Hossain, M.S., Sagor, M.S., Rostal, M., Giasuddin M., Flora, M.S., Epstein, J.H., Chawdhury, S. and Islam, A. 2020. Ecology of bat drinking behaviour and AMR patterns of Sal spp., Staphylococcus spp. and E. coli recovered from fecal droppings of bats and water in BD. Int. J. Infect. Dis. 101(S1), 119. Wall, T.L., Luczak, S.E. and Hiller-Sturmhöfel, S. 2016. Biology, genetics, and environment: underlying factors influencing alcohol metabolism. Alcohol. Res. 38(1), 59–68. Wibisono, F.J., Sumiarto, B., Untari, T., Effendi, M.H., Permatasari, D.A. and Witaningrum, A.M. 2021. Molecular identification of CTX Gene of extended spectrum beta-lactamase (ESBL) producing Escherichia coli on layer chicken in Blitar, Indonesia. J. Anim. Plant Sci. 31(4), 954–959 Widodo, A., Lamid, M., Effendi, M.H., Khairullah, A.R., Riwu, K.H.P., Yustinasari, L.R., Kurniawan, S.C., Ansori, A.N.M., Silaen, O.S.M. and Dameanti, F.N.A.E.P. 2022. Antibiotic sensitivity profile of multidrug-resistant (MDR) Escherichia coli isolated from dairy cow’s milk in Probolinggo, Indonesia. Biodiversitas 23(10), 4971–4976. Yanestria, S.M., Dameanti, F.N.A.E.P., Musayannah, B.G., Pratama, J.W.A., Witaningrum, A.M., Effendi, M.H. and Ugbo, E.N. 2022. Antibiotic resistance pattern of Extended-Spectrum β-lactamase (ESBL) producing Escherichia coli isolated from broiler farm environment in Pasuruan district, Indonesia. Biodiversitas 23(9), 4460–4465. | ||
| How to Cite this Article |
| Pubmed Style Mustika YR, Kinasih KN, Effendi MH, Puspitasari Y, Kurniawan SC, Khairullah AR, Samodra MEE, Hasib A, Agustin ALD, Moses IB, Silaen OSM. Molecular detection of extended-spectrum β-lactamase-producing Escherichia coli from bat caves on Lombok Island. Open Vet J. 2024; 14(2): 699-706. doi:10.5455/OVJ.2024.v14.i2.10 Web Style Mustika YR, Kinasih KN, Effendi MH, Puspitasari Y, Kurniawan SC, Khairullah AR, Samodra MEE, Hasib A, Agustin ALD, Moses IB, Silaen OSM. Molecular detection of extended-spectrum β-lactamase-producing Escherichia coli from bat caves on Lombok Island. https://www.openveterinaryjournal.com/?mno=178548 [Access: April 27, 2024]. doi:10.5455/OVJ.2024.v14.i2.10 AMA (American Medical Association) Style Mustika YR, Kinasih KN, Effendi MH, Puspitasari Y, Kurniawan SC, Khairullah AR, Samodra MEE, Hasib A, Agustin ALD, Moses IB, Silaen OSM. Molecular detection of extended-spectrum β-lactamase-producing Escherichia coli from bat caves on Lombok Island. Open Vet J. 2024; 14(2): 699-706. doi:10.5455/OVJ.2024.v14.i2.10 Vancouver/ICMJE Style Mustika YR, Kinasih KN, Effendi MH, Puspitasari Y, Kurniawan SC, Khairullah AR, Samodra MEE, Hasib A, Agustin ALD, Moses IB, Silaen OSM. Molecular detection of extended-spectrum β-lactamase-producing Escherichia coli from bat caves on Lombok Island. Open Vet J. (2024), [cited April 27, 2024]; 14(2): 699-706. doi:10.5455/OVJ.2024.v14.i2.10 Harvard Style Mustika, Y. R., Kinasih, . K. N., Effendi, . M. H., Puspitasari, . Y., Kurniawan, . S. C., Khairullah, . A. R., Samodra, . M. E. E., Hasib, . A., Agustin, . A. L. D., Moses, . I. B. & Silaen, . O. S. M. (2024) Molecular detection of extended-spectrum β-lactamase-producing Escherichia coli from bat caves on Lombok Island. Open Vet J, 14 (2), 699-706. doi:10.5455/OVJ.2024.v14.i2.10 Turabian Style Mustika, Yolla Rona, Kurnia Nisa Kinasih, Mustofa Helmi Effendi, Yulianna Puspitasari, Shendy Canadya Kurniawan, Aswin Rafif Khairullah, Muhammad Esa Erlang Samodra, Abdullah Hasib, Alfiana Laili Dwi Agustin, Ikechukwu Benjamin Moses, and Otto Sahat Martua Silaen. 2024. Molecular detection of extended-spectrum β-lactamase-producing Escherichia coli from bat caves on Lombok Island. Open Veterinary Journal, 14 (2), 699-706. doi:10.5455/OVJ.2024.v14.i2.10 Chicago Style Mustika, Yolla Rona, Kurnia Nisa Kinasih, Mustofa Helmi Effendi, Yulianna Puspitasari, Shendy Canadya Kurniawan, Aswin Rafif Khairullah, Muhammad Esa Erlang Samodra, Abdullah Hasib, Alfiana Laili Dwi Agustin, Ikechukwu Benjamin Moses, and Otto Sahat Martua Silaen. "Molecular detection of extended-spectrum β-lactamase-producing Escherichia coli from bat caves on Lombok Island." Open Veterinary Journal 14 (2024), 699-706. doi:10.5455/OVJ.2024.v14.i2.10 MLA (The Modern Language Association) Style Mustika, Yolla Rona, Kurnia Nisa Kinasih, Mustofa Helmi Effendi, Yulianna Puspitasari, Shendy Canadya Kurniawan, Aswin Rafif Khairullah, Muhammad Esa Erlang Samodra, Abdullah Hasib, Alfiana Laili Dwi Agustin, Ikechukwu Benjamin Moses, and Otto Sahat Martua Silaen. "Molecular detection of extended-spectrum β-lactamase-producing Escherichia coli from bat caves on Lombok Island." Open Veterinary Journal 14.2 (2024), 699-706. Print. doi:10.5455/OVJ.2024.v14.i2.10 APA (American Psychological Association) Style Mustika, Y. R., Kinasih, . K. N., Effendi, . M. H., Puspitasari, . Y., Kurniawan, . S. C., Khairullah, . A. R., Samodra, . M. E. E., Hasib, . A., Agustin, . A. L. D., Moses, . I. B. & Silaen, . O. S. M. (2024) Molecular detection of extended-spectrum β-lactamase-producing Escherichia coli from bat caves on Lombok Island. Open Veterinary Journal, 14 (2), 699-706. doi:10.5455/OVJ.2024.v14.i2.10 |