| Research Article | ||
Open Veterinary Journal, (2024), Vol. 14(1): 389–397 Original Research Bacteriological evaluation and advanced SYBR-green multiplex real-time PCR assay for detection of minced meat adulterationSoad H. El-Sheikh1, Reham M. Abdel Whab1, Rania A. ElDaly2, Mona T. Raslan3, Hanan A. Fahmy4 and Azza S. El-Demerdash5*1Department of Food Hygiene, Agriculture Research Centre (ARC), Animal Health Research Institute (AHRI), Zagazig, Egypt 2Department of Botany and Microbiology, Faculty of Science, Arish University, Al-Arish, Egypt 3Department of Food Hygiene, Agriculture Research Centre (ARC), Animal Health Research Institute (AHRI), Giza, Egypt 4Department of Biotechnology, Agricultural Research Centre, Animal Health Research Institute, Giza, Egypt 5Laboratory of Biotechnology, Department of Microbiology, Agriculture Research Centre (ARC), Animal Health Research Institute (AHRI), Zagazig, Egypt *Corresponding Author: Azza Salah El-Demerdash. Laboratory of Biotechnology, Department of Microbiology, Agriculture Research Centre (ARC), Animal Health Research Institute (AHRI), Zagazig, Egypt. Email: dr.azzasalah [at] yahoo.com Submitted: 01/10/2023 Accepted: 15/12/2023 Published: 31/01/2024 © 2024 Open Veterinary Journal
ABSTRACTBackground: Minced meat is a valuable source of nutrients, but it is vulnerable to contamination by microorganisms commonly present in the environment. In addition, there is a risk of adulteration with cheaper meat sources, which can be harmful to consumers. Aim: It is crucial to identify meat adulteration with distinct microbiological analysis for legal, economic, religious, and public health purposes. Methods: A total of 100 minced meat samples were collected from several markets in Sharkia Governorate, Egypt. These samples were then subjected to bacteriological testing and an advanced multiplex PCR method. This method enables the detection of bovine, equine, porcine, and dog species in meat samples with just one step. Results: The adulterated samples had a higher total bacterial count and pH values compared to pure bovine meat. These differences in bacterial count and pH values were statistically significant, with p-values of 0.843 (log10) and 0.233, respectively. The frequency of Escherichia coli occurrence was 13%, and the O111 serotype was predominant in the adulterated samples. Listeria monocytogenes and Staphylococcus aureus were isolated with prevalence rates of 3% and 29%, respectively. Besides, the SYBR-green multiplex real-time PCR assay used in this study detected adulteration with dog, equine, and porcine meats in the examined samples at rates of 9%, 5%, and 4%, respectively. Conclusion: This method provides a sensitive and specific approach to detect issues related to well-being and safety. Keywords: Bacteriological examination, Meat adulteration, Multiplex PCR assay, pH values. IntroductionMeat is a nutritious food for humans due to its high protein content, essential amino acids, vitamins, fats, minerals, and other nutrients. On the other hand, it might also offer the perfect conditions for the development of different species. This is because it has a high moisture content, minerals, nitrogenous substances, and a trace amount of fermentable carbohydrates, such as glycogen. In addition, meat’s pH encourages the majority of bacteria to grow and reproduce (Alahakoon et al., 2015; El-Demerdash et al., 2023b). Raw meat and minced meat can contain Escherichia coli (E. coli), Listeria monocytogenes (L. monocytogenes), and Staphylococcus aureus (S. aureus), which pose significant health risks to individuals who consume contaminated food. These bacteria have the ability to cause a wide range of illnesses, varying from mild discomfort to severe and even life-threatening complications (Osman et al., 2017; El-Demerdash and Raslan, 2019; Essawi et al., 2020; Dawwam et al., 2022; Ali et al., 2023). Escherichia coli is a significant member of the Enterobacteriaceae family and has been linked to food poisoning cases involving creamed fish, undercooked or poorly cooked meat, and poultry. Beef appears to be the main food source for this organism’s infection. Entero-hemorrhagic E. coli (EHEC) has been linked to infant and young children’s diarrhea outbreaks. Acute and severe abdominal discomfort, watery diarrhea, and later bloody diarrhea are the typical signs of an EHEC infection (Lee et al., 2009). Listeria monocytogenes is a foodborne pathogen that can cause listeriosis, a dangerous disease primarily affecting immunocompromised individuals, neonates, and pregnant women. Symptoms of listeriosis include fever, aches in the muscles, headaches, a stiff neck, disorientation, and convulsions. If contracted during pregnancy, listeriosis may lead to miscarriage, stillbirth, or infection in the newborn child (Ryser and Marth, 2007; Tarazi et al., 2021). In addition, minced meat can provide a favorable environment for the growth of S. aureus bacteria. Food poisoning caused by S. aureus typically manifests as symptoms such as diarrhea, vomiting, nausea, cramping in the abdomen, fever, and headache that usually occur three to 6 hours after consuming contaminated food. In severe cases, S. aureus food poisoning can lead to dehydration, electrolyte imbalance, and even death (Le Loir et al., 2003; Bhargava et al., 2011; Hegab et al., 2020). As a result, due to improper handling and insufficient control measures against these pathogens, raw meat poses a serious risk to human health. The adulteration of meat species, which not only threatens public health but also aids in the spread of harmful foodborne pathogens, is another serious problem that meat consumers must contend with. Manufacturers sometimes substitute cheaper alternatives for the primary meat ingredients to further their economic goals. These practices have adverse effects on customers, violate social and religious norms, and pose health hazards (Spink et al., 2019). In addition, some people refrain from consuming pork and horse meat for moral, spiritual, or humanitarian reasons. As a consequence, according to Haunshi et al. (2009), these consumer groups seek methods to identify various forms of meat (including mouse, pork, chicken, and horse) in food products or staff. A common method for determining meat species is DNA analysis. Because DNA is more stable than proteins, it is appreciated for identifying species. Due to its simplicity, speed, and specificity, polymerase chain reaction (PCR) applications have been widely used (Kesmen et al., 2007; El-Demerdash et al., 2023a). According to several studies (Abuelnaga et al., 2020; Shalaby et al., 2021; Megahed et al., 2023), multiplex PCR is a powerful approach that can simultaneously amplify several templates, lower the detection rate, and get beyond the restrictions of a single PCR that only targets one type of meat at a time. For the purpose of avoiding health and safety hazards, the study’s goal was to ascertain the microbial burden and identify the main pathogens in minced meat samples. In addition, the study sought to evaluate an advanced SYBR-green multiplex real-time PCR test as a sensitive and accurate method for detecting adulteration in minced meat across different marketplaces in Sharkia, Egypt. Material and MethodsSamples collectionA total of 100 fresh and frozen minced meat samples (50 each) were purchased from various markets in Sharkia Governorate, Egypt, from January 2023 to April 2023. All samples were collected as soon as possible, transferred to an icebox container, handled aseptically, and immediately transported to the laboratory for further examination. pH measurementsIn a blender, 10 ml of neutralized distilled water was added to approximately 10 g of sample. After 10 minutes of constant shaking, the homogenate was allowed to stand at room temperature. A Bye model 6020 electrical pH meter (Bye, USA) was used to calculate the pH value in accordance with Pearson (1984). Microbiological examinationSamples preparationFollowing Da Silva et al. (2018), 10 g of each sample were taken and put into a sterile plastic bag. To create a dilution of 1/10, 90 ml of 0.1% sterile buffered peptone water (Oxoid CM9) was aseptically added. The mixture was made in a blender at a high speed of not less than 2,000 rpm for no longer than 2.5 minutes. Before applying the following technique, the contents of the jar were mixed by shaking. To produce a dilution of 10−2, 1 ml of the initial suspension (10−1) was aseptically transferred along with 9 ml of sterile peptone water (0.1%), into a sterile test tube. Additional 10-fold decimal dilutions were prepared from this dilution up to an acceptable countable dilution. Total bacterial countThe conventional plate count method (ISO 18593, 2018) for total bacterial counts (log CFU/cm2) by agar plating was used. Dilutions were properly mixed before being pipetted in 0.1 ml portions onto the plate count agar solid media surfaces in pre-labeled Petri dishes. A glass rod that had been bent was used to disseminate the inoculum throughout the entire surface after it had been sterilized by being dipped in 95% ethanol and then swiftly flamed to burn off the ethanol. The plates were then turned over and left to sit for 24 hours at 37°C. A new spreader was used for each dilution (at low dilutions) to avoid any adverse effects from product residue carrying during the flame-sterilization procedure. To ensure effective inoculum absorption, the agar surface was freshly made. All colonies in plates containing 25–250 colonies were enumerated and the findings were recorded. The colony counts obtained were averaged and multiplied by 10 and then by the suitable dilution factor (10−1–10−6). The outcomes were revealed to calculate the log number of colonies. If plates did not have between 25 and 250 colonies, the dilution count was documented, along with the number of colonies discovered. Isolation and identification of three major pathogens (E. coli, L. monocytogenes, and S. aureus)All of the collected samples were subjected to microbial isolation and identification following the method explained by Quinn et al. (2011) and ISO 18593 (2018). Table 1. Oligonucleotide primers and target genes used in this study.
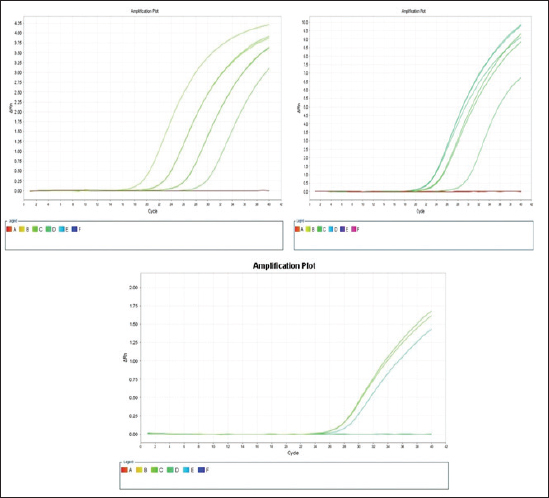
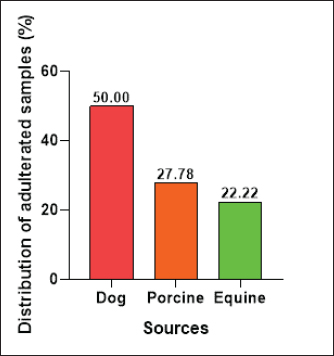
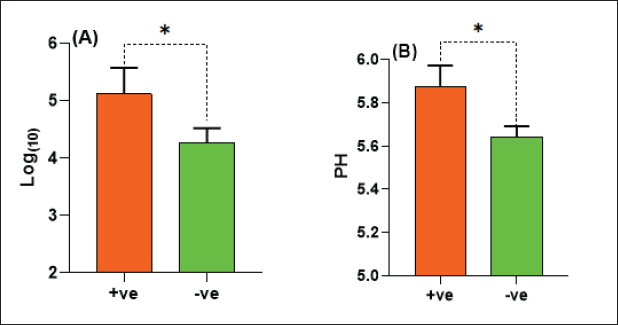
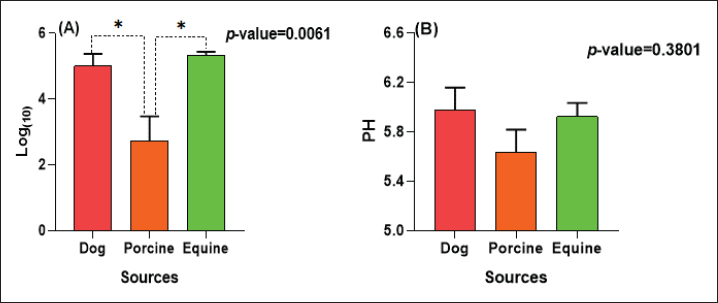
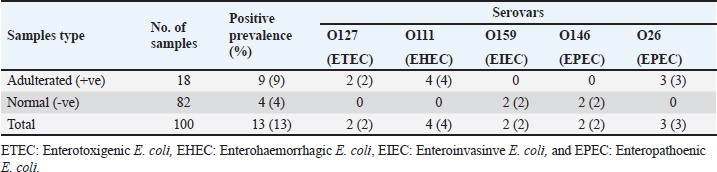
The E. coli confirmed isolates were serotyped using known antisera (Sifin) as described by Lee et al. (2009) in the Serology Unit, Animal Health Research Institute, Dokki, Egypt. Molecular assayValidation of PCRThe PCR primers used in this study were originally designed for single-conventional PCR techniques. Their specificity and sensitivity were previously confirmed (Hamouda and Abdelrahim, 2022). These primers were further tested for multiplex real-time applications in the Biotechnology Unit, Animal Health Research Institute, Zagazig Branch, Egypt. Willowfort (UK) supplied the primers which are listed in Table 1. Genomic DNA extractionFollowing the manufacturer’s instructions for the QIAamp DNA Mini kits (Qiagen, Germany, GmbH, Catalogue No. 51304), DNA was extracted from each blinded sample. SYBR-green multiplex real-time PCR assayThe amplifications were carried out using a Step OnePlusTM apparatus from Applied Biosystems (Foster City, California, United States). Fifteen μl of master mix (iQTM SYBR Green Supermix; Bio-Rad, USA) was mixed with 5 μl of the extracted DNA. 10 μl of iQTM SYBR green supermix, 1 μl of forward and reverse primers, and 3 μl of deionized water made up the master mix. The thermal profile consisted of 5 minutes of activation at 95°C, 40 cycles of denaturation at 95°C for 15 seconds, 30 seconds of 60°C annealing, and 30 seconds of extension at 55°C. After evaluating the SYBR green fluorescence intensity and melting curve studies, a threshold cycle (Ct) less than 35 and a particular melting temperature (Tm) showed a successful result. Statistical analysisMS Excel, developed by Microsoft Corporation in Redmond, WA, USA, was utilized to modify the data. The Levene and Shapiro–Wilk tests were employed to ascertain the consistency and normalcy of variance (Razali and Wah, 2011). Significant variances among the adulterated and normal samples for total bacterial count and pH were examined using a t-test (Proc T-test; Stokes et al., 2012). In addition, the significant differences between the different sources of adulterated samples were tested using a one-way ANOVA (PROC ANOVA) with a significance level set at α=0.05. Results were expressed as means ± SE. If a significant effect was found, Tukey’s test was used to compare the means in pairs. The GraphPad Prism software 5.0 (GraphPad, USA) was used to create the figures. The p-value, which indicates statistical significance, was found to be less than 0.05. Ethical approvalNot needed for this study. ResultsFigure 1 shows SYBR green real-time PCR amplification plots for porcine, dog, and equine meat samples examined to detect the percentage of adulteration. The plots display the fluorescence signal over time for each sample. A higher fluorescence signal indicates a greater presence of DNA. Based on these plots, the total adulteration percentage is 18% (n=18/100). The dog sample exhibits the strongest fluorescence signal, suggesting that it contains the highest amount of dog DNA. The equine sample displays a weaker fluorescence signal, indicating a lower amount of dog DNA. The porcine sample has the weakest fluorescence signal, indicating that it contains the least amount of DNA. Regarding the distribution of adulterated samples, 50% of the studied samples were sourced from dogs followed by 27.7% from porcine and 22.2% from equine (Fig. 2). The present results indicate that the adulterated samples have higher total bacterial count and pH values compared to the normal (pure bovine) samples. The variances in bacterial count and pH values are statistically significant, with p-values of 0.843 (log10) and 0.233, respectively (p-values=0.0227 and 0.0489; Fig. 3A and B). Interestingly, there are significant differences in total bacterial count between the samples from dogs and equine compared to those from porcine (p < 0.05; Fig. 4A). In contrast, no significant differences are observed for pH values among the three studied sources (p > 0.05; Fig. 4B). Table 2 displays the positive prevalence of various serovars of E. coli in adulterated and normal meat samples. The most frequently occurring serovars in adulterated meat samples were O127, O111, and O26.
Fig. 1. Porcine, dog, and equine SYBR green real-time PCR amplification plots; control positive is represented by the upper curve, and control negative is represented by the lower linear curve. Positive control is parallel to all rest curves.
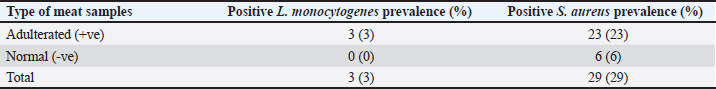
Fig. 2. Distribution of adulterated samples (%) between the different sources. Table 3 provides data on the incidence of S. aureus and L. monocytogenes isolates in the minced meat samples that were examined. The table shows that 3% of the minced meat samples were positive for L. monocytogenes, while 29% tested positive for S. aureus. The results also reveal that the prevalence of E. coli, L. monocytogenes, and S. aureus in adulterated meat samples was higher compared to normal meat samples. This indicates that adulterated meat samples are more prone to containing these pathogens than normal meat samples. DiscussionThe prevalence of adulterated minced meat samples varies depending on the country or region being studied. However, several studies have reported significant rates of adulteration in minced meat samples. For instance, a study conducted in Egypt discovered that 6% of beef minced meat samples were adulterated with either chicken or pork (Abuelnaga et al., 2021). In other wise, a study in India found that 11% of minced meat samples contained nonmeat ingredients, such as soy protein and starch (Moirangthem et al., 2022).
Fig. 3. Total bacterial count and pH values in adulterated (+ve) and normal samples (-ve).
Fig. 4. Total bacterial count and pH values in different sources of adulterated samples. Table 2. Incidence of E. coli isolates and their serovars isolated from minced meat samples.
Table 3. Occurrence rate of S. aureus and L. monocytogenes isolates in examined minced meat samples.
Adulterated minced meat can pose various risks to consumers. One concern is that consumers may be deceived about the actual content of the minced meat if it is mixed with cheaper meats like chicken or pork. Furthermore, adulterated minced meat may contain harmful bacteria or other contaminants (Han et al., 2020). There are several reasons why minced meat may be adulterated. One motive is to lower production costs. Adulterators may add cheaper meats or nonmeat ingredients to minced meat to reduce expenses. Another reason is to deceive consumers. Adulterators may sell adulterated minced meat as a more expensive meat variety, such as beef (Setiadi et al., 2022). This work presents the development and application of an advanced multiplex PCR assay for the simultaneous detection of dog, porcine, equine, and bovine species in minced meat. In addition, this study validates the lack of any contaminant pathogens in the examined samples. This investigation recorded the occurrence of adulteration in 18% of the examined samples, with 9% containing dog meat, 5% equine meat, and 4% porcine meat. Five samples of minced beef that were tested by Margawat and Ridwan (2011) were found to be contaminated with pork flesh. According to the researchers, variations in the time and temperature settings for each PCR step, as well as variations in the concentration of reaction chemicals may be to blame for the inconsistencies in PCR results. Sakalar and Abasiyanik (2011) conducted another investigation that found that 40% of commercially labeled meat products contained meat species that were not listed on the labels. In addition, Ha et al. (2017) detected three samples that tested positive for pork out of 35 processed meat products in Korea, while Ghovvati et al. (2009) found no traces of porcine residuals in any of the meat they examined. Of interest, the results suggest that adulterated minced meat samples are more likely to be contaminated with bacteria compared to normal samples. In addition, the type of adulterant used can also impact the bacterial count of the sample due to several factors. First, adulterants can alter the nutritional composition of meat, which in turn affects the growth of bacteria. Second, the moisture content of each type of meat plays a crucial role in bacterial growth. Adulterants that alter the moisture content can have a significant impact on the bacterial count. Finally, adulterants can affect the storage conditions of meat, indirectly influencing the bacterial count (Momtaz et al., 2023). For instance, samples adulterated with dog or equine meat have higher bacterial counts than samples adulterated with porcine meat. Furthermore, the higher pH values found in adulterated minced meat samples may be due to the addition of alkaline substances, such as baking soda or sodium bicarbonate. These substances can be added to adulterate minced meat to improve its appearance and extend its shelf life. However, they can also create an environment that is more favorable to the growth of bacteria. This highlights the potential risks associated with consuming adulterated minced meat as these samples may be contaminated with harmful bacteria that can cause food poisoning (Saleem et al., 2022). The findings obtained showed that 13% of the examined minced meat samples had E. coli and the incidence ratio was higher in adulterated samples compared to normal ones. This is concerning because E. coli can cause food poisoning, which can lead to a range of symptoms, including diarrhea, vomiting, and cramps. In some cases, food poisoning from E. coli can be severe and even life-threatening. There are a number of reasons why adulterated minced meat samples may be more likely to be contaminated with E. coli. One reason is that adulterated meat may come from animals that are not healthy or that have been slaughtered in unsanitary conditions. Another reason is that adulterated meat may be handled or processed in a way that increases the risk of contamination with bacteria (Sharif et al., 2018). It is important to note that not all E. coli isolates are harmful. In fact, most strains are harmless and play a crucial role in maintaining a healthy digestive system and gut health. These strains aid in the breakdown of food and the production of essential vitamins and nutrients that our bodies require (Martinson and Walk, 2020). However, some strains and serovars can produce toxins that can cause food poisoning (Rey et al., 2003). The obtained enterotoxigenic E. coli (O127), which produces heat-labile enterotoxin (LT) and heat-stable enterotoxin (ST), is believed to be a significant cause of diarrheal diseases in adults and infants, especially in tropical regions with inadequate hygiene. According to Karmali (1989), these strains are the main reason why travelers’ diarrhea occurs in many nations. This aligns with the finding of Mellor et al. (2016), who also reported a 2% prevalence rate for O127, a particularly dangerous serovar of E. coli that can lead to severe food poisoning, including hemolytic uremic syndrome, which can be fatal (Goldwater, 2007). Enterohaemorrhagic (O 111) was found in a ratio of 4%. It may be transmitted to human reservoirs either via the ingestion of contaminated food or water or by contact with STEC-positive animals (Shiga toxigenic E. coli) or with their environment (Miko et al., 2009). Enteroinvasive (O159) was found in a ratio of 2% and Enteropathogenic (O146 and O26) was detected in a ratio of 5%. Infantile diarrhea is most frequently caused by EPEC, which typically leads to symptoms appearing within 12–36 hours. These symptoms include fever, nausea, vomiting, and watery diarrhea which may contain mucus but usually does not result in significant blood (Cabrera-Sosa and Ochoa, 2020). These findings are consistent with the serological results reported by Abuelnaga et al. (2021) and El-Demerdash et al. (2018). Overall and unfortunately, the serovars obtained are pathogenic and pose great hazards to public health. Furthermore, the presence of predominantly toxigenic Gram-positive bacteria (S. aureus and L. monocytogenes) in adulterated meat with these precents, as opposed to bovine meat, suggests potential contamination and improper handling practices during the adulteration process (Bintsis, 2017). Typically, these pathogens are not found in bovine meat during handling. These pathogens present a significant health risk to consumers, as they can cause a range of foodborne illnesses, varying in severity. Staphylococcus aureus can lead to skin infections, pneumonia, and bacteremia, while L. monocytogenes can result in meningitis, listeriosis, and complications during pregnancy (Ali and Alsayeqh, 2022). Therefore, consumers should be aware of the risks associated with consuming adulterated minced meat. They can mitigate these risks by purchasing from reputable suppliers, ensuring that minced meat is cooked thoroughly, and avoiding cross-contamination by keeping minced meat separate from other foods during preparation and cooking. ConclusionThe recent study’s findings on microbial contamination highlight the necessity of establishing food safety standards in situ. Advanced multiplex PCR has been determined to be crucial and should be widely employed to identify adulteration in animal-derived products and meat. To develop a program that educates citizens about the religious and cultural aspects of the food they consume, governments should establish stronger regulations through various religious, political, educational, and scientific organizations. To effectively detect meat adulteration, food control laboratories should utilize this technique to guarantee prompt and precise outcomes. AcknowledgmentNone. Conflict of interestAll authors declare that there is no conflict of interest. FundingThis research received no external funding. Dat availabilityThe manuscript contains all of the data used. Author contributionsDesign and forming an idea, ASE-D, SHE, and HAF; methodology, ASE-D, SHE, RMA, RAE, and MTR; authentication, ASE-D; formal evaluation, ASE-D; investigation, ASE-D; data management, ASE-D; writing—original draft preparation, ASE-D and RMA; writing—review and editing, ASE-D. The published version of the manuscript has been read and approved by all authors. ReferencesAbdel-Rahman, S.M., El-Saadani, M.A., Ashry, K.M. and Haggag, A.S. 2009. Detection of adulteration and identification of cat’s, dog’s, donkey’s and horse’s meat using species-specific PCR and PCR-RFLP techniques. Aust. J. Basic. Appl. Sci. 3(3), 1716–1719. Abuelnaga, A.S.M., ABD EL-RAZIK, K.A.E.-H., Ata, N.S., Hedia, R.H., ELGABRY, E.A.-E., Soliman, M.M.H. and Marie, H.S.A.W. 2020. Bacteriological assessment and multiplex-PCR test for the detection of meat adulteration of different animal species. Food. Sci. Technol. 41, 98–104. Abuelnaga, A.S.M., El-Razik, K.A.E.H.A., Soliman, M.M.H., Ibrahim, H.S., Abd-Elaziz, M.M.M., Elgohary, A.H., Hedia, R.H. and Elgabry, E.A.-E. 2021. Microbial contamination and adulteration detection of meat products in Egypt. World. Vet. J. 11(4), 735–744. Alahakoon, A.U., Jayasena, D.D., Ramachandra, S. and Jo, C. 2015. Alternatives to nitrite in processed meat: Up to date. Trends. Food. Sci. Technol. 45(1), 37–49. Ali, N.M., Mohamed, G.A.E. and El-Demerdash, A.S. 2023. Impact of oral administration of chitosan--nanoparticles on oxidative stress index and gut microbiota of heat stressed broilers. J. Adv. Vet. Res. 13(6), 997–1003. Ali, S. and Alsayeqh, A.F. 2022. Review of major meat-borne zoonotic bacterial pathogens. Front. Public. Health. 10, 1045599. Bhargava, K., Wang, X., Donabedian, S., Zervos, M., da Rocha, L. and Zhang, Y. 2011. Methicillin-resistant staphylococcus Aureus in retail meat, Detroit, Michigan, USA. Emerg. Infect. Dis. 17(6), 1135–1137. Bintsis, T. 2017. Foodborne pathogens. AIMS Microbiol. 3(3), 529–563. Cabrera-Sosa, L. and Ochoa, T.J. 2020. Escherichia coli diarrhea. In Hunter’s tropical medicine and emerging infectious diseases. Elsevier, pp: 481–485. Dawwam, G.E., Al-Shemy, M.T. and El-Demerdash, A.S. 2022. Green synthesis of cellulose nanocrystal/ZnO bio-nanocomposites exerting antibacterial activity and downregulating virulence toxigenic genes of food-poisoning bacteria. Sci. Rep. 12(1), 1–18. El-Demerdash, A.S., Aggour, M.G., El-Azzouny, M.M. and Abou-Khadra, S.H. 2018. Molecular analysis of integron gene cassette arrays associated multi-drug resistant Enterobacteriaceae isolates from poultry. Cell. Mol. Biol. 64(5), 149–156. El-Demerdash, A.S., Bakry, N.R., Aggour, M.G., Elmasry, S.S., Mowafy, R.E. and Erfan, A. 2023a. Bovine mastitis in Egypt: bacterial etiology and evaluation of diagnostic biomarkers. Int. J. Vet. Sci. 12, 60–69. El-Demerdash, A.S., Mohamady, S.N., Megahed, H.M. and Ali, N.M. 2023b. Evaluation of gene expression related to immunity, apoptosis, and gut integrity that underlies Artemisia’s therapeutic effects in necrotic enteritis-challenged broilers. Biotech 13(6), 181. El-Demerdash, A.S. and Raslan, M.T. 2019. Molecular Characterization of Listeria Monocytogenes isolated from different animal-origin food items from Urban and Rural Areas. Adv. Anim. Vet. Sci. 7(2), 51–56. Essawi, W.M., El-Demerdash, A.S., El-Mesalamy, M.M. and Abonorag, M.A. 2020. Validation of camel’s fetal fluids as antimicrobial agents. Curr. Microbiol. 77(8), 1399–1404. Ghovvati, S., Nassiri, M.R., Mirhoseini, S.Z., Moussavi, A.H. and Javadmanesh, A. 2009. Fraud identification in industrial meat products by multiplex PCR assay. Food. Control. 20(8), 696–699. Goldwater, P.N. 2007. Treatment and prevention of enterohemorrhagic Escherichia coli infection and hemolytic uremic syndrome. Expert. Rev. Anti. Infect. Ther. 5(4), 653–663. Ha, J., Kim, S., Lee, J., Lee, S., Lee, H., Choi, Y., Oh, H. and Yoon, Y. 2017. Identification of pork adulteration in processed meat products using the developed mitochondrial DNA-based primers. Korean. J. Food. Sci. Anim. Resour. 37(3), 464. Hamouda, A. and Abdelrahim, K.A. 2022. Molecular assay (Polymerase Chain Reaction based technology) for identification of cheating and adulteration of marketable Canned beef, Handmade sausage, Kofta and Hawawshi samples in Qalubia Governorate, Egypt. Benha. Vet. Med. J. 42(2), 14–18. Han, F., Huang, X., H. Aheto, J., Zhang, D. and Feng, F. 2020. Detection of beef adulterated with pork using a low-cost electronic nose based on colorimetric sensors. Foods 9(2), 193. Haunshi, S., Basumatary, R., Girish, P.S., Doley, S., Bardoloi, R.K. and Kumar, A. 2009. Identification of chicken, duck, pigeon and pig meat by species-specific markers of mitochondrial origin. Meat. Sci. 83(3), 454–459. Hegab, O.W., Abdel-Latif, E.F. and Moawad, A.A. 2020. Isolation of enterotoxigenic Staphylococcus aureus harboring seb gene and enteropathogenic Escherichia coli (serogroups O18, O114, and O125) from soft and hard artisanal cheeses in Egypt. Open. Vet. J. 10(3), 297–307. ISO 18593. 2018. Microbiology of the food chain-horizontal methods for surface sampling. Geneva, Switzerland: International Organization for Standardization. Karmali, M.A. 1989. Infection by verocytotoxin-producing Escherichia coli. Clin. Microbiol. Rev. 2(1), 15–38. Kesmen, Z., Sahin, F. and Yetim, H. 2007. PCR assay for the identification of animal species in cooked sausages. Meat. Sci. 77(4), 649–653. Lee, G.Y., Jang, H.I., Hwang, I.G. and Rhee, M.S. 2009. Prevalence and classification of pathogenic Escherichia coli isolated from fresh beef, poultry, and pork in Korea. Int. J. Food. Microbiol. 134(3), 196–200. Le Loir, Y., Baron, F. and Gautier, M. 2003. Staphylococcus aureus and food poisoning. Genet. Mol. Res. 2(1), 63–76. Maede, D. 2006. A strategy for molecular species detection in meat and meat products by PCR-RFLP and DNA sequencing using mitochondrial and chromosomal genetic sequences. Euro. Food. Res. Technol. 224(2), 209–217. Margawat, E.T. and Ridwan, M. 2011. A sensitive method for identification of porcine contaminant in unprocessed food by PCR Amplification. Technique. Biota. 16, 342–347. Martinson, J.N. V, and Walk, S.T. 2020. Escherichia coli residency in the gut of healthy human adults. EcoSal. Plus. 9(1), 1110–1128. Megahed, M.M.M., El-Nagar, A.M.A., El-Demerdash, A.S., Ayoub, M.A. and Tolba, H.M.N. 2023. Evaluation and development of diagnostic tools for rapid detection of Riemerella anatipestifer and Pasteurella multocida in ducks. J. Adv. Vet. Anim. Res. 10(2), 211. Mellor, G.E., Fegan, N., Duffy, L.L., McMillan, K.E., Jordan, D. and Barlow, R.S. 2016. National survey of Shiga toxin--producing Escherichia coli serotypes O26, O45, O103, O111, O121, O145, and O157 in Australian beef cattle feces. J. Food. Prot. 79(11), 1868–1874. Miko, A., Pries, K., Haby, S., Steege, K., Albrecht, N., Krause, G. and Beutin, L. 2009. Assessment of Shiga toxin-producing Escherichia coli isolates from wildlife meat as potential pathogens for humans. Appl. Environ. Microbiol. 75(20), 6462–6470. Moirangthem, S., Laskar, S.K., Das, A., Upadhyay, S., Hazarika, R.A., Mahanta, J.D. and Sangtam, H.M. 2022. Effect of incorporation of soy protein isolate and inulin on quality characteristics and shelf-life of low--fat duck meat sausages. Anim. Biosci. 35(8), 1250. Momtaz, M., Bubli, S.Y. and Khan, M.S. 2023. Mechanisms and health aspects of food adulteration: a comprehensive review. Foods 12(1), 199. Osman, K., Alvarez-Ordóñez, A., Ruiz, L., Badr, J., ElHofy, F., Al-Maary, K.S., Moussa, I.M.I., Hessain, A.M., Orabi, A., Saad, A. and Elhadidy, M. 2017. Antimicrobial resistance and virulence characterization of Staphylococcus aureus and coagulase-negative staphylococci from imported beef meat. Ann. Clin. Microbiol. Antimicrob. 16(1), 1–10. Pearson, D. 1984. Chemical analysis of foods. Edinburgh, London, United Kingdom: Churchill Livingstones. Quinn, P.J., Markey, B.K., Leonard, F.C., Hartigan, P., Fanning, S. and Fitzpatrick, Es. 2011. Veterinary microbiology and microbial disease. Hoboken, NJ: John Wiley & Sons. Razali, N.M. and Wah, Y.B. 2011. Power comparisons of shapiro-wilk, kolmogorov-smirnov, lilliefors and anderson-darling tests. J. Statis. Model. Anal. 2(1), 21–33. Rey, J., Blanco, J.E., Blanco, M., Mora, A., Dahbi, G., Alonso, J.M., Hermoso, M., Hermoso, J., Alonso, M.P., Usera, M.A., González, E.A., Bernárdez, M.I. and Blanco, J. 2003. Serotypes, phage types and virulence genes of Shiga-producing Escherichia coli isolated from sheep in Spain. Vet. Microbiol. 94(1), 47–56. Ryser, E.T. and Marth, E.H. 2007. Listeria, listeriosis, and food safety. CRC press. Sakalar, E., and Abasiyanik, M.F. 2011. Qualitative analysis of meat and meat products by multiplex polymerase chain reaction (PCR) technique. Afr. J. Biotechnol. 10(46), 9379–9386. Saleem, A., Sahar, A., Pasha, I. and Shahid, M. 2022. Determination of adulteration of chicken meat into minced beef mixtures using front face fluorescence spectroscopy coupled with chemometric. Food. Sci. Anim. Resour. 42(4), 672. Setiadi, I.C., Hatta, A.M., Koentjoro, S., Stendafity, S., Azizah, N.N. and Wijaya, W.Y. 2022. Adulteration detection in minced beef using low-cost color imaging system coupled with deep neural network. Front. Sustain. Food. Syst. 6, 1073969. Shalaby, A.G., Bakry, N.R. and El-Demerdash, A.S. 2021. Virulence attitude estimation of Pasteurella multocida isolates in embryonated chicken eggs. Arch. Microbiol. 203(10), 6153–6162. Sharif, M.K., Javed, K. and Nasir, A. 2018. Foodborne illness: threats and control. In Foodborne diseases, pp: 501–523. Da Silva, N., Taniwaki, M.H., Junqueira, V.C.A., Silveira, N., Okazaki, M.M. and Gomes, R.A.R. 2018. Microbiological examination methods of food and water: a laboratory manual. CRC Press. Spink, J., Hegarty, P.V., Fortin, N.D., Elliott, C.T. and Moyer, D.C. 2019. The application of public policy theory to the emerging food fraud risk: Next steps. Trends. Food. Sci. Technol. 85, 116–128. Stokes, M.E., Davis, C.S., and Koch, G.G. 2012. Categorical data analysis using SAS. Cary, NC: SAS Institute. Tarazi, Y., El-Sukhon, S., Al-Rahbi, A., and Ismail, Z.B. 2021. Molecular characterization and in vivo pathogenicity study of Listeria monocytogenes isolated from fresh and frozen local and imported fish in Jordan. Open. Vet. J. 11(3), 517–524. Tasara, T., Schumacher, S., and Stephan, R. 2005. Conventional and real-time PCR--based approaches for molecular detection and quantitation of bovine species material in edible gelatin. J. Food. Prot. 68(11), 2420–2426. | ||
| How to Cite this Article |
| Pubmed Style El-Sheikh SH, Whab RMA, ElDaly RA, Raslan MT, Fahmy HA, El-Demerdash AS. Bacteriological evaluation and advanced SYBR-green multiplex real-time PCR assay for detection of minced meat adulteration. Open Vet J. 2024; 14((1) (Zagazig Veterinary Conference)): 389-397. doi:10.5455/OVJ.2024.v14.i1.35 Web Style El-Sheikh SH, Whab RMA, ElDaly RA, Raslan MT, Fahmy HA, El-Demerdash AS. Bacteriological evaluation and advanced SYBR-green multiplex real-time PCR assay for detection of minced meat adulteration. https://www.openveterinaryjournal.com/?mno=178806 [Access: May 08, 2024]. doi:10.5455/OVJ.2024.v14.i1.35 AMA (American Medical Association) Style El-Sheikh SH, Whab RMA, ElDaly RA, Raslan MT, Fahmy HA, El-Demerdash AS. Bacteriological evaluation and advanced SYBR-green multiplex real-time PCR assay for detection of minced meat adulteration. Open Vet J. 2024; 14((1) (Zagazig Veterinary Conference)): 389-397. doi:10.5455/OVJ.2024.v14.i1.35 Vancouver/ICMJE Style El-Sheikh SH, Whab RMA, ElDaly RA, Raslan MT, Fahmy HA, El-Demerdash AS. Bacteriological evaluation and advanced SYBR-green multiplex real-time PCR assay for detection of minced meat adulteration. Open Vet J. (2024), [cited May 08, 2024]; 14((1) (Zagazig Veterinary Conference)): 389-397. doi:10.5455/OVJ.2024.v14.i1.35 Harvard Style El-Sheikh, S. H., Whab, . R. M. A., ElDaly, . R. A., Raslan, . M. T., Fahmy, . H. A. & El-Demerdash, . A. S. (2024) Bacteriological evaluation and advanced SYBR-green multiplex real-time PCR assay for detection of minced meat adulteration. Open Vet J, 14 ((1) (Zagazig Veterinary Conference)), 389-397. doi:10.5455/OVJ.2024.v14.i1.35 Turabian Style El-Sheikh, Soad H., Reham M. Abdel Whab, Rania A. ElDaly, Mona T. Raslan, Hanan A. Fahmy, and Azza S. El-Demerdash. 2024. Bacteriological evaluation and advanced SYBR-green multiplex real-time PCR assay for detection of minced meat adulteration. Open Veterinary Journal, 14 ((1) (Zagazig Veterinary Conference)), 389-397. doi:10.5455/OVJ.2024.v14.i1.35 Chicago Style El-Sheikh, Soad H., Reham M. Abdel Whab, Rania A. ElDaly, Mona T. Raslan, Hanan A. Fahmy, and Azza S. El-Demerdash. "Bacteriological evaluation and advanced SYBR-green multiplex real-time PCR assay for detection of minced meat adulteration." Open Veterinary Journal 14 (2024), 389-397. doi:10.5455/OVJ.2024.v14.i1.35 MLA (The Modern Language Association) Style El-Sheikh, Soad H., Reham M. Abdel Whab, Rania A. ElDaly, Mona T. Raslan, Hanan A. Fahmy, and Azza S. El-Demerdash. "Bacteriological evaluation and advanced SYBR-green multiplex real-time PCR assay for detection of minced meat adulteration." Open Veterinary Journal 14.(1) (Zagazig Veterinary Conference) (2024), 389-397. Print. doi:10.5455/OVJ.2024.v14.i1.35 APA (American Psychological Association) Style El-Sheikh, S. H., Whab, . R. M. A., ElDaly, . R. A., Raslan, . M. T., Fahmy, . H. A. & El-Demerdash, . A. S. (2024) Bacteriological evaluation and advanced SYBR-green multiplex real-time PCR assay for detection of minced meat adulteration. Open Veterinary Journal, 14 ((1) (Zagazig Veterinary Conference)), 389-397. doi:10.5455/OVJ.2024.v14.i1.35 |