| Research Article | ||
Open Veterinary Journal, (2024), Vol. 14(1): 350-359 Original Research Expression of recombinant Florida clade 2 hemagglutinin in baculovirus expression system: A step for subunit vaccine development against H3N8 equine influenza virusAhmed S. Atwa1, Lamis Gomaa2, Wael Elmenofy2,3, Haitham M. Amer1 and Basem M. Ahmed1*1Department of Virology, Faculty of Veterinary Medicine, Cairo University, Giza, Egypt 2Agricultural Genetic Engineering Research Institute, Agricultural Research Center, Giza, Egypt 3Department of Arid Land Agriculture, College of Agricultural and Food Sciences, King Faisal University, Al-Ahsa 31982, Saudi Arabia *Corresponding Author: Basem M. Ahmed. Department of Virology, Faculty of Veterinary Medicine, Cairo University, Giza, Egypt. Email: basem-ahmed [at] cu.edu.eg Submitted: 01/10/2023 Accepted: 15/12/2023 Published: 31/01/2024 © 2024 Open Veterinary Journal
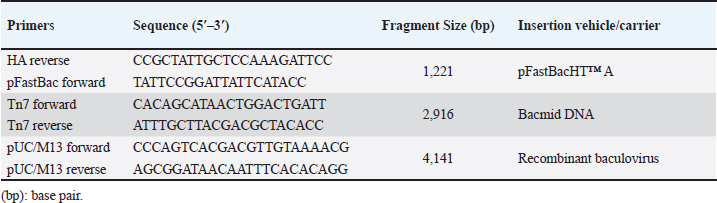
AbstractBackground: Equine influenza (EI) is a transmissible viral respiratory sickness of the Equidae family. Two viruses, H7N7 and H3N8 caused EI; however, H7N7 has not been detected for decades. H3N8 has circulated and bifurcated into Eurasian and American lineages. The latter subsequently diversified into Kentucky, South America, and Florida sub-lineages. Florida clade 1 (FC1) and Florida clade 2 (FC2) strains are the only circulating EI viruses (EIVs) in the meantime. Immunization is considered the major means for the prevention and control of EI infection. Using disparate technologies and platforms, several vaccines have been developed and commercialized. According to the recommendations of the World Organization for Animal Health (WOAH), all commercial vaccines shall comprise representatives of both FC1 and FC2 strains. Unfortunately, most of the commercially available vaccines were not updated to incorporate a representative of FC2 strains. Aim: The purpose of this research was to develop a new EI vaccine candidate that incorporates the hemagglutinin (HA) antigen from the currently circulating FC2. Methods: In this study, we report the expression of the full-length recombinant HA gene of FC2 in the baculovirus expression system. Results: The HA recombinant protein has been proven to maintain its biological characteristics by hemadsorption (HAD) and hemagglutination tests. Moreover, using a reference-specific serum, the specificity of the HA has been confirmed through the implementation of immunoperoxidase and western immunoblotting assays. Conclusion: In conclusion, we report the expression of specific biologically active recombinant HA of FC2, which would act as a foundation for the generation of an updated EI subunit or virus vector vaccine candidates. Keywords: Baculovirus expression system, equine influenza virus H3N8, Florida clade 2, hemagglutinin, recombinant vaccine. IntroductionEquine influenza (EI) is a remarkably contagious viral sickness that influences the upper respiratory tract of all members of the family Equidae (WOAH, 2019). The infection usually results in a mild self-limiting disease and rarely leads to mortalities. However, epizootics of the illness have remarkable economic impacts including the burden of fighting the outbreak alongside the costs of postponing or canceling race and show events (Powell et al., 1995, Smyth et al., 2011). Moreover, the capacity for inter-species transmission of influenza viruses is a remarkable concern that requires attention. A case in point is the incidence of canine influenza in the United States, which was conclusively trailed by EI virus (EIV) H3N8 transmission from equine to canine species (Klivleyeva et al., 2022). EI occurs as a consequence of EIV, which originates from the family Orthomyxoviridae, genus Alphainlfuenzavirus (Lefkowitz et al., 2018, Walker et al., 2022). The EIV genome comprises eight negative-sense RNA segments of single strands that, in contrast to less common strains, translate for ten fundamental proteins and multiple auxiliary nonessential polypeptides (Hao et al., 2020). Hemagglutinin (HA), the translation product of the fourth segment, is a surface glycoprotein that acts as a neutralizing antigen and is considered one of the most substantial pathogenicity determinants (Rott, 1992, Böttcher-Friebertshäuser et al., 2014). HA protein consists of three identical subunits (i.e., trimer) that comprise an ectodomain, a transmembrane domain, and an endodomain (Gamblin and Skehel, 2010). H7N7 and H3N8 are the two identified subtypes of EIV. The first mentioned was isolated in Czechoslovakia in 1958 (Sovinova et al., 1958) and had been circulating till its complete extinction in the late 1970s (Tůmová, 1980). Conversely, H3N8 emerged in, Florida, the United States in 1963 (Waddell et al., 1963), spread through various geographic regions, and dominated the globe thereafter (Wilson, 1993). Based on the phylogenetic analysis of the (HA1) portion of the HA gene, H3N8 started to diverge in the 1980s into genetically and antigenically distinct strains, and two independent lineages (American and Eurasian) bifurcated (Daly et al., 1996). The American lineage was subsequently subdivided and the following sub-lineages resulted: Kentucky, South America, and Florida; the latter was predominant all over the globe (Lai et al., 2001). The evolution of H3N8 had never stopped at this point, yet it took a step further when Florida sub-lineage split off into Florida clades 1 (FC1) and 2 (FC2) with divergent HA sequences (Lai et al., 2001). Such antigenic variation resulted in outbreaks even in vaccinated populations (Gildea et al., 2018) Hence, a recommendation was made by an expert surveillance panel (ESP) that EIV vaccines should encompass members of Florida clades 1 and 2 (Cullinane et al., 2010, WOAH, 2020). The first EI epizootic in Egypt, which took place in 1989, was documented in the Monufia governorate in 1989 involving horses, donkeys, and mules. The serum samples collected from infected animals demonstrated seroconversion with the H7N7 (Ismail et al., 1990, Abdalla et al., 1993). A further outbreak was described in Upper Egypt between January and April 2000 (Abd El-Rahim and Hussein, 2004) EIV hit Egypt again in 2008 and the circulating virus was typed as FC1 (Ahmed et al., 2011a, 2011b) Recently, an epizootic of EIV H3N8 of FC2 was reported in the Arabian racehorses in Egypt (Ahmed et al., 2022). This epizootic confirmed the necessity of revising the available EIV vaccines to comply with the ESP guidelines. Various commercial vaccines and vaccine candidates were developed to control EI in equines including modified live vaccines (MLVs) (Townsend et al., 2001, Youngner et al., 2001, Tabynov et al., 2014), whole inactivated vaccines (Paillot et al., 2013, Pavulraj et al., 2021), subunit (Paillot and Prowse, 2012), DNA vaccines (Soboll et al., 2003, Landolt et al., 2010) these antibody responses differ substantially in that particle mediated DNA vaccination does not induce an immunoglobulin A (IgA, recombinant canarypox vector vaccines (Toulemonde et al., 2005; Paillot et al., 2006), recombinant equine herpesvirus vaccine (Van de Walle et al., 2010), and reverse genetics-based vaccines (Ohta et al., 2021, 2022). The majority of commercially available vaccines in Egypt do not include representatives of FC2 strains. Hence, an urge to develop a vaccine candidate including FC2 has risen. However, the efforts exerted to isolate the FC2 virus in embryonated chicken eggs (ECEs) turned out to be unsuccessful (Ahmed et al., 2022). Baculovirus excels as an expression system due to various merits and distinctive features including the remarkable ability to incorporate large sequences of DNA inserts, ease of development and selection of recombinant baculoviruses, superior safety results from failure of baculoviruses to induce pathogenicity in nontarget host species, besides the nonsophisticated technology that allows site-specific recombination through transposition and flexible propagation of insect cell lines in both adherent and suspension cultures (Luckow and Summers, 1988, Maeda, 1989, Murhammer, 1991, Felberbaum, 2015). Currently, multiple commercial vaccines and therapeutics have been developed utilizing baculovirus vectors for use in humans, including but not limited to Cervarix®, Flublok®, and Flublok® Quadrivalent, and Nuvaxovid/Covovax®, which protect from cervical cancer, seasonal influenza, and COVID-19, respectively (Felberbaum, 2015, Hong et al., 2022). In addition, Porcilis Pesti®, Porcilis PCV®, Circumvent PCV®, BAYOVAC CSF E2®, and Ingelvac CircoFLEX® have been approved for veterinary use. In the current study, we were able to produce a recombinant HA FC2 antigen using baculovirus as an expression system, aiming to develop an effective H3N8 FC2 subunit vaccine. Materials and MethodsCells and antibodiesSpodoptera frugiperda (Sf9) insect cell line was gifted by courtesy of Dr. A. Abd El Wahed, University of Leipzig, Germany, for use in the generation and propagation of the recombinant baculoviruses. EX-CELL® 420 serum-free medium (SFM) with L-glutamine, which is supplemented with 3% (v/v) heat-inactivated fetal bovine serum (FBS), was utilized to culture Sf9 cells at 27°C with the addition of penicillin (100 U/ml), streptomycin (100 μg/ml), and amphotericin B (0.25 μg/ml) (Sigma-Aldrich, Taufkirchen, Germany). Cells were maintained in SFM without supplements during transfection with the bacmid DNA. Reference antibodies, prepared in equine species, against the HA antigen of EIV H3N8, were provided by courtesy of Dr. M.R. Gadalla, Freie Universität Berlin, Germany. Generation of the recombinant bacmidA consensus sequence of the full-length HA gene (1728 bp) of EIV H3N8 was generated by the alignment of three partial HA sequences of lately sequenced FC2 Egyptian strains (GenBank® accession numbers: MK089850; MK089827; MK089810), besides the reference prototype FC2 strain (A/equine/Richmond/1/07) using BioEdit software, version 7.2.5 (Ibis Biosciences, Carlsbad, CA). The obtained consensus sequence was synthesized by Biomatik Corporation (Ontario, Canada) and cloned into pFastBacHT™ A vector (ThermoFisher Scientific, Waltham, MA) downstream the polyhedrin promoter, and in frame with the initiation codon ATG and the N-terminal 6X Histidine tag. Insertion of the HA gene in the correct orientation was checked by PCR employing pFastBac forward and HA reverse primers (Table 1) and restriction endonuclease digestion via HindIII and SalI (New England Biolabs, Ipswich, MA). MAX Efficiency® DH10Bac™ chemically competent E. coli cells (ThermoFisher Scientific) were transformed by 1 ng (in 100 μl) of the purified recombinant DNA plasmid vector according to the manufacturer’s instructions. Three different volumes (10, 50, and 100 μl) of the transformation reaction, to optimize the recombinant DH10Bac™ E. coli growth condition, were plated on Luria-Bertani (LB) agar containing 50 μg/ml kanamycin, 7 μg/ml gentamicin, 10 μg/ml tetracycline, 100 μg/ml X-gal, and 40 μg/ml IPTG (Sigma-Aldrich). After incubation at 37°C for 48 hours, a suitable number of well-separated white colonies were picked up and propagated in selective LB broth containing kanamycin, gentamicin, and tetracycline. Recombinant bacmid DNA was extracted from the transformed DH10Bac™ cells using QIAprep® Spin Maxiprep Kit (Qiagen, Hilden, Germany), quantified using Qubit® dsDNA BR Assay Kit (ThermoFisher Scientific), and verified for insertion of the full-length HA gene in correct orientation by PCR using Tn7 forward and Tn7 reverse primers (Table 1). Table 1. Oligonucleotide primers used for PCR verification of HA gene insertion.
Generation of the recombinant baculovirusRecombinant baculoviruses were generated by transfection of Sf9 cells at 80% confluence in 6-well tissue culture (TC) plates. Two different volumes (25 μl and 50 μl) of purified bacmid DNA (188 ng/μl) were mixed with (8 μl) of Cellfectin™ II reagent (1 mg/ml) (Invitrogen), 8% diluted in SFM, and incubated for 15–30 minutes at room temperature. The DNA-lipid mixture was added dropwise onto the cells and plates were incubated at 27°C till CPE developed. The supernatant medium was collected 48 hours ( early phase) and 96–120 hours (late phase) post-transfection; both fractions are considered P1 stock of recombinant baculoviruses. The clarified P1 harvest was further propagated in Sf9 cells to amplify the virus stock and to obtain the highest virus titers for expression studies. The high-titer virus stock was therefore verified for integration of the full-length HA gene by PCR using pUC/M13 forward and pUC/M13 reverse primers (Table 1) and titrated by plaque assay according to the Bac-to-Bac® manual (Invitrogen, 2015). Hemadsorption (HAD) and HA assaysHAD and HA assays were performed for the detection of the recombinant HA protein in expression studies. HAD was executed according to (Nakajima et al., 2003) with some modifications as follows: 10-fold serial dilutions of P3 virus stock were prepared, and five dilutions (10-4 to 10-8) were used for inoculation of Sf9 cells at 80% confluence in 6-well TC plates (two wells/dilution). After 3 days of incubation at 27°C, the culture medium was discarded, and cells were washed twice with sterile PBS. One milliliter of freshly prepared 5% chicken RBCs was added to all wells of the plate including the cell control wells. The plates were incubated for 15 minutes at room temperature, with frequent gentle shaking to ensure well distribution of RBCs. After discarding of RBC solution, three-cycle washing of cells with PBS (5 minutes each) was performed, and plates were examined using the inverted microscope. HA assay was performed in 96-well U-shaped bottom plates according to the standard methods. Briefly, Sf9 cells infected with P3 virus stock were harvested and disrupted by three-cycle freezing and thawing. Two-fold serial dilutions of cell lysates were prepared across the wells of plate columns, and equal volumes of freshly prepared 1% chicken RBCs were added to each dilution. The plates were incubated for 20 minutes at room temperature, and the protein concentration was expressed as the reciprocal of the highest dilution showing complete hemagglutination. ImmunoperoxidaseThe ability of recombinant baculoviruses to express antigenic HA protein of EIV H3N8 was further confirmed by immunoperoxidase staining of infected SF9 cells using the standard protocol of (Haegeman et al., 2020) with few modifications. In brief, Sf9 cells, cultured in six-well plates at 80% confluence, were infected with the recombinant baculoviruses at a multiplicity of infections (MOI) of 0.1. Following incubation at 27°C for 48 hours, the culture medium was discarded, and cells were washed twice with PBS, dried at 37°C for 1 hour, and frozen at −80°C for 30 minutes. Infected and control cells were fixed with 4% formaldehyde for 10 minutes and permeabilized with a mixture of methanol and hydrogen peroxide 30% (30:1) (Sigma-Aldrich). After removal of the fixatives and washing of cells twice with PBS, the equine anti-HA primary antibodies (diluted 1:256 in PBS) were incubated with cells at room temperature for 1 hour with gentle agitation. Cells were further washed three times with PBS-Tween, and HRP-conjugated anti-equine antibodies prepared in rabbit, diluted 1:1000 in PBS (Sigma-Aldrich) were added and incubated at room temperature for 1 hour. After another cycle of washing with PBS-Tween, cells were stained with DAB substrate solution (Sigma-Aldrich) and examined for immunostaining using the inverted microscope. Western immunoblottingIn-vitro expression of EIV H3N8 HA protein was additionally confirmed by Western immunoblotting assay. SF9 cells were infected with the recombinant baculovirus at MOI of 10 and incubated at 27°C for 120 hours. Expressed proteins were harvested 48 and 120 hours post-infection using lysis buffer (1% Triton, 50 mM Tris-Hcl, pH 7.4, 150 mM NaCl, 1 mM EDTA, 2 mM AEBSF, 1 mM Phosphoramidon, 130 mM Bestatin, 14 mM E-64, 1 mM Leupeptin, 0.2 mM Aprotinin, 10 mM Pepstatin A). Cell lysates were heated with 4X SDS sample buffer at 95°C for 10 minutes and were separated into 10% SDS-polyacrylamide gels. Separated proteins were transferred to 0.45 μm nitrocellulose membrane (BioRad, Hercules, CA) using a transblot semidry blot transfer cell (BioRad, Hercules, CA). The membrane was blocked in TBS-Tween20 (TBS-T) containing 5% Bovine serum albumin overnight and then incubated for 2 hours at room temperature with 1:1,000 dilutions of equine anti-HA primary antibodies and mouse anti-β-actin antibodies (Fagus Antibody Services, Oxfordshire, U.K.). Unbound antibodies were washed five times using TBS-T. HRP-conjugated anti-equine and anti-mouse antibodies (diluted 1:1,000) were incubated with the membrane for 1 hour at room temperature before another cycle of washing was applied. The reaction was developed by enhanced chemiluminescence (ECL) according to the manufacturer’s instructions (ThermoScientific) and images were captured using the ChemiDoc imaging system (BioRad). Ethical approvalNot needed for this study. ResultsConstruction of recombinant bacmids carrying full-length HA geneThe synthetic construct that includes a consensus sequence of the full-length HA gene of EIV H3N8 (FCL2 Egyptian strains) was successfully subcloned into the transfer vector pFastBacHT™ A (Fig. 1). Clones carrying the full-length HA gene in the correct orientation were verified by PCR utilizing forward vector-specific and reverse gene-specific primers. A specific amplification fragment of 1,221 bp was only identified in positive clones (Fig. 2A). Restriction endonuclease digestion also elucidated two fragments of 4,789 and 1,707 bp in properly cloned plasmids. Transformation of DH10Bac™ chemically competent E. coli with pFastBacHT™ A-HA clones has enabled the generation of recombinant bacmids with full-length HA gene. Colonies with recombinant bacmids were selected by propagation in LB media containing a proper combination of antibiotics, and colormetrically (white colonies). HA gene insertion was further verified by PCR using Tn7 primers that generated an amplification product of 2916 bp.
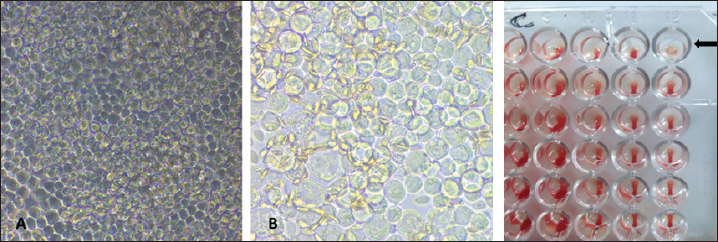
Fig. 1. Illustration of the transfer vector plasmid pFastBacHT™. Production and titration of recombinant baculovirus stocksBaculoviruses expressing the HA gene of EIV H3N8 were produced in Sf9 cell culture transfected with the recombinant DNA bacmids with the aid of Cellfectin. Transfected cells showed signs of infection by budded recombinant baculoviruses starting from 24 hours in the form of increased cell diameter (25%–50%) and enlarged nuclei. After 48–72 hours of transfection, cells cease growing as compared to cell control with the development of granular manifestation and monolayer detachment. Later, most cells lyse and exhibit signs of monolayer clearance (Fig. 3). P1 stock that contains the recombinant baculoviruses was collected at two time points; 48 and 96 hours post-transfection. Both stocks were mixed together and utilized for propagation in Sf9 for the preparation of high-titer P2 stock. The insertion of the HA gene in the correct orientation was confirmed by PCR employing pUC/M13 forward and reverse primers (Fig. 2B). P2 virus stock titer was determined as 1×106 pfu/ml.
Fig. 2. (A) HA gene integration and orientation confirmed by PCR using pFastBac-forward and HA-reverse primers and (B) PCR confirmation of insertion and orientation of HA gene into the DNA bacmid using Tn7 forward and Tn7 reverse primers. M: Marker Promega Corporation 100bp DNA Ladder, N: Negative, 1: sample No. 1, 2: sample No. 2.
Fig. 3. Development of CPE in Sf9-infected cells under an inverted microscope (10X). (A) Cell control. (B) Early phase CPE. (C) Late phase CPE. Characterization of the expressed recombinant HA proteinFunctional analysisTo express the HA protein, insect cells were infected with a high-titer virus stock. Several factors that influence the expression conditions were tested and optimized including cell line, culture media, MOI, and time of protein harvesting. The expressed protein was characterized for its functional activity (i.e., hemagglutination) using both HAD and plate hemagglutination assays. HAD was performed on Sf9 cells infected by different dilutions of the recombinant baculovirus (108 to 106 pfu/ml). Clumps of chicken RBCs surrounding Sf-9 cells were evident in the entire monolayer sheet of cells in all dilutions compared to control noninfected cells (Fig. 4A). Similarly, proteins expressed in cell lysates were able to hemagglutinate chicken RBCs in solution (1%) as determined by plate hemagglutination test till the dilution of 5 × 107 pfu/ml (Fig. 4B).
Fig. 4. (A) HAD of avian RBCs to the recombinant baculovirus Sf9-infected cells (right), (A) the control noninfected Sf9 cells. (B) Sf9-infected cells with 10-7 virus dilution. (C) Hemagglutination assay of lysates obtained from baculovirus-infected Sf9 cells; the black arrow indicates the positive result of agglutination. ImmunogenicityThe expressed HA protein was further investigated for its immunogenicity using standard equine anti-HA antibodies in two assays: immuno-peroxidase and western immunoblotting. Infected Sf-9 cells at two different MOIs (0.1 and 0.01) were able to develop brown–yellowish batches of immuno-peroxidase staining of variable sizes scattered all over the monolayer sheet of cells. No similar batches were evident in noninfected control cells (Fig. 5). Two specific protein bands of 30 and 70 kDa were detected in infected Sf-9 cultures but not in control noninfected cells in the Western immunoblotting assay. Both bands were visualized in 48- and 120-hours cultures with higher band intensity in the latter (figures were not included for suboptimal quality).
Fig. 5. IMPA: (A) Noninfected Sf9 control cells. (B) and (C) Sf9 cells expressing the recombinant HA stained with DAB substrate solution. DiscussionAlthough H3N8 FC2 EIV strains have been detected in numerous countries and the WOAH recommends FC2-related strains inclusion in EI vaccines, most commercial vaccines, as far as we know, have not been updated to incorporate FC2 strains. The only vaccine that does contain the A/equine/Richmond/1/07 strain is Prestige® by Merck. As a result, there is a discrepancy between the circulating virus and available vaccine strains, which renders immunized animals at risk of either infection or subclinical infection (Newton et al., 2006, Martella et al., 2007, Damiani et al., 2008, Bryant et al., 2009, Oladunni et al., 2021) that, in turn, leads to economic loss and silent spread of the virus, respectively. Modified live vaccines tend to mimic the immune response that occurs as a consequence of infection with a wild-type (WT) virus. Notwithstanding, there are considerable concerns about the vaccination against influenza viruses with MLV as this may result in reassortment with WT viruses. Moreover, the production of inactivated EI vaccines in ECE could be a time-consuming process, especially with viruses like influenza as they mutate frequently. In addition, the mass production of vaccines in ECE necessitates relatively huge facilities. The baculovirus expression system in insect cells was employed for decades for the production of recombinant proteins (Smith et al., 1983, Luckow and Summers, 1988) and the development of several therapeutics and vaccines (Felberbaum, 2015, Hong et al., 2022). Furthermore, the baculovirus expression system could be used for specific antigen generation, which is further utilized in monoclonal antibodies’ production (Liberti et al., 2023). This popularity could be connected with a myriad of merits including the straightforward scalability during the manufacturing process. Besides, it confers the rapid update of highly mutational viruses’ vaccines as influenza viruses, as the process of vaccine virus seed preparation is both time-consuming and costly. The recombinant proteins are expressed downstream the polyhedrin promoter of Autographa californica multiple nucleopolyhedrovirus (AcMNPV); a strong promoter that aids considerable levels of recombinant protein expression (Ciccarone et al., 1998). The site-specific transposition (Luckow et al., 1993) as well as the blue–white screening of E. coli colonies with recombinant bacmids (Pennock et al., 1984) facilitated the recombination and selection processes required for the production of recombinant baculoviruses. The HA antigens of discrete influenza viruses have been expressed in baculovirus vectors several times (Possee et al., 1986; Weyer and Possee, 1991; Yang et al., 2007; Sim et al., 2016); it is worth mentioning that the tested antigens showed strong immunogenicity in vivo, which shows the potential for the development of HA subunit vaccine. Furthermore, the intranasal inoculation of BALB/c mice with recombinant baculovirus expressing HA antigen elicited robust innate immune response, which protected the inoculated animals from fatal challenge with WT influenza virus (Abe et al., 2003), concluding a possibility of using baculovirus as a vector vaccine for boosting nonspecific immune response. Hence, to produce a subunit EI vaccine in the baculovirus expression system, we have constructed a transfer vector that comprehends the entire HA gene which was the result of a consensus sequence obtained from the Egyptian strains’ sequences and the sequence of the prototype reference strain A/equine/Richmond/1/07. The insertion of the synthetic HA gene into the DNA bacmid of baculovirus was confirmed by PCR and restriction digestion mapping. Both hemagglutination and HAD exclusively occur through a biologically active HA protein. Nevertheless, the HAD requires the integration of the biologically active HA antigen into the cell’s cell membrane. The incorporation of the HA is facilitated by its own transmembrane domain, whereas the adsorption to the RBCs is aided by the interaction between the RBCs’ sialic acid receptors and the HA trimer’s receptor binding site. Merely, proper HA protein folding allows for the trimerization of the HA antigen and subsequently its competent biological activity. Thus, we deduce the expression of full-length biologically active HA trimer with appropriate conformation. These results align with the findings of (Holtz et al., 2003, Wang et al., 2006), who stated that hemagglutination and HAD activities are indications of the appropriate folding of the HA antigen. Furthermore, research carried out by (Li et al., 2015) revealed that the folding of HA in the Sf9 cell line follows a comparable pathway to what could be identified in the cell lines of mammalian origin, yet occurs in a faster and more efficient pattern. Furthermore, HA is expressed as monomers, then these monomers form trimers and get integrated into the plasma cell membrane. In addition, (Davis et al., 1992, Kakker et al., 1999) stated that the protein processing and post-translational modifications are not distinct from eukaryotic cells. The immunoperoxidase assay (IMPA) was previously used to detect, type, and sub-type influenza viruses, where MAbs generated against the HA antigen of a specific subtype solely interacted with the homologous antigen subtype (Ziegler et al., 1995), which illustrates the high specificity of IMPA. Therefore, we utilized this technique to detect the HA antigen expressed by the baculovirus expression system, and we could successfully identify the recombinant HA antigen in Sf9 cells. This specific antigen–antibody interaction also highlights the proper folding of the antigen and the capability of using it as an immunogen to vaccinate against H3N8 EIV. Unfortunately, the absence of in vivo testing of the recombinant HA in laboratory animals, not to mention target host species, has limited the sufficient study of antigen immunogenicity. The in vivo testing requires the scaling up of virus cultures and the purification of the target protein, and both need specific equipment that was lacking at the time of the study. Nonetheless, the expressed HA would be subjected to extended study in the near future. In conclusion, these results collectively support the capability of the HA expressed in the baculovirus expression system to serve as an immunogenic antigen in a subunit or a baculovirus vector vaccine against H3N8 FC2 EIV infection. AcknowledgmentsAuthors are grateful to the Department of Virology and AGERI members for their help during this work. The presented results are the core of the first author master’s thesis and is a part of the grant funded by Cairo University entitled ‘preparation and experimental evaluation of an equine influenza H3N8 vaccine based on rDNA technology’, PI Dr. Basem Ahmed. Conflict of interestThe authors affirm that they have no conflict of interest to disclose. Author contributionsAll authors have contributed significantly to the manuscript, participated in revisions, and provided consent for the submitted version. AA: Conducted experimental work and contributed to manuscript writing. LG: Contributed to the cell culture work. WE: Contributed to the design of the experiments. HA: Played a remarkable role in conceptualizing the study and writing the manuscript. BA: Responsible for plasmid design and experimental design, and contributed to the conceptualization of the study. Data availabilityThe manuscript contains all the necessary data to support the study’s findings. Top of form. ReferencesAbd El-Rahim, I.H.A. and Hussein, M. 2004. An epizootic of equine influenza in Upper Egypt in 2000. OIE Rev Sci Tech 23, 921–930. Abdalla, M.A., Taleb, Z.A. and Ebid, M.H. 1993. Characterization of serum lysosomal enzymatic activities. III. Effect of infectious influenza in Egyptian equines. DTW. Deutsche tierarztliche Wochenschrift 100, 147–148. Abe, T., Takahashi, H., Hamazaki, H., Miyano-Kurosaki, N., Matsuura, Y. and Takaku, H. 2003. Baculovirus induces an innate immune response and confers protection from lethal influenza virus infection in mice. J. Immunol. 171, 1133–1139. Ahmed, B., Ahmed, H. and El-Sanousi, A. 2011a. Isolation and characterization of H3N8 equine influenza virus associated with 2008 EI outbreak in Egypt. Egypt. J. Virol. 8, 1–12. Ahmed, B., Ahmed, H. and El-Sanousi, A. 2011b. Phylogenetic analysis of the Egyptian equine influenza virus (A/Equine/Egypt/VRLCU/2008 (H3N8)). Egypt. J. Virol. 8, 53–72. Ahmed, B.M., Bayoumi, M.M., Farrag, M.A., Elgamal, M.A., Daly, J.M. and Amer, H.M. 2022. Emergence of equine influenza virus H3Nx Florida clade 2 in Arabian racehorses in Egypt. Virol. J. 19, 1–10. Böttcher-Friebertshäuser, E., Garten, W., Matrosovich, M. and Klenk, H.D., 2014. The hemagglutinin: a determinant of pathogenicity. Influenza Pathogen. Control 1, 3–34. Bryant, N.A., Rash, A.S., Russell, C.A., Ross, J., Cooke, A., Bowman, S., Macrae, S., Lewis, N. S., Paillot, R., Zanoni, R., Meier, H., Griffiths, L.A., Daly, J.M., Tiwari, A., Chambers, T.M., Newton, J.R. and Elton, D.M. 2009. Antigenic and genetic variations in European and North American equine influenza virus strains (H3N8) isolated from 2006 to 2007. Vet. Microbiol. 138, 41–52. Ciccarone, V.C., Polayes, D.A. and Luckow, V.A. 1998. Generation of recombinant baculovirus DNA in E. coli using a Baculovirus shuttle vector. Method Mol. Med. 13, 213–235. Cullinane, A., Elton, D. and Mumford, J. 2010. Equine influenza—surveillance and control. Influenza Other Resp. Viruses 4, 339–344. Daly, J.M., Lai, A.C.K., Binns, M.M., Chambers, T.M., Barrandeguy, M. and Mumford, J.A. 1996. Antigenic and genetic evolution of equine H3N8 influenza A viruses. J. Gen. Virol. 77, 661–671. Damiani, A.M., Scicluna, M.T., Ciabatti, I., Cardeti, G., Sala, M., Vulcano, G., Cordioli, P., Martella, V., Amaddeo, D. and Autorino, G.L. 2008. Genetic characterization of equine influenza viruses isolated in Italy between 1999 and 2005. Virus Res. 131, 100–105. Davis, T.R., Trotter, K.M., Granados, R.R. and Wood, H.A. 1992. Baculovirus expression of alkaline phosphatase as a reporter gene for evaluation of production, glycosylation and secretion. Nature Biotechnol. 10, 1148–1150. Felberbaum, R.S. 2015. The baculovirus expression vector system: a commercial manufacturing platform for viral vaccines and gene therapy vectors. Biotechnol. J. 10, 702–714. Gamblin, S.J. and Skehel, J.J. 2010. Influenza hemagglutinin and neuraminidase membrane glycoproteins. J. Biol. Chem. 285, 28403–28409. Gildea, S., Garvey, M., Lyons, P., Lyons, R., Gahan, J., Walsh, C. and Cullinane, A. 2018. Multifocal equine influenza outbreak with vaccination breakdown in thoroughbred racehorses. Pathogens 7, 1–16. Haegeman, A., De Leeuw, I., Mostin, L., Van Campe, W., Aerts, L., Vastag, M. and De Clercq, K. 2020. An immunoperoxidase monolayer assay (IPMA) for the detection of lumpy skin disease antibodies. J. Virol. Methods 277, 113800–113800. Hao, W., Wang, L. and Li, S. 2020. Roles of the non-structural proteins of influenza A virus. Pathogens 9, 812. Holtz, K., Anderson, K. and Cox, M.J. 2003. Production of a recombinant influenza vaccine using the baculovirus expression vector system. BioProcess. J. 2, 65–73. Hong, M., Li, T., Xue, W., Zhang, S., Cui, L., Wang, H., Zhang, Y., Zhou, L., Gu, Y., Xia, N. and Li, S. 2022. Genetic engineering of baculovirus-insect cell system to improve protein production. Front. Bioengin. Biotechnol. 10, 1–15. Ismail, T.M., Sami, A.M., Youssef, H.M. and Abou Zaid, A.A. 1990. An outbreak of equine influenza type 1 in Egypt in 1989. Vet. Med. J. Giza 38, 195–206. Kakker, N.K., Mikhailov, M.V., Nermut, M.V., Burny, A. and Roy, P. 1999. Bovine Leukemia virus gag particle assembly in insect cells: formation of chimeric particles by domain-switched Leukemia/Lentivirus gag polyprotein. Virology 265, 308–318. Klivleyeva, N., Glebova, T., Shamenova, M. and Saktaganov, N. 2022. Influenza A viruses circulating in dogs—a review of the scientific literature. Open Vet. J. 12, 676–676. Lai, A.C.K., Chambers, T.M., Holland, R.E., Morley, P.S., Haines, D.M., Townsend, H.G.G. and Barrandeguy, M. 2001. Diverged evolution of recent equine-2 influenza (H3N8) viruses in the Western Hemisphere. Arch. Virol. 146, 1063–1074. Landolt, G.A., Hussey, S.B., Kreutzer, K., Quintana, A. and Lunn, D.P. 2010. Low-dose DNA vaccination into the submandibular lymph nodes in ponies. Vet. Rec. 167, 302–304. Lefkowitz, E.J., Dempsey, D.M., Hendrickson, R.C., Orton, R.J., Siddell, S.G. and Smith, D.B. 2018. Virus taxonomy: the database of the International Committee on Taxonomy of Viruses (ICTV). Nucleic Acids Res. 46, D708-D717. Li, X., Van Oers, M.M., Vlak, J.M. and Braakman, I. 2015. Folding of influenza virus hemagglutinin in insect cells is fast and efficient. J. Biotechnol. 203, 77–83. Liberti, R., Colabella, C., Anzalone, L., Severi, G., Paolo, A., Casciari, C., Casano, A., Giammarioli, M., Cagiola, M., Feliziani, F. and Giuseppe, A. 2023. Expression of a recombinant ASFV P30 protein and production of monoclonal antibodies. Open Vet. J. 13, 358–364. Luckow, V.A., Lee, S.C., Barry, G.F. and Olins, P.O. 1993. Efficient generation of infectious recombinant baculoviruses by site-specific transposon-mediated insertion of foreign genes into a baculovirus genome propagated in Escherichia coli. J. Virol. 67, 4566–4579. Luckow, V.A. and Summers, M.D. 1988. Trends in the development of baculovirus expression vectors. Bio/Technol. 6, 47–55. Maeda, S. 1989. Expression of foreign genes in insects using baculovirus vectors. Ann. Rev. Entomol. 34, 351–372. Martella, V., Elia, G., Decaro, N., Di Trani, L., Lorusso, E., Campolo, M., Desario, C., Parisi, A., Cavaliere, N. and Buonavoglia, C. 2007. An outbreak of equine influenza virus in vaccinated horses in Italy is due to an H3N8 strain closely related to recent North American representatives of the Florida sub-lineage. Vet. Microbiol. 121, 56–63. Murhammer, D.W. 1991. Review and patents and literature—the use of insect cell cultures for recombinant protein synthesis: engineering aspects. Appl. Biochem. Biotechnol. 31, 283–292. Nakajima, K., Nobusawa, E., Tonegawa, K. and Nakajima, S. 2003. Restriction of amino acid change in influenza A virus H3HA: comparison of amino acid changes observed in nature and in vitro. J. Virol. 77, 10088–10098. Newton, J.R., Daly, J.M., Spencer, L. and Mumford, J.A. 2006. Description of the outbreak of equine influenza (H3N8) in the United Kingdom in 2003, during which recently vaccinated horses in Newmarket developed respiratory disease. Vet. Rec. 158, 185–192. Ohta, M., Bannai, H., Kambayashi, Y., Tamura, N., Tsujimura, K., Yamayoshi, S., Kawaoka, Y. and Nemoto, M. 2021. Growth properties and immunogenicity of a virus generated by reverse genetics for an inactivated equine influenza vaccine. Equine Vet. J. 54, 139–144. Ohta, M., Kambayashi, Y., Mita, H., Kuroda, T., Bannai, H., Tsujimura, K., Yamanaka, T., Garvey, M., Cullinane, A., Yamayoshi, S., Kawaoka, Y. and Nemoto, M. 2022. Protective efficacy of a reverse genetics-derived inactivated vaccine against equine influenza virus in horses. Vaccine 40, 6362–6366. Oladunni, F.S., Oseni, S.O., Martinez-Sobrido, L. and Chambers, T.M. 2021. Equine influenza virus and vaccines. Viruses 13, 1–36. Paillot, R., Kydd, J.H., Sindle, T., Hannant, D., Edlund Toulemonde, C., Audonnet, J.C., Minke, J.M. and Daly, J.M. 2006. Antibody and IFN-γ responses induced by a recombinant canarypox vaccine and challenge infection with equine influenza virus. Vet. Immunol. Immunopathol. 112, 225–233. Paillot, R. and Prowse, L. 2012. ISCOM-matrix-based equine influenza (EIV) vaccine stimulates cell-mediated immunity in the horse. Vet. Immunol. Immunopathol. 145, 516–521. Paillot, R., Prowse, L., Montesso, F., Huang, C.M., Barnes, H. and Escala, J. 2013. Whole inactivated equine influenza vaccine: efficacy against a representative clade 2 equine influenza virus, IFNgamma synthesis and duration of humoral immunity. Vet. Microbiol. 162, 396–407. Pavulraj, S., Bergmann, T., Trombetta, C.M., Marchi, S., Montomoli, E., Alami, S.S.E., Ragni-Alunni, R., Osterrieder, N. and Azab, W. 2021. Immunogenicity of Calvenza-03 EIV/EHV® vaccine in horses: comparative in vivo study. Vaccines 9, 166. Pennock, G.D., Shoemaker, C. and Miller, L.K. 1984. Strong and regulated expression of Escherichia coli beta-galactosidase in insect cells with a baculovirus vector. Mol. Cell. Biol. 4, 399–406. Possee, R.D., Kuroda, K., Hauser, C., Rott, R., Klenk, H.D., Doerfler, W. and Possee, R.D. 1986. Cell-surface expression of influenza virus haemagglutinin in insect cells using a baculovirus vector. Virus Res. 5, 43–59. Powell, D., Watkins, K., Li, P. and Shortridge, K. 1995. Outbreak of equine influenza among horses in Hong Kong during 1992. Vet. Rec. 136, 531–536. Rott, R. 1992. The pathogenic determinant of influenza virus. Vet. Microbiol. 33, 303–310. Sim, S.-H., Kim, J.Y., Seong, B.L., Nguyen, H.H. and Chang, J. 2016. Baculovirus displaying hemagglutinin elicits broad cross-protection against influenza in mice. PLoS One 11, e0152485. Smith, G.E., Summers, M.D. and Fraser, M.J. 1983. Production of human beta interferon in insect cells infected with a baculovirus expression vector. Mol. Cell. Biol. 3, 2156–2165. Smyth, G.B., Dagley, K. and Tainsh, J. 2011. Insights into the economic consequences of the 2007 equine influenza outbreak in Australia. Austr. Vet. J. 89, 151–158. Soboll, G., Horohov, D.W., Aldridge, B.M., Olsen, C.W., Mcgregor, M.W., Drape, R.J., Macklin, M.D., Swain, W.F. and Lunn, D.P. 2003. Regional antibody and cellular immune responses to equine influenza virus infection, and particle mediated DNA vaccination. Vet. Immunol. Immunopathol. 94, 47–62. Sovinova, O., Tumova, B., Pouska, F. and Nemec, J. 1958. Isolation of a virus causing respiratory disease in horses. Acta Virologica 2, 52–61. Tabynov, K., Kydyrbayev, Z., Ryskeldinova, S., Assanzhanova, N. and Sansyzbay, A. 2014. Duration of the protective immune response after prime and booster vaccination of yearlings with a live modified cold-adapted viral vaccine against equine influenza. Vaccine 32, 2965–2971. Toulemonde, C.E., Daly, J., Sindle, T., Guigal, P.M., Audonnet, J.C. and Minke, J.M. 2005. Efficacy of a recombinant equine influenza vaccine against challenge with an American lineage H3N8 influenza virus responsible for the 2003 outbreak in the United Kingdom. Vet. Rec. 156, 367–371. Townsend, H.G.G., Penner, S.J., Watts, T.C., Cook, A., Bogdan, J., Haines, D.M., Griffin, S., Chambers, T., Holland, R.E., Whitaker-Dowling, P., Youngner, J.S. and Sebring, R.W. 2001. Efficacy of a cold-adapted, intranasal, equine influenza vaccine: Challenge trials. Equine Vet. J. 33, 637–643. Tůmová, B. 1980. Equine influenza—a segment in influenza virus ecology. Comp. Immunol., Microbiol. Infect. Dis. 3, 45–59. Van De Walle, G.R., May, M.A., Peters, S.T., Metzger, S.M., Rosas, C.T. and Osterrieder, N. 2010. A vectored equine herpesvirus type 1 (EHV-1) vaccine elicits protective immune responses against EHV-1 and H3N8 equine influenza virus. Vaccine 28, 1048–1055. Waddell, G., Teigland, M. and Sigel, M. 1963. A new influenza virus associated with equine respiratory disease. J. Amer. Vet. Med. Assoc. 587–590. Walker, P.J., Siddell, S.G., Lefkowitz, E.J., Mushegian, A.R., Adriaenssens, E.M., Alfenas-Zerbini, P., Dempsey, D.M., Dutilh, B.E., García, M.L., Curtis Hendrickson, R., Junglen, S., Krupovic, M., Kuhn, J.H., Lambert, A.J., Łobocka, M., Oksanen, H.M., Orton, R.J., Robertson, D.L., Rubino, L., Sabanadzovic, S., Simmonds, P., Smith, D. B., Suzuki, N., Van Doorslaer, K., Vandamme, A.-M., Varsani, A. and Zerbini, F.M. 2022. Recent changes to virus taxonomy ratified by the International Committee on Taxonomy of Viruses (2022). Arch. Virol. 167, 2429–2440. Wang, K., Holtz, K.M., Anderson, K., Chubet, R., Mahmoud, W. and Cox, M.M.J. 2006. Expression and purification of an influenza hemagglutinin—One step closer to a recombinant protein-based influenza vaccine. Vaccine 24, 2176–2185. Weyer, U. and Possee, R.D. 1991. A baculovirus dual expression vector derived from the autographa californica nuclear polyhedrosis virus polyhedrin and p10 promoters: co-expression of two influenza virus genes in insect cells. J. Gen.Virol. 72, 2967–2974. Wilson, W.D. 1993. Equine influenza. Vet Clin North America Equine Pract. 9, 257–282. Woah. 2019. Equine Influenza. World Organization for Animal Health (WOAH)." should be "WOAH. 2019. Terrestrial Manual (online), Chapter 3.6.7. Equine influenza (infection with equine influenza virus). World Organization for Animal Health (WOAH), Paris, France. WOAH. 2020. OIE expert surveillance panel on equine influenza vaccine composition—WOAH Bulletin. Yang, D.-G., Chung, Y.-C., Lai, Y.-K., Lai, C.-W., Liu, H.-J. and Hu, Y.-C. 2007. Avian influenza virus hemagglutinin display on baculovirus envelope: cytoplasmic domain affects virus properties and vaccine potential. Mol. Ther. 15, 989–996. Youngner, J.S., Whitaker-Dowling, P., Chambers, T.M., Rushlow, K.E. and Sebring, R. 2001. Derivation and characterization of a live attenuated equine influenza vaccine virus. Amer. J. Vet. Res. 62, 1290–1294. Ziegler, T., Hall, H., Sánchez-Fauquier, A., Gamble, W.C. and Cox, N.J. 1995. Type- and subtype-specific detection of influenza viruses in clinical specimens by rapid culture assay. J. Clin. Microbiol. 33, 318–321. | ||
| How to Cite this Article |
| Pubmed Style Atwa AS, Gomaa L, Elmenofy W, Amer HM, Ahmed BM. Expression of recombinant Florida clade 2 hemagglutinin in baculovirus expression system: A step for subunit vaccine development against H3N8 equine influenza virus. Open Vet J. 2024; 14((1) (Zagazig Veterinary Conference)): 350-359. doi:10.5455/OVJ.2024.v14.i1.32 Web Style Atwa AS, Gomaa L, Elmenofy W, Amer HM, Ahmed BM. Expression of recombinant Florida clade 2 hemagglutinin in baculovirus expression system: A step for subunit vaccine development against H3N8 equine influenza virus. https://www.openveterinaryjournal.com/?mno=179183 [Access: May 08, 2024]. doi:10.5455/OVJ.2024.v14.i1.32 AMA (American Medical Association) Style Atwa AS, Gomaa L, Elmenofy W, Amer HM, Ahmed BM. Expression of recombinant Florida clade 2 hemagglutinin in baculovirus expression system: A step for subunit vaccine development against H3N8 equine influenza virus. Open Vet J. 2024; 14((1) (Zagazig Veterinary Conference)): 350-359. doi:10.5455/OVJ.2024.v14.i1.32 Vancouver/ICMJE Style Atwa AS, Gomaa L, Elmenofy W, Amer HM, Ahmed BM. Expression of recombinant Florida clade 2 hemagglutinin in baculovirus expression system: A step for subunit vaccine development against H3N8 equine influenza virus. Open Vet J. (2024), [cited May 08, 2024]; 14((1) (Zagazig Veterinary Conference)): 350-359. doi:10.5455/OVJ.2024.v14.i1.32 Harvard Style Atwa, A. S., Gomaa, . L., Elmenofy, . W., Amer, . H. M. & Ahmed, . B. M. (2024) Expression of recombinant Florida clade 2 hemagglutinin in baculovirus expression system: A step for subunit vaccine development against H3N8 equine influenza virus. Open Vet J, 14 ((1) (Zagazig Veterinary Conference)), 350-359. doi:10.5455/OVJ.2024.v14.i1.32 Turabian Style Atwa, Ahmed S., Lamis Gomaa, Wael Elmenofy, Haitham M. Amer, and Basem M. Ahmed. 2024. Expression of recombinant Florida clade 2 hemagglutinin in baculovirus expression system: A step for subunit vaccine development against H3N8 equine influenza virus. Open Veterinary Journal, 14 ((1) (Zagazig Veterinary Conference)), 350-359. doi:10.5455/OVJ.2024.v14.i1.32 Chicago Style Atwa, Ahmed S., Lamis Gomaa, Wael Elmenofy, Haitham M. Amer, and Basem M. Ahmed. "Expression of recombinant Florida clade 2 hemagglutinin in baculovirus expression system: A step for subunit vaccine development against H3N8 equine influenza virus." Open Veterinary Journal 14 (2024), 350-359. doi:10.5455/OVJ.2024.v14.i1.32 MLA (The Modern Language Association) Style Atwa, Ahmed S., Lamis Gomaa, Wael Elmenofy, Haitham M. Amer, and Basem M. Ahmed. "Expression of recombinant Florida clade 2 hemagglutinin in baculovirus expression system: A step for subunit vaccine development against H3N8 equine influenza virus." Open Veterinary Journal 14.(1) (Zagazig Veterinary Conference) (2024), 350-359. Print. doi:10.5455/OVJ.2024.v14.i1.32 APA (American Psychological Association) Style Atwa, A. S., Gomaa, . L., Elmenofy, . W., Amer, . H. M. & Ahmed, . B. M. (2024) Expression of recombinant Florida clade 2 hemagglutinin in baculovirus expression system: A step for subunit vaccine development against H3N8 equine influenza virus. Open Veterinary Journal, 14 ((1) (Zagazig Veterinary Conference)), 350-359. doi:10.5455/OVJ.2024.v14.i1.32 |