| Research Article | ||
Open Veterinary Journal, (2024), Vol. 14(1): 534-544 Original Research Molecular characterization of equine herpes viruses type 1 and 4 among Arabian horse populations in Egypt during the period between 2021 and 2022Ahmed A.H. Ali1, Fatma Abdallah1*, Omayma A. Shemies2, Gamilat Kotb1 and Maged R. Nafea21Department of Virology, Faculty of Veterinary Medicine, Zagazig University, Zagazig, Egypt 2Agricultural Research Center (ARC), Animal Health Research Institute (AHRI), Dokki, Egypt *Corresponding Author: Fatma Abdallah. Department of Virology, Faculty of Veterinary Medicine, Zagazig University, Zagazig, Egypt. Email: mm.fatma [at] yahoo.com Submitted: 01/10/2023 Accepted: 15/12/2023 Published: 31/01/2024 © 2024 Open Veterinary Journal
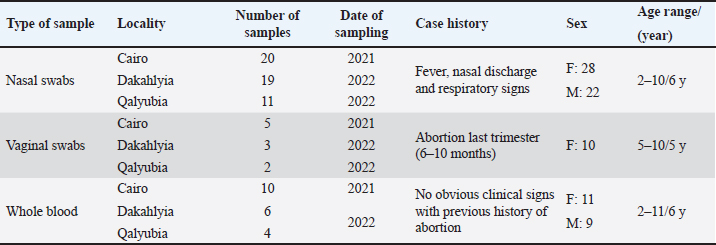
AbstractBackground: Equine herpesvirus type 1 (EHV-1) is a major cause of abortion and respiratory disease. Equine herpesvirus type 4 (EHV-4), on the other hand, is exclusively associated with respiratory disease in horse populations worldwide, particularly in Egypt and Arabian countries. Aim: This study aims to investigate the circulation of EHV-1 and EHV-4 in the Arabian horse population through molecular detection and genetic characterization of EHV-1 and/or EHV-4 that may threaten the stability of horse industry. Methods: A total of 80 samples including 50 nasal swabs, 10 vaginal swabs and 20 whole blood samples were collected from vaccinated and registered pure-bred Arabian adult horses from different studs in the governorates of northern Egypt (Cairo, Dakahlyia and Qalyubia) from 2021 to 2022. The collected samples were screened using consensus PCR for detection of EHVs using specific primers targeting DNA polymerase gene. The positive samples were subjected to conventional PCR for detection of EHV-1 and/or EHV-4using specific primers targeting glycoprotein (gB) gene. EHV-1 and EHV-4 amplicons were partially sequenced and phylogenetically analyzed using Sanger method. Results: Consensus PCR revealed that 48 out of 80 samples were positive for EHVs with percentage of 60%. Typing of the selected positive samples using conventional PCR showed that 29 out of 80 were positive for EHV-1 with percentage 36.25%, while 24 out of 80 samples were positive for EHV-4 with percentage 30%. Mixed infections with both viruses were detected in five samples. The amplified products were sequenced using Sanger method and submitted to GenBank under accession number OM362231MG-1 for EHV-1 strain and OM362232 MG-4 for EHV-4 strain. Sequence analysis and alignments of the amplified fragments of the EHV-1 and EHV-4 glycoprotein B (gB) gene to that of GenBank-derived reference strains revealed a high degree of similarity. According to the phylogenetic tree, the obtained sequences of EHV-1 and 4 in the current study showed homogeneity with local Egyptian and foreign EHV-1 and 4 strains and heterogeneity with EHV-2 and 5. Conclusion: The current investigation showed that molecular methods are appropriate assays for an efficient and accurate diagnosis of EHVs. Furthermore, it supports earlier research findings about the prevalence of EHV-1 and 4 in Arabian horse populations in Egypt. Keywords: Alphaherpesvirus, Arabian horse, Sequencing, Phylogenetic tree, Egypt. IntroductionEHVs are a group of viruses belonging to the Herpesviridae family including nine EHVs that assigned to Alphaherpesvirinae and Gammaherpesvirinae subfamilies and none to Betaherpesvirinae subfamily (ICTV, 2019). To date, there are nine herpesviruses have been identified comprising viruses belong to the Alphaherpesvirinae subfamily [six viruses: EHV-1, EHV-3, EHV-4, EHV-6, EHV-8 and EHV-9] or the Gammaherpesvirinae subfamily [three viruses; EHV-2, EHV-5 and EHV-7] (Davison et al., 2009). Alphaherpesviruses are ubiquitous viruses that affect many mammals; including EHV-1and EHV-4 which are closely related and are considered the most common equine respiratory tract viruses, with significant economic impact on the equine industry worldwide. The majority of adult horses were reported to be probably infected with EHV-1and/or EHV-4 during their lifetime (Gonzalez-Medina and Newton, 2015). EHV-1 and EHV-4 are the most common causes of respiratory disease (Hussey and Landolt, 2015). EHV-1 results in major welfare and financial impacts due to interruption of athletic programs, loss of replacement stock, abortions, neonatal deaths, respiratory disorders, and neurological disorders (myeloencephalopathy). This can lead to deaths and widespread movement restrictions which subsequently disrupts breeding and training schedules, causing management difficulties at training centers, racetracks, and horse racing events (Hussey et al., 2013; Ma et al., 2013). The survival of EHV-1 and EHV-4 in horse populations is related to their ability to establish lifelong latency in affected animals which act as carriers of virus with ability to disseminate viruses following stressors resulting in outbreak of the virus. The foals were recognized as the main source of early infection as they can get infected with EHV-4 by two years of age so that there is an urgent need for proper quarantine and control measures (Allen et al., 2004). EHV-1 and EHV-4 are serologically identified in donkeys with the risk of spreading to neighboring countries. Natural occurrence and experimental infection with EHV-1 were reported in donkeys (Lara et al., 2017). In Egypt, the horse population represents approximately 0.02% of the total equine population that requires regular monitoring of their health status. Arabian horses in particular are of great economic importance to horse breeders and are considered one of the most important horse breeds. The loss of valuable horses around the world has negative implications for the equine breeding and sporting industry (Amer et al., 2011). EHV-1 and 4 have been identified as viral agents that cause respiratory disease and/or abortion in Arabian mares (Warda, 2003). Few monitoring studies have been conducted but the continuous circulation of EHVs, particularly EHV-1 and EHV4 has been reported (Al-Shammari et al., 2016; Azab et al., 2019). The diagnosis of EHVs depends on a variety of detection methods in clinical samples (OIE, 2008). Early intervention measures to limit the spread of the virus are primarily based on rapid and sensitive diagnosis. Basic molecular analysis assay is an urgent approach for understanding the molecular epidemiology of EHV in the locality and clarification of the identity of the virus which is important in future control and prevention of EHV (Al-Shammari et al., 2016). Recently, several PCR-based assays were established for detection and typing of EHVs (Elia et al., 2006). Semi-nested and nested PCR assays were applied for the diagnosis of EHVs (Amer et al., 2011; Al-Shammari et al., 2016). The Effective control of EHV outbreaks requires rapid identification of EHV-1 infection, immediate restriction and maintenance of horse movement (Gonzalez-Medina and Newton, 2015). An improved biosecurity along with vaccines has resulted in the reduction of the abortion rates (Goodman et al., 2012). All currently available vaccines result in unsatisfactory responses. However, vaccination is thought to be partially responsible for reducing the severity of EHV by reducing the frequency of abortions and controlling viral shedding (Hussey et al., 2011; Wagner et al., 2017). The present work aimed to investigate the circulation of EHV-1 and EHV-4 in the Arabian horse population of northern Egypt during the period between 2021–2022 through molecular detection and genetic characterization based on nucleic acid amplification assays as well as phylogenetic analysis. Materials and MethodsSample collection and area of the studyA total of 80 samples from 80 animals including; 50 nasal swabs, 10 vaginal swabs and 20 whole blood samples were collected from vaccinated pure-bred registered Arabian horses from different Equine studs at Cairo, Dakahlyia and Qalyubia governorates during the period from 2021 to 2022. Nasal swabs were collected from animals suffering from fever and respiratory disorders, vaginal swabs were collected from aborted mares (Fig. 1), and whole blood samples were collected from apparently healthy mares with a previous history of abortion as illustrated in (Table 1). Reference strainsEHV-1 and EHV-4 positive controls used in the current study for validation of diagnostic assays are; EHV-1 strain (Giza-ARRI-USC-VM.EG19, 2021, Egypt) under accession number OL505458.1 and EHV-4 strain (AHRI-RV18, 2020, Egypt) under accession number MW039139.1.
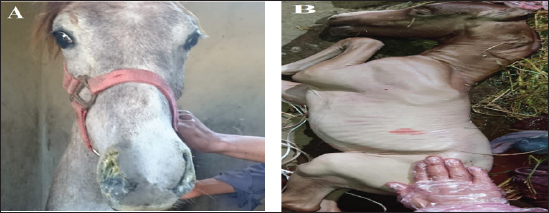
Fig. 1. The selected animals for sampling (A) Horses suffered from respiratory manifestations with nasal discharges, (B) aborted fetus at the last trimester (6–10 months) from the Arabian mares subjected for vaginal swabbing. Table 1. Details of collected samples from different localities during the period 2021–2022.
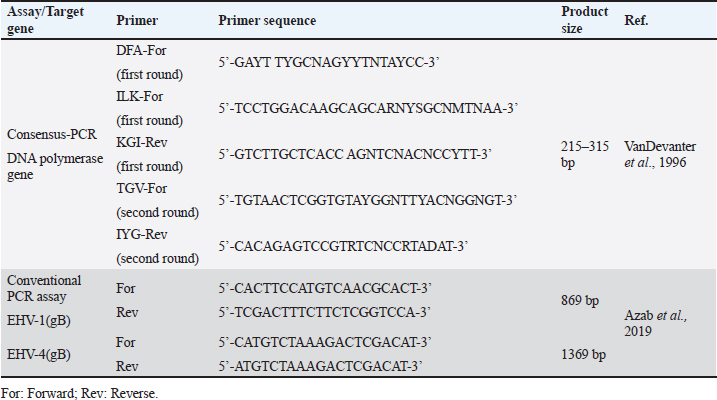
Sample processing and preparationThe processing and preparation of samples was applied in coincidence with World Organization for Animal Health guidelines (OIE, 2015). Nasal and vaginal swabs were placed in 3 ml of phosphate buffer saline (PBS) supplemented with antibiotic-antimycotic solution (100 IU/ml penicillin, 100 μg/ml streptomycin, and 0.25 μg/ml mycostatin B). The solution was collected in a clean sterile tube after vigorous mixing and swabs were discarded. The tubes were immediately frozen and kept at −80°C till examined. Buffy coat separation from collected whole blood samples was performed using Ficoll method. An equal amount of PBS was added to the tube containing blood on anticoagulant (EDTA). 3 ml of Ficoll®400solution (Sigma) (leukocytes separation reagent) was added to a tube. Non-coagulated blood PBS mix was added by pasteur pipette on the wall of the ficoll tube gradually until an insulating layer formation. Complete evacuation of the remaining non-clotted blood in ficoll tube then centrifuged at 3,000 rpm/30 minutes. Three layers are formed. The second layer is evacuated in another tube and completed with PBS then centrifuged at 2,000 rpm/15 minutes. The supernatant was discarded and the sediment collected. Drops of distilled water added on the sedimented cells and mixing by 8-figure for hemolysis of RBCs. Washing of sediment is repeated and then centrifuged at 4,000 rpm for 15 minutes till clear supernatant obtained. After every wash, a drop of sediment was put on a glass slide and examined under a microscope to confirm that sediment became clear from RBCs and only WBCs were present. The buffy coat will be used later for the detection of EHVs. Total viral DNA extractionEHV DNA was extracted from prepared nasal and vaginal swabs using (QIAamp DNA Mini Kit, QIAGEN, Germany), while from whole blood samples using (QIAamp DNA blood Mini Kit, QIAGEN, Germany). Consensus herpesvirus PCRScreening of equine herpesviruses in the extracted DNA was done by consensus PCR using general primers targeting DNA polymerase gene according to Van Devanter (1996) as listed in (Table 2). The primary mixture was prepared as follows; 10 μl master mix, 0.2 μl Primer (DFA), 0.2 μl Primer (ILK), 0.2 μl Primer (KG1), 5 μl Template DNA and nuclease-free water added up to 20 μl. The secondary mixture was prepared as follows; 10 μl master mix, 0.2 μl Primer (TGV), 0.2 μl Primer (IG), 4 μl PCR product of the first cycle and nuclease-free water added up to 20 μl. Primary and secondary mixtures were subjected to amplification in a thermocycler (Biometra, Germany) under a thermal profile for two cycles as follows: a cycle of initial denaturation at 95°C for 5 minutes followed by 40 cycles including (denaturation at 95°C for 30 seconds, annealing 46°C for 60 seconds, and extension at 72°C for 60 seconds) and one cycle of final extension at 72°C for 10 minutes. Conventional polymerase chain reaction assayTyping of DNA extracts was done by conventional PCR using specific primers targeting (gB) for detection of EHV-1 and/or EHV-4 as shown in Table 2. PCR mixture was prepared as follows; 12.5 μl Emerald Amp GT PCR mastermix (2× premix), 1 μl Forward primer (20 pmol), 1 μl Reverse primer (20 pmol), and 5 μl Template DNA PCR grade water was added up to 25 μl. The mixture was placed in a thermocycler. The thermal profile for EHV-1 included 35 cycles for amplification of EHV-1(gB) gene as follows: primary denaturation at 95°C for 5 minutes, secondary denaturation at 94°C for 30 seconds, annealing at 55°C for 40 seconds, extension at 72°C for 50 seconds and final extension 72°C for 10 minutes. The EHV-4 thermal profile also includes 35 cycle for amplification of EHV-4 (gB) as follows: primary denaturation at 95°C for 5 minutes, secondary denaturation at 94°C for 30 seconds, annealing at 55°C for 40 seconds, extension at 72°C for 1.2 minutes, and final extension at 72°C for 12 minutes. Table 2. List of primers used in the current study.
Table 3. Results of consensus PCR including year and positive samples.
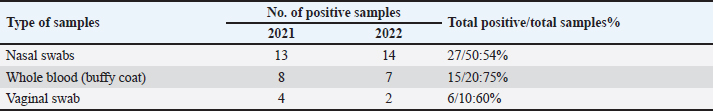
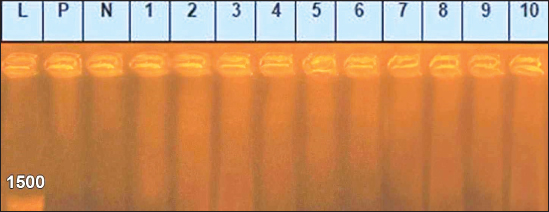
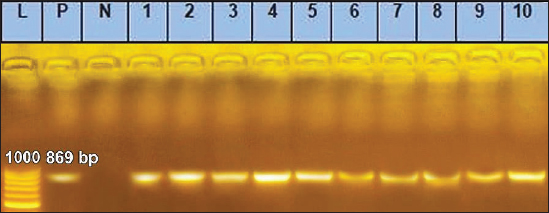
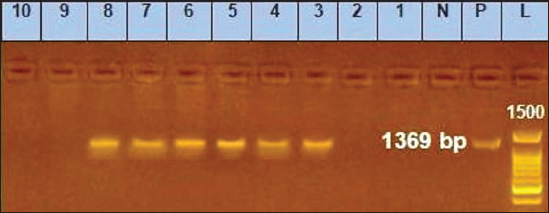
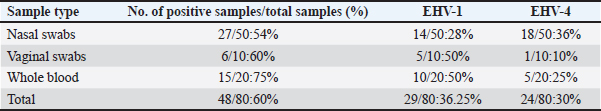
Sanger DNA sequencing of EHV-1 and EHV-4Amplified DNA fragments of EHV-1 and EHV-4 were selected and excised from agarose gel. The PCR products were then purified using the QiAquick gel extraction kit (Qiagen, Germany) (Altschul et al., 1990). The purified products were then sequenced by (Applied Bio Systems, Amp 9600 PCR system) and the resulting sequences were submitted to GenBank under accession number OM362231MG-1 or OM362232MG-4. Phylogenetic analysis of the EHV-1 and EHV-4 sequencesEHV-1 and EHV-4 obtained DNA sequences were submitted to Genbank and compared to other EHV-1 and EHV-4 isolates published in the Genbank database by using the Basic local alignment search tool of National Center for Biotechnology Information for demonstration of sequence similarity (Altschul et al., 1990). Finally, analysis of the obtained sequences was conducted and phylogenetic trees were designed using MEGA-11 software through neighbor-joining method to calculate the identity percentage and help in subtyping of EHV-1 and EHV-4. Nucleotide sequence identities and divergences were calculated utilizing Meg Align software (DNA STAR® Laser gene® version 7.2, USA). Ethical approvalAll sampling and examination procedures were performed with permission of Animal health research institute (AHRI) and Faculty of Veterinary Medicine, Zagazig University, Egypt, that complies with all legal requirements in Egypt. ResultsConsensus herpesvirus PCRScreening of the extracted EHVs DNA by consensus PCR in different samples showed EHVs in 48 out of 80 samples with a percentage of 60% including 54% (27/50) nasal swabs, 75% (15/20) whole blood samples, and 60% (6/10) vaginal swabs as summarized in (Table 3). The positive PCR products of field EHVs are at the size of approximately 250 bp (Fig. 2), whereas for EHV-1, the positive PCR products are at the size of approximately 869 bp (Fig. 3), and for EHV-4, the positive PCR products are at the size of approximately and 1369 bp (Fig. 4).
Fig. 2. Agarose gel electrophoresis of consensus PCR amplified DNA products for DNA polymerase gene of field EHVs, (positive PCR products at the size of approximately 250 bp). Lane L: 100 bp DNA marker, Lane P: positive control, Lane N: negative Control. 1–6: field samples.
Fig. 3. Agarose gel electrophoresis of conventional PCR amplified DNA products by for EHV-1, (Positive PCR products are at the size of approximately 869 bp). Lane L: 100 bp DNA marker, Lane P: positive control, Lane N: negative Control. 1–10: filed samples.
Fig. 4. Agarose gel electrophoresis of conventional PCR amplified DNA products by for EHV-4, (positive PCR products are at the size of approximately and 1,369 bp). Lane L: 100 bp DNA marker, Lane P: positive control, Lane N: negative control, 1–10: filed samples. Molecular typing using conventional PCR assayEHV-1 was found in 29 out of 80 samples with a percentage of 36.25% including 28%3 (14/50) nasal swabs, 50% (5/10) vaginal swabs, and 50% (10/20) whole blood samples. While, EHV-4 was identified in 24 out of 80 samples with percentage of 30% including 36% (18/50) nasal swabs,10% (1/10) vaginal swabs, and 25% (5/20) whole blood samples. Co-infection with EHV-1 and EHV-4 was detected in five samples as seen in (Table 4). Table 4. Result of typing of positive samples by conventional PCR.
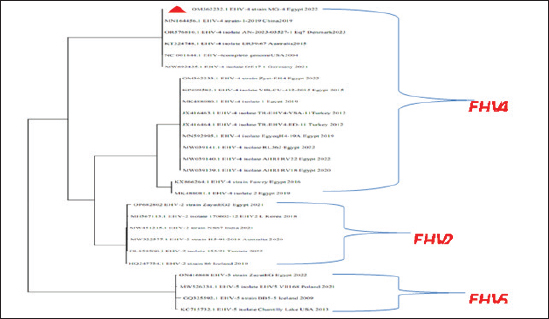
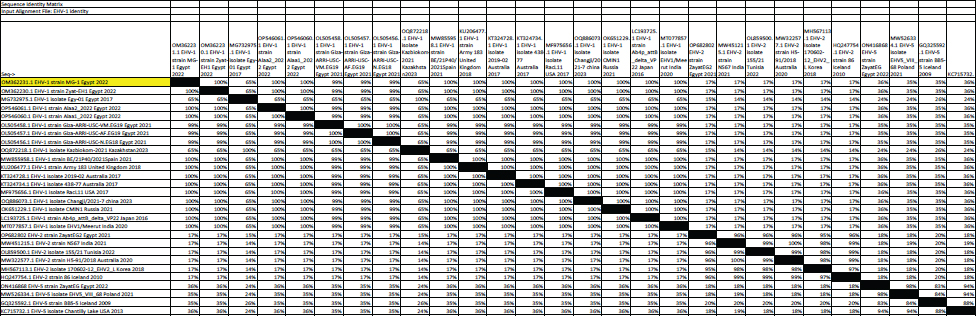
Fig. 5. Phylogenetic tree of EHV-1, 2, 5 (gB gene) and EHV-1 local strain of the present study labeled with red triangle as OM362231 MG-1. Sequencing of EHV-1 and EHV-4 gB geneThe nucleotide sequences of gB gene of the two isolates MG-1 strain, Egypt, 2022 and MG-4 strain, Egypt, 2022 were submitted to the GenBank database under the accession numbers OM362231MG-1 and OM362232 MG-4 for EHV-1 and EHV-4 isolates respectively. Phylogenetic analysis of EHV-1 gB genePhylogenetic analysis of the obtained sequence of EHV-1 (MG-1 strain) with other reference EHV-1, EHV-2, and EHV-5 sequences from GenBank showed that phylogenetic tree divided into two clusters. All Egyptian EHV-1 strains including MG-1and foreign EHV-1 strains were aligned in the first cluster, while the second cluster includes EHV-2 and EHV-5 strains. The phylogenetic tree revealed that EHV-1(MG-1 strain) from the current study was closely related to EHV-1 strains from other countries as seen in Figure 5. The identity percent of the obtained MG-1 strain with other Egyptian strains was 100%, while MG-1 strain showed 65%–100% identity with EHV-1 foreign strains,17% identity with EHV-2 strains and 35%–36% identity with EHV-5 strains. The genetic relationship between the current MG-1 strain and the reference EHV-1 strains indicates ancestral relation of the virus, while low identity percent with EHV-2 and EHV-5 reflects the genetic difference between alphaherpesviruses (EHV-1) and gammaherpresviruses (EHV-2 and EHV-5) as illustrated in (Table 5). Phylogenetic analysis of EHV-4 gB genePhylogenetic analysis of the obtained sequence of EHV-4 (MG-4 strain, Egypt, 2022) with other reference EHV-4, EHV-2, and EHV-5 sequences from GenBank revealed that phylogenetic tree divided into two clusters. All Egyptian and foreign EHV-4 strains including MG-4, Egypt, 2022 strain in addition to EHV-2 strains were aligned in the first cluster, while the second cluster include EHV-5 strains. The phylogenetic tree revealed that obtained EHV 4 strain (MG-4, Egypt, 2022) from the current study was closely related to other EHV-4 strains from other parts of the world (Fig. 6). The identity percent of the obtained MG-4 strain with other Egyptian strains was 100%, while MG-4 strain showed 40%–42% identity with EHV-2 strains and 11%–13% identity with EHV-5 strains (Table 6). The genetic relationship between MG-4 strain and the reference EHV-4 strains indicates ancestral relation of the virus, while low identity percent with EHV-2 and EHV-5 reveals the genetic difference between alphaherpesvirus (EHV-4) and gammahepres virus (EHV-2 and EHV-5). Table 5. The nucleotide diversity percentage between GenBank EHV 4, 2, 5 reference strains and local strain EHV-4 strain MG-4-2022 of the current study.
DiscussionEHVs are common viruses affecting equine populations and cause significant economic losses (Dunowska, 2014). The equine industry triggers are of great interest all over the world, especially in Egypt and Arabian countries which play an important role in the national income. EHV-1 can cause abortions, loss of fetuses, death of horses due to encephalomyelitis, loss of semen of infected stallions, loss of infected newborn foals, and loss of training time and performance opportunities during convalescence and quarantine (Salib et al., 2016). EHV-1 and EHV-4 are endemic in Egypt representing a continuous threat for horses and objective regular surveillance measures were recommended to investigate the presence of both viruses in various horse populations around Egypt (Azab et al., 2019). EHV 1 and 4 represent a high risk to equine populations, so that requires surveillance and prevention due to persistent and latent nature of the virus subsequently the absence of clinical signs and high economic losses (Kydd et al., 2012). Balasuriya et al. (2015) and Radalj et al. (2018) documented that the molecular methods have advantages of rapidity and accuracy in detection and identification of EHV nucleic acid in clinical samples in compared to the traditional methods such as virus isolation, immunohistochemical, and direct immunofluorescence method. Furthermore, standard virological methods are often very laborious and time-consuming which sheds the light on the requirement for the application of nucleic acid amplification-based detection techniques, such as consensus PCR, which has already been applied in this current study, as a screening assay for EHV, while Conventional PCR was used for EHV typing. A total of 80 samples including 50 nasal swabs, 10 vaginal swabs, and 20 whole blood samples were collected from adult, non-vaccinated showcasing horses from different Equine studs at Cairo, Dakahlyia, and Qalyubia governorates during the years 2021–2022. The collected samples were screened by consensus PCR using primers targeting DNA polymerase gene for detection of EHVs which found 60% positive samples. These results confirmed that EHVs are still circulating in Egypt’s northern governorates, with significant health impacts and economic losses due to the lack of vaccination programs and frequent virus reactivation, which could threaten the stability of the Egyptian horse industry in agreement with Salib et al. (2016), Azab et al. (2019), and Ali et al. (2020). Molecular screening and typing of samples using consensus PCR and conventional PCR assays as accurate, rapid, and innovative diagnostic approaches resulted in the control of EHV outbreaks as previously documented by Azab et al. (2019) and Amer et al. (2011). Connolly et al. (2011) mentioned that gB is the most conserved glycoprotein among EHVs types. Conventional PCR assay using specific primers targeting a highly conserved region of glycoprotein (gB) was valuable for molecular typing of EHVs in the collected samples due to high sensitivity and specificity for detection of EHV-1 and EHV-4 nucleic acids to demonstrate the circulation of both viruses among equine populations as previously reported by Amer et al. (2011); Al-Shammari et al. (2016) and Azab et al. (2019).
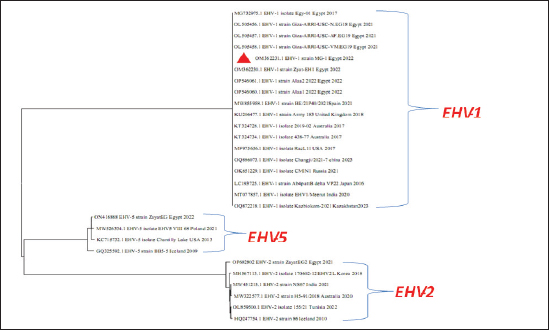
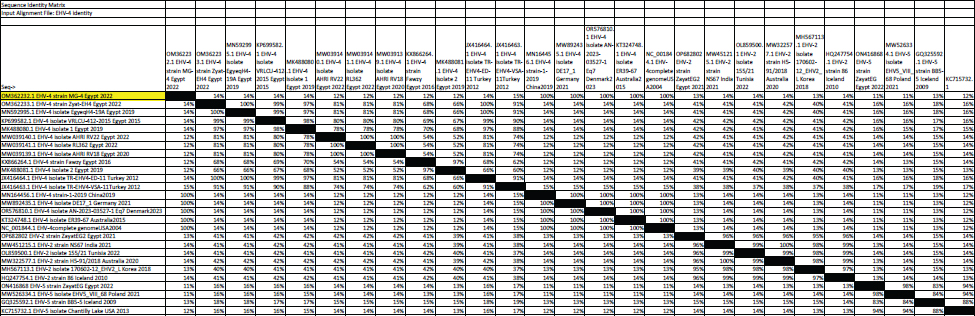
Fig. 6. Phylogenetic tree of EHV-4, 2, 5(gB gene) and EHV-4 local strain of the present study labeled with red triangle as OM362232 MG-4. Screening results revealed that EHV-1 and EHV-4 were identified in 36% and 30% of total of collected samples, respectively. This reflected that both viruses are still circulating among equine populations and EHV-1 is still the most prevalent type of EHVs in Egypt as previously mentioned by Van Maanen et al. (2000). Typing of EHVs in vaginal swabs from aborted mares showed that EHV-1 (Equine virus abortion) was more predominant as 50% than EHV-4 as 10% and this meant that EHV-1 is the main causative agent of abortion cases in mares with severe economic drawbacks in agreement with Azab et al. (2019) and Khattab et al. (2022). While, typing of EHVs in nasal swabs revealed that EHV-4 (rhinopneumonitis) was detected in 36% more than EHV-1 which was identified as 28% which reflected that both viruses are incriminated in respiratory dysfunctions in horses affecting the performance of showcasing and race horses as well as hindering the equine events which represent a source of income as previously mentioned by Gonzalez-Medina and Newton (2015) and Hussey and Landolt (2015). Whole blood samples collected from apparently healthy-vaccinated adult mare with history previous history of abortion were examined with conventional PCR assay which showed 50% and 25% positive samples for EHV-1and EHV-4, respectively which meant that EHVs have the ability for the establishment of latency and that EHV-1 and EHV-4 could be detected in apparently healthy animals as a result of reactivation process from latently infected horses due to external environmental and management stressor factors with sporadic shedding of the virus and this in agreement with Gilkerson et al. (2015). Co-infection with EHV-1 and EHV-4 was identified in five examined samples which reflects the probability of the simultaneous infection of the same horse with EHV-1 and 4 as mentioned by Azab et al. (2019). Phylogenetic analysis of the obtained EHV-1 sequences from the present study showed that they were homogeneous and clustered with previously detected Egyptian EHV-1 strains, indicating complete identity. Furthermore, this sequence is similar to other foreign EHV-1 strains with very low nucleotide substitutions, indicating high stability of the genomic DNA without gB gene diversity (Pusterla et al., 2007, 2009). There was a remarkable similarity between the phylogenetic analysis of the EHV-4 sequence and the previously detected EHV-4 Egyptian strains available on GenBank except for strains except for strains Egypt fawzy_2016 and Egypt isolate-2-2019. EHV-4 whole genome sequencing is recommended during outbreaks of abortion due to the probability of modification of the pathogenesis and behavior of the virus as described by Khattab et al. (2022). There was high heterogeneity and genetic diversity between alphaherpesviruses (EHV-1 and EHV-4) of the present study and gammaherpesviruses (EHV-2 and EHV-5) (Fortier et al., 2010). It is worth mentioning that there are few reports on EHV-2 and EHV-5 in Egypt. Table 6. The nucleotide diversity percentage between Genbank EHV 4, 2, 5 reference strains and local strain EHV-4 strain MG-4-2022 of the current study.
The outcomes of the phylogenetic analysis of the obtained EHVs sequences in the present work shed the light on the similarity with the local strains and other strains from different countries which may be attributed to the continuous movement of horses through borders to participate in horse events as global races or showcases that give the opportunity for the incidence of infections by foreign EHVs with recommendations for checking up all imported horses as well as horses participated in races upon return against EHVs. ConclusionThe current study confirmed the continuous circulation of EHV-1 (equine virus abortion) and EHV-4 (equine viral rhino pneumonitis) among Arabian horse populations in north Egypt leading to feverish respiratory disorders and abortions in these localities where intense horse populations with severe economic drawbacks. The application of consensus and conventional PCR assays are considered to be suitable diagnostic tools for use in veterinary laboratories to improve the detection and characterization of EHV-1 and EHV-4 prevalent in Egypt. The sequence information of the glycoprotein (gB) gene from field isolates of EHV-1 and EHV-4 is important for future effective vaccine development, while phylogenetic analysis is important for understanding the molecular epidemiology of EHV-1 and EHV-4. Reliable preventive measures must be conducted including official screening, strict quarantine measures, improved biosecurity and management considerations in combination with effective vaccine trials to reduce the dissemination and outbreak of these viruses through other governorates. Authors’ contributionAll authors contributed in this work. Maged Nafea wrote the manuscript under supervision of Ahmed A.H. Ali and Fatma Abduallah who analyzed the experimental data with discussion and writing the manuscript. Omayma A. Shemies has contributed to this work by designing and carrying out the experiments at AHRI. Gamilat K. Farag helped with sample preparation and data analysis. All authors reviewed and approved the final manuscript. Conflict of interestThe authors declare that there are no conflicts of interest. FundingNot external funding for this study. Data availabilityAll data are provided in the manuscript. ReferencesAli, A.A., Refat, N.A., algabri, N.A. and Sobh, M.S. 2020. Fetal lesions of EHV-1 in equine. Anais da Acade. Brasil. de Cien. 92, e20180837. Allen, G.P., Kydd, J.H., Slater, J.D. and Smith, K.C. 2004. Equid herpesvirus 1 and equid herpesvirus 4 infections. In Infectious diseases of livestock. Eds., Coetzer, J.A.W. and Tustin, R.C. Cape Town, South Africa: Oxford Press, pp: 829–859. Al-Shammari, Z., Ahmed, B., Haroun, M., Afify, A., Elsanousi, A.A. and Shalaby, M. 2016. A first molecular phylogeny of an Egyptian equine herpesvirus-4 strain derived from a fetal Arabian horse. J. of Vet. Sci. Med. Diag. 5(1), 2–4. Altschul, S.F., Gish, W., Miller, W., Myers, E.W. and Lipman, D.J. 1990. Basic local alignment search tool. J. Mol Biol. 215(3), 403–410. Amer, H.M., Shaltout, A.K., EL-Sabag, I.M., El-Sanousi, A.A. and Shalaby, M.A. 2011. Prevalence of equine herpes viruses 1, 2 and 4 in Arabian horse population in Egypt. Afri. J. Microbiol. Res. 5, 4805–4811. Azab, W., Bedair, S., Abdelgawad, A., Eschke, K., Farag, G.K., Abdel-raheim, A. and Ahmed A.H. 2019. Detection of equid herpesviruses among different Arabian horse populations in Egypt. Vet. Med. Sci. 5(3), 361–371. Balasuriya, U.B., Crossley B.M. and Timoney, P.J. 2015. A review of traditional and contemporary assays for direct and indirect detection of equid herpesvirus 1 in clinical samples. J. Vet. Diag. Invest. 27(6), 673–687. Connolly, S.A., Jackson, J.O., Jardetzky, T.S. and Longnecker, R. 2011. Fusing structure and function: a structural view of the herpes virus entry machinery. Nat. Rev. Microbiol. 9, 369–381. Davison, A.J., Eberle, R., Ehlers, B., Hayward, G.S., Mcgeoch, D.J., Minson, A.C. and Thiry, E. 2009. The order herpesvirales. Archives Virol. 154, 171–177. Dunowska, M. 2014. A review of equid herpesvirus 1 for the veterinary practitioner. Part A: Clinical presentation, diagnosis and treatment. New Zealand Vet. J. 62, 37–41. Elia, G., Decaro, N., Martella, V., Campolo, M., Desario, C., Lorusso, E., Cirone, F. and Buonavoglia, C. 2006. Detection of equine herpesvirus type 1 by real time PCR. J. Virol. Methods 133, 70–75. Fortier, G., Van Erck, E., Pronost, S., Lekeux, P. and Thiry, E. 2010. Equine gammaherpesviruses: pathogenesis, epidemiology and diagnosis. Vet. J. 186(2), 148–156. Gilkerson, J.R., Bailey, K.E., Diaz-méndez, A. and Hartley, C.A. 2015. Update on viral diseases of the equine respiratory tract. Vet. Clin. Equine Pract. 31(1), 91–104. Gonzalez-medina, S. and Newton, J.R. 2015. Equine herpesvirus-1: dealing practically but effectively with an ever present threat. Equine Vet. J. 47, 142–144. Goodman, L.B., Wimer, C., Dubovi, E.J., Gold, C. and Wagner, B. 2012. Immunological correlates of vaccination and infection for equine herpesvirus 1. Clini. Vaccine Immun. 19(2), 235–241. Hussey, G.S., Hussey, S.B., Wagner, B., Horohov, D.W., Van de Walle, G.R., Osterrieder, N. and Lunn, D.P. 2011. Evaluation of immune responses following infection of ponies with an EHV-1 ORF1/2 deletion mutant. Vet. Res. 42, 1–12. Hussey, G.S., Goehring, L.S., Lunn, D.P., Hussey, S.B., Huang, T., Osterrieder, N. and Slater, J. 2013. Experimental infection with equine herpesvirus type 1 (EHV-1) induces chorioretinal lesions. Vet. Res. 44, 1–15. Hussey, G.S. and Landolt, G.A. 2015. Equine alphaherpesviruses. In Robinson’s current therapy in equine medicine, 7th ed., Amsterdam, The Netherlands: Elsevier, pp: 158–161. International Committee on Taxonomy of Viruses ICTV. 2019. Virus taxonomy: 2019 release. EC 52, Online meeting, October 2019. Kydd, J.H., Slater, J., Osterrieder, N., Lunn, D.P., Antczak, D.F., Azab, W., Balasuriya, U., Barnett, C., Brosnahan, M., Cook, C., Damiani, A., Elton, D., Frampton, A., Gilkerson, J., Goehring, L., Horohov, D., Maxwell, L., Minke, J., Morley, P., Nauwynck, H., Newton, R., Perkins, G., Pusterla, N., Soboll-Hussey, G., Traub-Dargatz, J., Townsend, H., Van de Walle, G.R. and Wagner, B. 2012. Third international havemeyer workshop on equine herpesvirus type 1. Equine. Vet. J. 44(5), 513–517. Khattab, O.M., Abdelmegeedhk, M.M., Hamdyme, H.N., Hamed, A., Fahmy, H. A., Ibrahim, E. and Ahmed, E.M. 2022. Equine herpes virus 4 (EHV4) investigation in aborted Egyptian Mares; molecular detection, isolation, and phylogeny for viral glycoprotein B. Adv. Anim. Vet. Sci. 10(9), 1907–1915. Lara, M.C.S.H., Villalobos, E.M.C., Cunha, E.M.S., Oliveira, J.V., Castro, V. and Nassar, A.F.C. 2017. Occurance of viral diseases in donkeys (Equusasinus) in Sao Paulo State, Brazil. Braz. J. Vet. Res. Anim. Sci. 54(2), 154–158. Ma, G., Azab, W. and Osterrieder, N. 2013. Equine herpesviruses type 1 (EHV-1) and 4 (EHV-4). Masters of co-evolution and a constant threat to equids and beyond. Vet. Microbiol. 167(1–2), 123–134. Office International des Epizooties (OIE). 2008. Manual of diagnostic tests and vaccines for terrestrial animals (mammals, birds and bees). 6th ed. Editions of the OIE biological standards commission, Paris, France. Office International des Epizooties (OIE). 2015. Manual of diagnostic tests and vaccines for terrestrial animals. Chapter 2.5.9. Equine rhinopneumonitis. Pusterla, N., Mapes, S. and Wilson, W.D. 2007. Comparison of the diagnostic sensitivity of nasopharyngeal and Nasal Swabs and use of viral loads for the molecular diagnosis of equine herpesvirus-1 infection. American Association of Equine Practitioners, Orlando, Florida, USA, 1–5 December, 2007, pp: 220–224. Pusterla, N., Hussey, S.B., Mapes, S., Leutenegger, C.M., Madigan, J.E., Ferraro, G.L. and Lunn, D.P. 2009. Comparison of four methods to quantify Equid herpesvirus 1 load by real-time polymerase chain reaction in nasal secretions of experimentally and naturally infected horses. J. Vet. Diag. Invest. 21(6), 836–840. Radalj, A., Nišavić, J., Krnjaić, D., Valčić, M., Jovanović, T., Veljović, L. and Milić, N. 2018. Detection and molecular characterization of equine herpesviruses 1, 2, and 5 in horses in the Republic of Serbia. Acta Vet.-Brno, 87(1), 27. Salib, F., Kalad, M., Hassan, H., and Said, S. 2016. Using indirect ELISA and PCR for the diagnosis of equine herpes virus-1 (EHV-1) infection in Egypt. J. Vet. Med. Res. 23(1), 117–124. VanDevanter, D.R., Warrener, P., Bennett, L., Schultz, E.R., Coulter, S., Garber, R L. and Rose, T.M. 1996. Detection and analysis of diverse herpesviral species by consensus primer PCR. J. Clin. Microbiol. 34(7), 1666. Van Maanen, C., De Boer-Luijtze, E. and Terpstra, C. 2000. Development and validation of a monoclonal antibody blocking ELISA for the detection of antibodies against both equine herpesvirus type 1 (EHV1) and equine herpesvirus type 4 (EHV4). Vet. Microbiol. 71(1–2), 37–51. Wagner, B., Perkins, G., Babasyan, S., Freer, H., Keggan, A., Goodman, L.B. and Björnsdóttir, S. 2017. Neonatal immunization with a single IL-4/antigen dose induces increased antibody responses after challenge infection with equine herpesvirus type 1 (EHV-1) at weanling age. PLoS One 12(1), e0169072. Warda, S.M. 2003. Studies on equine abortion problem in Egypt. Zagazig University, Benha Branch, Egypt. | ||
| How to Cite this Article |
| Pubmed Style Ali AA, Abdallah F, Shemies OA, Kotb GA, Nafea MR. Molecular characterization of equine herpes viruses type 1 and 4 among Arabian horse populations in Egypt during the period between 2021-2022. Open Vet J. 2024; 14((1) (Zagazig Veterinary Conference)): 534-544. doi:10.5455/OVJ.2024.v14.i1.48 Web Style Ali AA, Abdallah F, Shemies OA, Kotb GA, Nafea MR. Molecular characterization of equine herpes viruses type 1 and 4 among Arabian horse populations in Egypt during the period between 2021-2022. https://www.openveterinaryjournal.com/?mno=182421 [Access: May 08, 2024]. doi:10.5455/OVJ.2024.v14.i1.48 AMA (American Medical Association) Style Ali AA, Abdallah F, Shemies OA, Kotb GA, Nafea MR. Molecular characterization of equine herpes viruses type 1 and 4 among Arabian horse populations in Egypt during the period between 2021-2022. Open Vet J. 2024; 14((1) (Zagazig Veterinary Conference)): 534-544. doi:10.5455/OVJ.2024.v14.i1.48 Vancouver/ICMJE Style Ali AA, Abdallah F, Shemies OA, Kotb GA, Nafea MR. Molecular characterization of equine herpes viruses type 1 and 4 among Arabian horse populations in Egypt during the period between 2021-2022. Open Vet J. (2024), [cited May 08, 2024]; 14((1) (Zagazig Veterinary Conference)): 534-544. doi:10.5455/OVJ.2024.v14.i1.48 Harvard Style Ali, A. A., Abdallah, . F., Shemies, . O. A., Kotb, . G. A. & Nafea, . M. R. (2024) Molecular characterization of equine herpes viruses type 1 and 4 among Arabian horse populations in Egypt during the period between 2021-2022. Open Vet J, 14 ((1) (Zagazig Veterinary Conference)), 534-544. doi:10.5455/OVJ.2024.v14.i1.48 Turabian Style Ali, Ahmed A.H., Fatma Abdallah, Omayma A. Shemies, Gamilat A.H. Kotb, and Maged R. Nafea. 2024. Molecular characterization of equine herpes viruses type 1 and 4 among Arabian horse populations in Egypt during the period between 2021-2022. Open Veterinary Journal, 14 ((1) (Zagazig Veterinary Conference)), 534-544. doi:10.5455/OVJ.2024.v14.i1.48 Chicago Style Ali, Ahmed A.H., Fatma Abdallah, Omayma A. Shemies, Gamilat A.H. Kotb, and Maged R. Nafea. "Molecular characterization of equine herpes viruses type 1 and 4 among Arabian horse populations in Egypt during the period between 2021-2022." Open Veterinary Journal 14 (2024), 534-544. doi:10.5455/OVJ.2024.v14.i1.48 MLA (The Modern Language Association) Style Ali, Ahmed A.H., Fatma Abdallah, Omayma A. Shemies, Gamilat A.H. Kotb, and Maged R. Nafea. "Molecular characterization of equine herpes viruses type 1 and 4 among Arabian horse populations in Egypt during the period between 2021-2022." Open Veterinary Journal 14.(1) (Zagazig Veterinary Conference) (2024), 534-544. Print. doi:10.5455/OVJ.2024.v14.i1.48 APA (American Psychological Association) Style Ali, A. A., Abdallah, . F., Shemies, . O. A., Kotb, . G. A. & Nafea, . M. R. (2024) Molecular characterization of equine herpes viruses type 1 and 4 among Arabian horse populations in Egypt during the period between 2021-2022. Open Veterinary Journal, 14 ((1) (Zagazig Veterinary Conference)), 534-544. doi:10.5455/OVJ.2024.v14.i1.48 |