| Original Article | ||
Open Vet. J.. 2023; 13(2): 171-178 Open Veterinary Journal, (2023), Vol. 13(2): 171–178 Original Research TaqMan probe-based qPCR method for specific detection and quantification of fowl adenovirus 8b challenge from chickens inoculated with live attenuated or inactivated virusChidozie C. Ugwu1,2, Mohd Hair-Bejo1,3*, Mat I. Nurulfiza3,4, Abdul R. Omar1,3 and Ideris Aini1,31Faculty of Veterinary Medicine, Universiti Putra Malaysia, Serdang, Malaysia 2Department of Animal Science and Technology, Federal University of Technology, Owerri, Nigeria 3Institute of Bioscience, Universiti Putra Malaysia, Serdang, Malaysia 4Faculty of Biotechnology and Biomolecular Sciences, Universiti Putra Malaysia, Serdang, Malaysia *Corresponding Author: Mohd Hair-Bejo. Faculty of Veterinary Medicine, Universiti Putra Malaysia, Serdang, Malaysia. Email: mdhair [at] upm.edu.my Submitted: 13/09/2022 Accepted: 11/01/2023 Published: 09/02/2023 © 2023 Open Veterinary Journal
AbstractBackground: Fowl adenovirus (FAdV) 8b and other serotypes cause inclusion body hepatitis (IBH) in chickens. Specific detection of aetiologic serotype in mixed infection and vaccine failure could be difficult. Aim: The objective of this study was to develop a TaqMan probe-based qPCR method for the detection and quantification of the FAdV 8b challenge virus. Methods: Forty-eight broiler chickens inoculated with live attenuated or inactivated FAdV 8b strains at day 1 of age either with or without booster at day 14 post-inoculation were used. The chickens were challenged with a pathogenic strain of FAdV 8b at day 28 of age. Liver and cloacal swabs were collected on days 7 and 14 post-challenge. Primers and probes were designed, specificity confirmed, and used to carry out qPCR amplification. Results: The assay amplified the FAdV DNA challenge virus, but not that of the live attenuated virus. It could detect FAdV 8b DNA as low as 0.001 ng/µl in liver and cloacal swab samples. Copy numbers obtained indicate virus load and shedding. Conclusions: It shows that a selective detection of FAdV 8b within serotype is possible. It can be useful for rapid detection and diagnosis of the disease, virus quantification and differentiation within species, determination of vaccination failure, and efficacy especially the virus load in the target organ and shedding. Keywords: Fowl adenovirus 8b, Real-time PCR, TaqMan probe, Virus detection, Virus quantification. IntroductionFowl adenoviruses (FAdVs) which belong to the family Adenoviridae and genus Aviadenovirus have been reported to be ubiquitous in avian species including chickens. The five species of FAdV (FAdV-A–FAdV-E) have been classified into 12 serotypes (1–7, 8a, 8b, 9–11) (Hess, 2000; Günes et al., 2012; Gupta et al., 2017). FAdV serotype 8b is the aetiologic agent of inclusion body hepatitis (IBH) reported worldwide such as USA (Mendelson et al., 1995), India (Mittal et al., 2014), Canada (Ojkic et al., 2008), Hungary (Kajan et al., 2013), Korea (Choi et al., 2012), Japan (Mase et al., 2009), China (Li et al., 2010), and Malaysia (Hair-Bejo, 2005; Norina et al., 2016). But serotypes 2, 7, 8a, and 11 are also known to cause the disease (Gomis et al., 2006; Philippe et al., 2007; Ojkic et al., 2008; Steer et al., 2011; Choi et al., 2012). IBH is an emerging disease of chickens associated with increasing flock mortality rate usually ranging from 10% to 30% but could be as low as 2% (Choi et al., 2012; Dar et al., 2012). The disease is characterized by sudden onset of mortality which peaks at 3–4 days of infection and ends on the fifth day but can continue up to 2–3 weeks. Sick chickens appear with ruffled feathers and die within 48 hours or may recover (Calnek et al., 1991). It affects broiler chickens usually at 3–7 weeks of age and affects other avian species like turkeys, geese, pheasants, and quails (Cowen, 1992). The disease has also been reported within a week after hatching in broilers (Asthana et al., 2013) as well as in pullets (Choi et al., 2012). Transmission is vertical through embryonated eggs or horizontal through direct contact with excreta of infected chicken or fomites (Pilkington et al., 1997). Horizontal transmission through the oral-fecal route is an attribute of this disease which makes virus shedding through the cloaca an important factor. Due to the sudden mortality associated with this disease, it is pertinent to develop rapid detection and diagnostic methods for use during outbreaks to aid prompt intervention, effective control, and possible eradication. Isolation of FAdV either through the chicken embryonated egg (CEE) or cell culture followed by electron microscopy analysis had been reported (Hair-Bejo, 2005) but they are less sensitive, laborious, and time-consuming requiring in some cases weeks for obtaining the results (McFerran and Smyth, 2000). More recently, conventional polymerase chain reaction (cPCR) especially targeting the hexon gene (Meulemans et al., 2001; Schachner et al., 2016; Mohamed Sohaimi et al., 2019; Ugwu et al., 2020) and also fiber gene (Pallister et al., 1996; Mohamed Sohaimi et al., 2019; Ugwu et al., 2020) has been utilized as the gold standard for definitive diagnosis of FAdV infection. Although this method is sensitive and reliable, it may require further analysis like restriction enzyme digestion for genotyping of these strains in cases of mixed infection (Raue and Hess, 1998) or nucleotide sequencing (Meulemans et al., 2001; Mase et al., 2009; Romanova et al., 2009; Ugwu et al., 2020). This is more pronounced in a multi-species infection and could even be more delicate in an outbreak involving vaccinated chickens where the vaccine and the pathogen are the same pathotype. Furthermore, qPCR even with its sensitivity is not able to quantify viruses in tissues or other samples which could be necessary for estimating the effectiveness of control measures like vaccination. To achieve this, PCR and titration are usually required but this combination despite being expensive, too laborious, and time-consuming may also require different levels of expertise and may not yield accurate results. Real-time PCR (RT-PCR) is widely used for the detection and quantification of diverse pathogens with high sensitivity, specificity, and accuracy and has been reported to be 10 times more sensitive than cPCR (Günes et al., 2012). An SYBR green-based RT-PCR assay for universal detection of all five species of FAdV has been described (Günes et al., 2012) while a TaqMan-based RT-PCR assay for specific detection of FAdV-4 was also developed (Wang et al., 2017). To our knowledge, TaqMan probe-based RT-PCR assay for the specific detection of FAdV 8b. This study was designed to develop a TaqMan-based RT-PCR assay for the specific detection and quantification of pathogenic FAdV 8b from chickens inoculated with live attenuated and/or inactivated FAdV 8b. Materials and MethodsVirusesThe UPM11142CELP5EP2 FAdV serotype 8b isolate which was earlier passaged 5× in chicken embryo liver (CEL) cells and 2× in CEE with a titer of 108 EID50/ml was used as a challenge virus in this study. The chickens in this experiment were inoculated earlier with live attenuated FAdV serotype 8b isolate UPM08136CELP20B1 (GenBank accession no MT561445) and/or inactivated FAdV serotype 8b isolate UPM08136CELP5B1 (GenBank accession no MT561443). Experimental designForty-eight, 28-day-old commercial broiler chicks were randomly divided into 3 main groups: A (1 and 2), B (1 and 2), and C (1 and 2) containing 16 chickens, respectively. Group A chicks were inoculated with live attenuated UPM08136CELP20B1 FAdV serotype 8b isolate (107.5 TCID50/ml at day 1 of age and further divided into sub-group A1 (without booster) A2 (with a booster at 14 dpi). Group B was inoculated with UPM08136CELP5B1 FAdV serotype 8b isolate (107.5 TCID50/ml) inactivated by binary ethylene imine method and coupled with Montanide 71VG adjuvant at day 1 and further divided into sub-group B1 (without booster) and B2 (with a booster at 14 dpi). Group C was the uninoculated control which was divided into sub-groups C1 (challenged) and C2 (unchallenged). At day 28 of age, the chickens in groups A1, A2, B1, B2, and C1 were challenged by inoculating them with 0.5 ml of UPM11142CELP5EP2 FAdV serotype 8b (108 EID50/ml) intramuscularly. Chickens in each group were kept in the same cage illuminated throughout the trial and temperature was maintained at about 24°C. All chicks were given feed and water ad libitum and monitored at least twice daily. Sample collectionEach chicken to be sampled was sacrificed by cervical dislocation. Liver samples and cloacal swabs were collected from four chickens in each group at days 7 and 14 post-challenged (dpc). Liver samples were stored in sterile sample containers at −20°C until use while swab samples were suspended in 2 ml of DMEM media (without supplements) and stored at −20°C for further processing. Sample preparation and DNA extractionPooled liver samples from each group and cloacal swab samples from each group at 7 dpc and 14 dpc were recovered from −20°C, thawed, and mixed thoroughly by vortexing. The samples were then centrifuged at 1,500 rpm for 5 minutes and 200 µl supernatant was used for DNA extraction. Total DNA was extracted using the innuPREP viral DNA kit (analytikjena, Germany) following the manufacturer’s recommendations. Primer and probe designPrimers (FAdV-Hex_143F/FAdV-Hex_143R) for FAdV 8b qPCR used in this study were designed by Matrioux (M) Sdn Bhd based on partial sequence of the hexon gene of UPM1137E15 after alignment with the sequence of UPM08136CELP20B1 and UPM11142CELP5EP2 FAdV 8b strains. The primers and probe (Table 1) were produced by Macrogen, Singapore, and had 55% and 59% GC content, and 25 and 28 nmol yields, respectively. The primer forward, reverse and probe had 6,080, 6,142, and 7,787.3 calculated MW, respectively. They were designed to detect only the UPM11142CELP5EP2 virus. Table 1. Primer and probe sequences used for quantitative PCR.
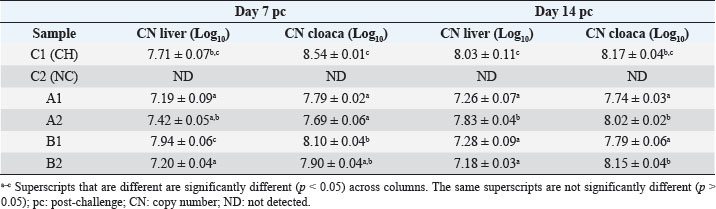
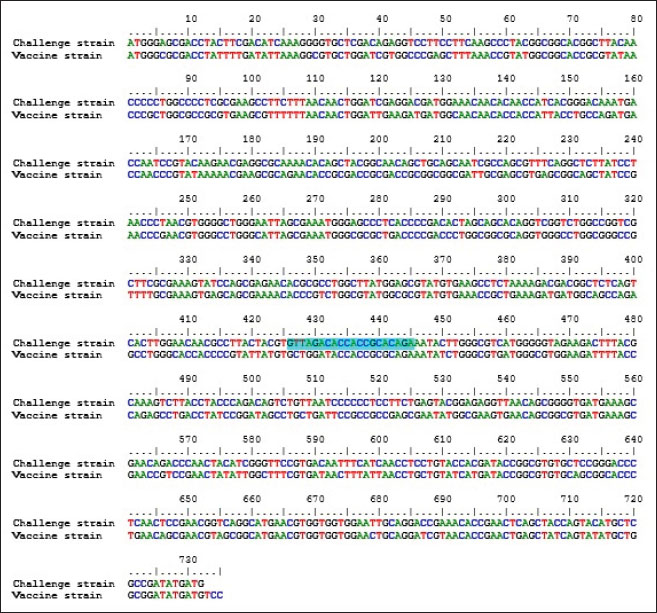
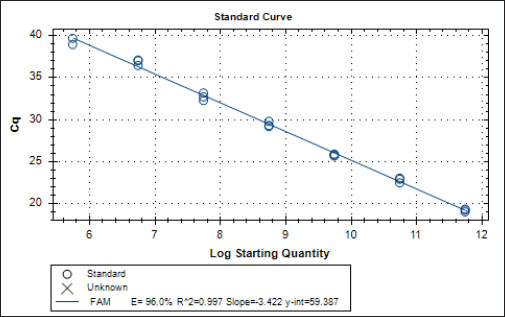
Specificity test for the qPCR primersConventional PCR was performed using FAdV-Hex_143F/R primers (Table 1) for amplification of 166 bp of the FAdV hexon gene. The DNA extracted from the UPM08136P20B1 live attenuated virus and the UPM11142CELP3EP2 challenge virus were used. The amplification conditions were 95°C for 2 minutes 1×, 95°C for 1 minute 35×, 50°C for 1 minute 35×, 72°C for 1 minute, 2 cycles 35× and 72°C for 1 minute 1×. The bands were separated in a 2% agarose gel electrophoresis using a 100 bp DNA ladder at 80 volts for 35 minutes. Standard curveFAdV positive control was used to generate the standard curve. An initial DNA concentration of 100 ng/µl was used and was diluted seven-fold from 100 to 0.0001 ng/µl. Optimization reactions were carried out to determine the best concentration for the primers and probe. The DNA copy number of the positive control was calculated using the formula below (Qiagen, 2014; Fajardo et al., 2017; Prediger, 2017). These dilutions were each amplified in triplicate in a total volume of 20 µl PCR reaction mix containing 10 µl of SensiFAST™ Probe No-ROX Kit (Bioline, London, UK), 0.8 µl of primer pair, 0.2 µl probe, 4.2 µl of nuclease-free water and 4 µl of the template. The non-template control was added in triplicate with nuclease-free water as a template. The qPCR amplification was carried out in CFX96™ Real-Time PCR Detection System (Bio-Rad, USA). The qPCR conditions were initial denaturation at 95°C for 2 minutes, 40 cycles of denaturation, and extension at 95°C for 5 seconds and 60°C for 20 seconds, respectively. qPCR amplification of extracted DNA of the liver and cloacal swab from chickensThe extracted DNA from the liver and cloacal swabs of chickens of each study group were checked for purity and concentration determined by using a spectrophotometer (UV-1601, PC, Shimadzu, Japan). Each pure DNA was used as a template for viral load determination by qPCR amplification with primer and probe as stated in Table 2. The qPCR reaction was carried out in triplicates in a total volume of 20 µl PCR reaction mix containing 10 µl of SensiFAST™ Probe No-ROX Kit (Bioline, London, UK), 0.8 µl of primer pair, 0.2 µl probe, 4.2 µl of nuclease-free water and 1 µl of the template. The non-template control was added in triplicate with nuclease-free water as a template. The qPCR amplification was carried out in CFX96™ Real-Time PCR Detection System (Bio-Rad, USA). The qPCR conditions were as described in the standard curve above. The Cq of each replicate was obtained and used to determine the copy number using a formula, and the average copy number of each sample recorded (Qiagen, 2014). Ethical approvalThis experiment was approved by the Universiti Putra Malaysia Institutional Animal Care and Use Committee and was conducted following international ethical standards. ResultsPrimers, probe, and specificityThe primers showed specificity to the UPM11142CELP5EP2 strain with a band corresponding to 166 bp and a negative band for the attenuated strain (Fig. 1). The primers and probe made use of mismatch positions in the conserved Li loop region of the hexon gene of FAdV 8b to differentiate between the two isolates (Fig. 2). Standard curveWith the aid of known FAdV 8b positive control, a standard curve amplification plot (Fig. 3) and standard curve (Fig. 4) were generated with an efficiency of 96.0%, Regression squared value of 0.997, slope value of 3.422, and y-intercept of 59.387. The plot shows the capacity of this assay to detect FAdV 8b from samples with DNA concentrations as low as 0.001 ng/µl. Detection and quantificationThere was positive FAdV 8b strain UPM11142CELP5EP2 in all the liver and cloacal samples of chickens that were inoculated and no detection among the C2 chickens that were uninoculated (Table 2). This indicates that the challenge virus replicated in the liver of the chickens and was shed in their feces. In the liver, at 7 dpi, the C1 chickens had significantly higher (p < 0.05) FAdV 8b copy number than chickens in A1 and B2 groups. At 14 dpi, the copy number of FAdV 8b in the C1 group was significantly higher (p < 0.05) than that of chickens in the A1, B1, and B2 groups and was higher (p > 0.05) that that of chickens in the A2 group. In the cloaca, at 7 dpi, the FAdV copy number of C1 chickens was significantly higher than that of chickens in the A1, A2, B1, and B2 groups, and at 14 dpi, the C1 group recorded significantly higher (p < 0.05) FAdV 8b copy number than that of A1 and B1, and higher (p > 0.05) than that of chickens in the A2 and B2 groups. Table 2. Copy number of FAdV UPM11142CELP5EP2 in the liver and cloaca of chickens at days 7 and 14 post-challenge.
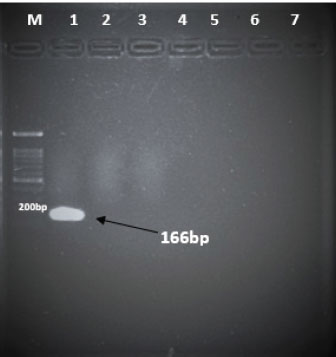
Fig. 1. Electrophoresis gel image of the hexon gene of FAdV isolates using the hexon-based qPCR primer pair qHexF/qHexR. Lane 1, UPM11142CELP3EP2; Lane 2, UPM08136P20B1 and Lane 3, Non-template. DiscussionThe TaqMan probe method described in this report was used successfully to detect and quantify FAdV 8b challenge virus in the liver and cloaca of chickens. Its specificity was shown by the inability of the primers to amplify the attenuated FAdV 8b UPM08136P20B1 isolate. It detected and quantified the pathogenic FAdV 8b UPM11142CELP5CEEP2 in the liver and cloaca indicating infection and shedding. It could detect FAdV 8b DNA as low as 0.001 ng/µl in the liver and cloacal swab samples which shows high sensitivity. Although various methods of diagnosis like virus isolation in CEE and cell culture, histological investigations to detect inclusion bodies, electron microscopy (Hair-Bejo, 2005; Adair and Fitzgerald, 2008), and immunofluorescence (Ugwu et al., 2020) could also be used to detect FAdVs in the tissues of suspected samples, they are time-consuming, tedious and may not be species-specific or give differential within serotype detection which is required for definitive diagnosis in cases of vaccine failure. Even cPCR which is usually relied on by researchers and clinicians for confirmatory diagnosis and detection (Raue and Hess, 1998; Meulemans et al., 2001; Mase et al., 2009; Kajan et al., 2011) is 10× less sensitive (Wang et al., 2017). Furthermore, cPCR may not be capable of viral load quantification (Watanabe et al., 2005) and may require titration for relative quantification, making qPCR assay the protocol of choice for the detection, diagnosis, and quantification of FAdV viruses. The SYBR Green-based real-time PCR method developed by Günes et al. (2012) could detect FAdVs in tissues and other samples but will still necessitate sequencing for serotype-specific identification. Although SYBR green method when used with proper materials, protocols, and better optimization could be comparable to the TaqMan probe method, the latter has better specificity because it only measures sequence-specific amplification offering advantages in comparison to the other methods on the issues of specificity (Tajadini et al., 2014). TaqMan probe is made up of DNA oligonucleotides that are dually labeled with fluorophores (quencher and reporter) (Mullis and Faloona, 1987; Mullis, 1990). The synergistic activity of the quencher which absorbs the signal during DNA amplification and the reporter ensures only a sequence-specific amplification (Tajadini et al., 2014). The inability of the primers to amplify the attenuated virus in the conventional PCR is an indication of the specificity of the primers to the challenge virus despite their being of the same species and serotype. This could be a result of nucleotide mismatches in the hexon gene of the two strains, as seen in the sequence alignment of the challenge and vaccine strains where the forward primer sequence was found in the challenge virus and not in the vaccine virus. A similar mismatch was explored in designing the primer used in the development of the Taqman assay for the specific detection of FAdV 4 (Wang et al., 2017). The results show a significantly higher FAdV 8b copy number in the liver of uninoculated control-challenged chickens than that of chickens inoculated with the attenuated FAdV, in the cloaca of all the inoculated chickens at 7 dpi; and in the liver of all the inoculated chickens at 14 dpi which may also be an indication that the qPCR method described herein could be efficient in within serotype detection of FAdV 8b.
Fig. 2. Sequence alignment of challenge and vaccine strain showing the forward primer in position 426–445 of the challenge virus and not in the vaccine virus. The reaction efficiency in this study was 96% with a regression squared value of 0.997 which is akin to 98% reaction efficiency and regression squared value of 0.999 obtained by Günes et al. (2012) in their SYBR Green-based assay and was higher than the 94.9% efficiency reported by Wang et al. (2017) in their TaqMan probe-based assay for the specific detection of FAdV 4. In this study, the assay was able to detect FAdV 8b inoculum distinct from live attenuated FAdV 8b strain. Similar specificity was reported by Wang et al. (2017) where their assay only detected FAdV 4 (KR5 strain) and could not detect FAdV 10 (C2B strain) which belong to the same species and have nearly identical hexon gene sequences. To our knowledge, this is the first report of a TaqMan probe-based qPCR assay with selective within-serotype FAdV serotype 8b detection. This RT-PCR assay could be useful in the investigative epidemiology of FAdV 8b outbreaks involving vaccinated chickens where the pathogenic virus is of the same or very closely related strain with the vaccine virus and could be modeled for use in other disease outbreaks as well. The Cq values obtained from each sample were within the range of the standard curve, and the amount of FAdV 8b in each sample was determined as the copy number of the FAdV 8b strain.
Fig. 3. Amplification plot of standard curve of probe-based qPCR of viral genome copy number in the liver and cloacal swab of chickens infected with FAdV 8b challenge virus. Seven-fold dilution (100–0.0001 ng).
Fig. 4. Standard curve of probe-based qPCR of viral genome copy number in the liver and cloacal swab of chickens infected with FAdV 8b challenge virus. Seven-fold dilution (100–0.0001 ng). The efficiency was 0.96 (96%), and the regression squared value was 0.997. ConclusionThis study reports the development of a probe-based qPCR assay designed to selectively detect the FAdV serotype 8b challenge virus from the liver and cloacal samples obtained from chickens previously inoculated with live attenuated and/or inactivated FAdV 8b and challenged with pathogenic FAdV 8b. The assay successfully detected and quantified the pathogenic strain and not the attenuated strain and could have higher sensitivity than conventional PCR. This assay has proven that selective within-species detection of the virus is possible and could be useful in the future for studying vaccine efficacy against its pathotype and in cases of vaccine failure. It will also be useful in establishing the efficacy of vaccines in reducing viral load in tissues and viral shedding. Author contributionsM.H.B. and A.R.O. conceived and designed the work; M.H.B. secured the funding; C.C.U. executed the experiments; C.C.U., M.I.N., and A.R.O. analyzed the data; C.C.U., M.I.N., A.R.O., and M.H.B. interpreted the data; C.C.U., M.I.N., A.R.O., M.H.B., and A.I. reviewed the manuscript. All authors read the article and agreed to publish the article. Conflicts of interestThe authors declare that there is no conflict of interest. ReferencesAdair, B.M. and Fitzgerald, S.D. 2008. Group 1 adenovirus infections. In Diseases of poultry, 12th ed. Eds., Saif, Y.M., Barners, H.J., Glisson, J.R., Fadly, A.M., McDougald, L.R. and Swayne, D.E. Ames, IA: Iowa State University Press, pp: 260–286. Asthana, M., Chandra, R. and Kumar, R. 2013. Hydropericardium syndrome: current state and future developments. Arch. Virol. 158, 921–931. Calnek, B.W., John Barnes, H., Beard, C.W., Reid, W.M. and H.W. Yoder, Jr. 1991. In Diseases of poultry, 9th ed. Ames, Iowa: Iowa State University Press, pp: 552–563. Choi, K.S., Kye, S.J., Kim, J.Y., Jeon, W.J., Lee, E.K., Park, K.Y. and Sung, H.W. 2012. Epidemiological investigation of outbreaks of fowl adenovirus infection in commercial chickens in Korea. Poult. Sci. 91, 2502–2506. Cowen, B.S. 1992. Inclusion body hepatitis-anaemia and hydropericardium syndromes: aetiology and control. Worlds Poult. Sci. J. 48(3), 247–254. Dar A., Gomis S., Shirley I., Mutwiri G., Brownlie R., Potter A., Gerdts V. and Tikoo S.K. 2012. Pathotypic and molecular characterization of a fowl adenovirus associated with inclusion body hepatitis in Saskatchewan chickens. Avian Dis. 56, 73–81. Fajardo, T.V.M., Vanni, M.F. and Nickel, O. 2017. Absolute quantification of viruses by TaqMan real-time RT-PCR in grapevines. Ciência Rural 47(6); doi:10.1590/0103-8478cr20161063. Gomis, S., Goodhope, A.R., Ojkic, A.D. and Willson, P. 2006. Inclusion body hepatitis as a primary disease in broilers in Saskatchewan, Canada. Avian Dis. 50, 550–555. Günes, A., Marek, A., Grafl, B., Berger, E. and Hess, M. 2012. Real-time PCR assay for universal detection and quantitation of all five species of fowl adenoviruses (FAdV-A to FAdV-E). J. Virol. Methods 183(2), 147–153. Gupta, A., Ahmed, K.A., Ayalew, L.E., Popowich, S., Kurukulasuriya, S., Goonewardene, K., Gunawardana, T., Karunarathna, R., Ojkic, D., Tikoo, S.K., Willson, P. and Gomis, S. 2017. Immunogenicity and protective efficacy of virus-like particles and recombinant fiber proteins in broiler-breeder vaccination against fowl adenovirus (FAdV)-8b. Vaccine 35(20), 2716–2722. Hair-Bejo M. 2005. Inclusion body hepatitis in a flock of commercial broiler chickens. J. Vet. Malaysia 17, 23–26. Hess, M. 2000. Detection and differentiation of avian adenoviruses: a review. Avian Pathol. 29, 195–206. Kajan, G.L., Kecskemeti, S., Harrach, B. and Benko, M. 2013. Molecular typing of fowl adenoviruses, isolated in Hungary recently, reveals high diversity. Vet. Microbiol. 167, 357–363. Kajan, G.L., Sameti, S. and Benkö, M. 2011. Partial sequence of the DNA-dependent DNA polymerase gene of fowl adenoviruses: a reference panel for a general diagnostic PCR in poultry. Acta Vet. Hungarica 59, 279–285. Li, H.Y., Yin, Y.B., Guo, Y.Y., Hu, Y.G., Xu, H.W., Wang, X.H. and Wang, S.C. 2010. Isolation and PCR identification of 12 strains of inclusion body hepatitis virus from clinical broilers. Chinese Vet. Sci. 40, 722–727. Mase, M., Mitake, H., Inoue, T. and Imada, T. 2009. Identification of group I-III avian adenovirus by PCR coupled with direct sequencing of the hexon gene. J. Vet. Med. Sci. 71, 1239–1242. McFerran, J.B. and Smyth, J.A. 2000. Avian adenoviruses. Rev. Sci. Tech. 19, 589–601. Mendelson, C., Nothelfer, H.B. and Monreal, G. 1995. Identification and characterization of an avian adenovirus isolated from a ‘spiking mortality syndrome’ field outbreak in broilers on the Delmarva Peninsula, USA. Avian Pathol. 24, 693–706. Meulemans, G., Boschmans, M., Berg, T.P. and Decaesstecker, M. 2001. Polymerase chain reaction combined with restriction enzyme analysis for detection and differentiation of fowl adenoviruses. Avian Pathol. 30, 655–660. Mittal, D., Jindal, N., Tiwari, A.K. and Khokhar, R.S. 2014. Characterization of fowl adenoviruses associated with hydropericardium syndrome and inclusion body hepatitis in broiler chickens. Virus Dis. 25, 114–119. Mohamed Sohaimi, N., Bejo, M.H., Omar, A.R., Ideris, A. and Mat Isa, N. 2019. Molecular characterization of fowl adenovirus isolates of Malaysia attenuated in chicken embryo liver cells and its pathogenicity and immunogenicity in chickens. PLoS One, 14(12), e0225863; doi:10.1371/journal.pone.0225863. Mullis, K.B. and Faloona, F.A. 1987. Specific synthesis of DNA in vitro via a polymerase-catalyzed chain reaction. Methods Enzymol. 155, 335–350. Mullis, K.B. 1990. The unusual origin of the polymerase chain reaction. Sci. Am. 262(4), 56–61, 64–65. Norina, L., Norsharina, A., Nurnadiah, A.H., Redzuan, I., Ardy, A. and Nor-Ismaliza, I. 2016. Avian adenovirus isolated from broiler affected with inclusion body hepatitis. Malaysian J. Vet. Res. 7(2), 121–126. Ojkic, D., Martin, E., Swinton, J., Vaillancourt, J.P., Boulianne, M. and Gomis, S. 2008. Genotyping of Canadian isolates of fowl adenoviruses. Avian Pathol. 37, 95–100. Pallister, J., Wright, P. and Sheppard, M. 1996. A single gene encoding the fiber is responsible for variations in virulence in the fowl adenoviruses. J. Virol. 70, 5115–5122. Philippe, C., Grgic, H., Ojkic, D. and Nagy, E. 2007. Serologic monitoring of a broiler breeder flock previously affected by inclusion body hepatitis and testing of the progeny for vertical transmission of fowl adenoviruses. Can. J. Vet. Res. 71, 98–102. Pilkington, P., Brown, T., Villegas, P., McMurray, B., Page, R.K., Rowland, G.N. and Thayer, S.G. 1997. Adenovirus-induced inclusion body hepatitis in four-day-old broiler breeders. Avian Dis. 41, 472–474. Prediger, E. 2017. Calculation: converting from nanogram to copy number. Integrated DNA echnologies. Available at: https://eu.idtdna.com/pages/education/decoded/article/calculations-converting-from-nanograms-to-copy-number (Accessed 17 September 2020). Qiagen. 2014. QuantiFast SYBR Green PCR Handbook. For fast, quantitative, real-time PCR and two-step RT-PCR using SYBR Green. Carlsbad, CA: Qiagen, 44 p. (Accessed: 20/08/2020). Raue, R. and Hess, M. 1998. Hexon-based PCRs combined with restriction enzyme analysis for rapid detection and differentiation of fowl adenoviruses and egg drop syndrome virus. J. Virol. Methods, 73, 211–217. Romanova, N., Corredor, J.C. and Nagy, E. 2009. Detection and quantitation of fowl adenovirus genome by a real-time PCR assay. J. Virol. Methods 159, 58–63. Schachner, A., Marek, A., Grafl, B. and Hess, M. 2016. Detailed molecular analyses of the hexon loop-1 and fibers of fowl aviadenoviruses reveal new insights into the antigenic relationship and confirm that specific genotypes are involved in field outbreaks of inclusion body hepatitis. Vet. Microbiol. 186, 13–20. Steer, P.A., Rourke, D.O., Ghorashi, S.A. and Noormohammadi, A.H. 2011. Application of high-resolution melting curve analysis for typing of fowl adenoviruses in field cases of inclusion body hepatitis. Aust. Vet. J. 89, 184–192. Tajadini, M., Panjehpour, M. and Javanmard, S. H. 2014. Comparison of SYBR Green and TaqMan methods in quantitative real-time polymerase chain reaction analysis of four adenosine receptor subtypes. Adv. Biomed. Res. 3, 85. Ugwu, C.C., Hair-Bejo, M., Nurulfiza, M.I., Omar, A.R. and Ideris, A. 2020. Propagation and molecular characterization of fowl adenovirus serotype 8b isolates in chicken embryo liver cells adapted on Cytodex™ 1 Microcarrier using stirred tank bioreactor. Processes 8(9), 1065. Wang, J., Wang, J., Chen, P., Liu, L. and Yuan, W. 2017. Development of a TaqMan-based real-time PCR assay for rapid and specific detection of fowl aviadenovirus serotype 4. Avian Pathol. 46(3), 338–343. Watanabe, M., Kohdera, U., Kino, M., Haruta, T., Nukuzuma, S., Suga, T., Akiyoshi, K., Ito, M., Suga, S. and Komada, Y. 2005. Detection of adenovirus DNA in clinical samples by SYBR Green real-time polymerase chain reaction assay. Pediatr. Int. 47, 286–291. | ||
| How to Cite this Article |
| Pubmed Style Ugwu CC, Bejo MH, Omar AR, Isa NM, Ideris A. TaqMan probe-based qPCR method for specific detection and quantification of Fowl adenovirus 8b challenge from chickens inoculated with live attenuated or inactivated virus. Open Vet. J.. 2023; 13(2): 171-178. doi:10.5455/OVJ.2023.v13.i2.4 Web Style Ugwu CC, Bejo MH, Omar AR, Isa NM, Ideris A. TaqMan probe-based qPCR method for specific detection and quantification of Fowl adenovirus 8b challenge from chickens inoculated with live attenuated or inactivated virus. https://www.openveterinaryjournal.com/?mno=112776 [Access: January 25, 2026]. doi:10.5455/OVJ.2023.v13.i2.4 AMA (American Medical Association) Style Ugwu CC, Bejo MH, Omar AR, Isa NM, Ideris A. TaqMan probe-based qPCR method for specific detection and quantification of Fowl adenovirus 8b challenge from chickens inoculated with live attenuated or inactivated virus. Open Vet. J.. 2023; 13(2): 171-178. doi:10.5455/OVJ.2023.v13.i2.4 Vancouver/ICMJE Style Ugwu CC, Bejo MH, Omar AR, Isa NM, Ideris A. TaqMan probe-based qPCR method for specific detection and quantification of Fowl adenovirus 8b challenge from chickens inoculated with live attenuated or inactivated virus. Open Vet. J.. (2023), [cited January 25, 2026]; 13(2): 171-178. doi:10.5455/OVJ.2023.v13.i2.4 Harvard Style Ugwu, C. C., Bejo, . M. H., Omar, . A. R., Isa, . N. M. & Ideris, . A. (2023) TaqMan probe-based qPCR method for specific detection and quantification of Fowl adenovirus 8b challenge from chickens inoculated with live attenuated or inactivated virus. Open Vet. J., 13 (2), 171-178. doi:10.5455/OVJ.2023.v13.i2.4 Turabian Style Ugwu, Chidozie Clifford, Mohd Hair Bejo, Abdul Rahman Omar, Nurulfiza Mat Isa, and Aini Ideris. 2023. TaqMan probe-based qPCR method for specific detection and quantification of Fowl adenovirus 8b challenge from chickens inoculated with live attenuated or inactivated virus. Open Veterinary Journal, 13 (2), 171-178. doi:10.5455/OVJ.2023.v13.i2.4 Chicago Style Ugwu, Chidozie Clifford, Mohd Hair Bejo, Abdul Rahman Omar, Nurulfiza Mat Isa, and Aini Ideris. "TaqMan probe-based qPCR method for specific detection and quantification of Fowl adenovirus 8b challenge from chickens inoculated with live attenuated or inactivated virus." Open Veterinary Journal 13 (2023), 171-178. doi:10.5455/OVJ.2023.v13.i2.4 MLA (The Modern Language Association) Style Ugwu, Chidozie Clifford, Mohd Hair Bejo, Abdul Rahman Omar, Nurulfiza Mat Isa, and Aini Ideris. "TaqMan probe-based qPCR method for specific detection and quantification of Fowl adenovirus 8b challenge from chickens inoculated with live attenuated or inactivated virus." Open Veterinary Journal 13.2 (2023), 171-178. Print. doi:10.5455/OVJ.2023.v13.i2.4 APA (American Psychological Association) Style Ugwu, C. C., Bejo, . M. H., Omar, . A. R., Isa, . N. M. & Ideris, . A. (2023) TaqMan probe-based qPCR method for specific detection and quantification of Fowl adenovirus 8b challenge from chickens inoculated with live attenuated or inactivated virus. Open Veterinary Journal, 13 (2), 171-178. doi:10.5455/OVJ.2023.v13.i2.4 |