| Review Article | ||
Open Vet. J.. 2020; 10(3): 239-251 doi: 10.4314/ovj.v10i3.1 Open Veterinary Journal, (2020), Vol. 10(3): 239–251 Review Article http://dx.doi.org/10.4314/ovj.v10i3.1 Animal coronaviruses and coronavirus disease 2019: Lesson for One Health approachUddab Poudel1, Deepak Subedi1*, Saurav Pantha1 and Santosh Dhakal21Institute of Agriculture and Animal Science (IAAS), Paklihawa Campus, Tribhuvan University, Siddharthanagar, Nepal 2W. Harry Feinstone Department of Molecular Microbiology and Immunology, Johns Hopkins Bloomberg School of Public Health, Baltimore, Maryland, USA *Corresponding Author: Deepak Subedi. Institute of Agriculture and Animal Science (IAAS), Paklihawa Campus, Tribhuvan University, Siddharthanagar, Nepal. Email: subedideepu26 [at] gmail.com Submitted: 10/06/2020 Accepted: 06/07/2020 Published: 25/07/2020 © 2020 Open Veterinary Journal
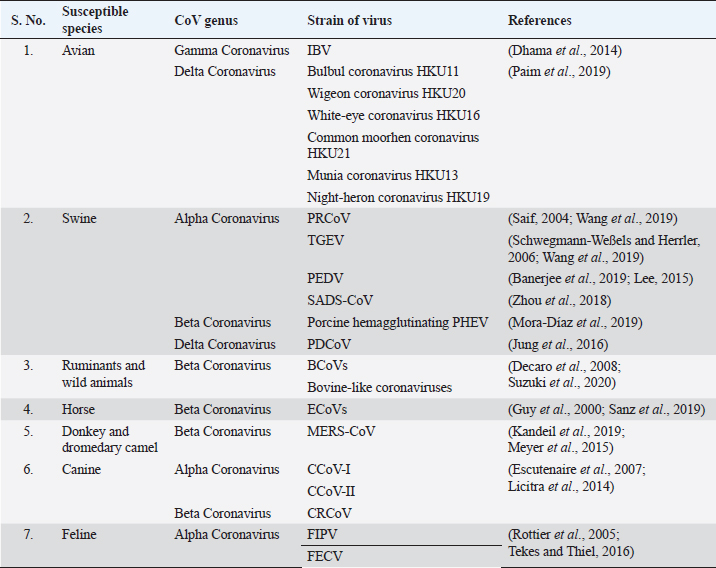
AbstractCoronaviruses are a group of enveloped, single-stranded, positive-sense RNA viruses that are broadly classified into alpha, beta, gamma, and delta coronavirus genera based on the viral genome. Coronavirus was not thought to be a significant problem in humans until the outbreak of severe acute respiratory syndrome in 2002, but infections in animals, including pigs, cats, dogs, and poultry, have been problematic for a long time. The outbreak of coronavirus disease 2019 in December 2019 in Wuhan, China, drew special attention towards this virus once again. The intermediate host of this novel coronavirus, severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), is yet to be determined, but it has a very close genomic relationship with the bat coronavirus (Bat-CoV), RaTG13 strain, and the pangolin coronaviruses. As veterinary medicine has a long-term experience dealing with coronaviruses, this could be helpful in better understanding and detecting the origin of SARS-CoV-2 and drive human medicine towards the development of vaccines and antiviral drugs through the collaborative and transdisciplinary approaches of One Health. Keywords: Animal Coronaviruses, COVID-19, One Health, SARS-CoV-2. IntroductionCoronaviruses (CoVs) (Subfamily Orthocoronavirinae, family Coronaviridae, order Nidovirales) are a group of large, enveloped, single-stranded, positive-sense RNA viruses having exceptional genetic variations (Masters, 2006). Due to mutations and various viral recombination events that have led to the continuous emergence of new viral strains with higher virulence property, the ability to spillover and expand to multiple host range exists (Buonavoglia et al., 2006; Masters, 2006). The structural morphology of virus appears to be moderately pleomorphic or roughly spherical with club-like projections formed by the spike protein and helically symmetrical nucleocapsid that encloses the single-stranded RNA when viewed under the electron microscope (Kolesnikova et al., 2003). The viral genome is linear and monopartite, consisting of 26–32 Kilobases with 51 terminal cap structures and 31 poly-A tails which act as the initial RNA for the infectious cycle, the template for viral replication and viral transcription, and also as a substrate for packaging of progeny virus (Masters, 2006; Pradesh et al., 2020). Based on the comparisons on the sequence of entire viral genomes, the International Committee for Taxonomy of Viruses (ICTV) classified CoVs into four main genera: Alphacoronavirus, Betacoronavirus, Gammacoronavirus, and Deltacoronavirus (Gorbalenya et al., 2004). During the early weeks of December 2019, several fatal cases of pneumonia with an unknown origin were reported in the city of Wuhan, Hubei Province of China, among those who had visited the wet seafood market which is the center for trade and marketing of other wild species (Lu et al., 2020). The suspected patients exhibited signs of respiratory illness, sneezing, coughing, dyspnea, severe chest pain, nausea, and diarrhea, and death was reported in comparatively older people (Gao, 2020; Lu et al., 2020). Genetic and molecular analysis revealed that the causative agent was the coronavirus, which was different from the previous human CoVs, and hence was provisionally named as novel CoV of 2019 (2019-nCov) by the World Health Organization (WHO) and the disease associated with it was called as coronavirus disease 2019 (COVID-19) (Du Toit, 2020). Later on, ICTV named the virus as severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) (Gorbalenya et al., 2020). Considering the rapid outbreak of COVID-19, on 30 January 2020, the disease was declared as a Public Health Emergency of International Concern (WHO, 2020a), which was ultimately declared as a global pandemic by the WHO on 11 March 2020 (WHO, 2020b). The rapid spread and high severity of the disease caused 11.87 million infections across 216 reported countries, where more than 545,000 people died as of 9 July 2020 (WHO, 2020c). The origin of the disease is supposed to be from the bat and the intermediate hosts are yet to be confirmed. With the potential chance of reverse spillover from humans to animals and pets, it is necessary to summon the representatives from different fields and counteract the situation with the collaborative One Health approach. This article highlights about CoVs in different animals, COVID-19 in animals and need of One Health approach for prevention and control of zoonotic infectious diseases. Previous experiences with animal CoVsThe spread of different zoonotic and vector-borne diseases to humans and the reverse spillover of pathogens, like West Nile virus and Chikungunya virus, depend on various environmental factors, like climate change, deforestation, and rapid urbanization (McMahon et al., 2018). The destruction of natural habitats of the disease reservoir animals maximizes their close association with humans which can lead to the emergence of new zoonotic diseases, like Ebola virus, Hendra virus, Nipah Virus, Hanta virus, and SARS-CoV-2 (Lorusso et al., 2020). Table 1. Coronaviruses that affect different animal species.
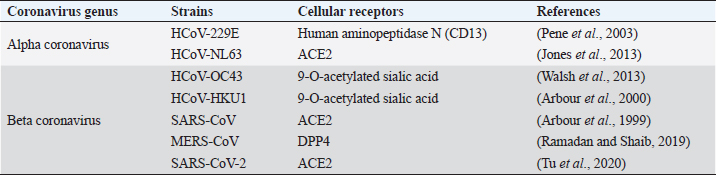
CoVs have the ability to infect different animals that may include humans, farm animals, companion animals, bats, laboratory animals, wild animals, and marine whales (Salata et al., 2020). Alphacoronaviruses and betacoronaviruses infect bats and other mammals only, whereas gammacoronaviruses and deltacoronaviruses infect birds, but some of them can also infect mammals and fishes as well (Table 1) (Cui et al., 2019). Infectious bronchitis virus (IBV), a member of gammacoronavirus, is the most common avian coronavirus that causes avian infectious bronchitis (IB) disease (Dhama et al., 2014). IB is responsible for significant morbidity and mortality in the poultry population, including pheasants, turkeys, and guinea fowls of all ages (Smith et al., 2015). Especially birds of younger age groups are highly susceptible, causing massive economic losses to the global poultry industry (Dhama et al., 2014; Smith et al., 2015). After the introduction of the virus in the birds through different routes, like feco–oral route and airborne transmission, the virus usually replicates in ciliated epithelial cells of the respiratory tract and causes severe respiratory infections, like coughing, nasal discharge, rales, gasping, swollen sinus, and also affects the digestive, reproductive, and urogenital tract, resulting in nephritis and egg drop syndrome (Weiss and Navas-Martin, 2005; Fan et al., 2018). Other CoVs from the deltacoronavirus genus include Bulbul coronavirus HKU11, Wigeon coronavirus HKU20, White-eye coronavirus HKU16, Common moorhen coronavirus HKU21, Munia coronavirus HKU13, and Night-heron coronavirus HKU19 that can severely infect birds with more complications (Paim et al., 2019). Even though there were different attempts for effective vaccine development against the CoVs, the rapid mutations and antigenic variations created a hindrance to successfully trigger the local immune response and cure infections (Saif, 2020; Zhang et al., 2020b). Viruses from alphacoronavirus, betacoronavirus, and deltacoronavirus affect the porcine population. Currently, six CoVs are circulating in the swine population. Alphacoronavirus infection in pigs includes four CoVs: porcine respiratory coronavirus (PRCoV), transmissible gastroenteritis virus (TGEV), porcine epidemic diarrhea virus (PEDV), and swine acute diarrhea syndrome coronavirus (SADS-CoV). Deltacoronavirus infection in pigs includes porcine deltacoronavirus (PDCoV), and betacoronavirus responsible for pig disease is porcine hemagglutinating encephalomyelitis virus (PHEV) (Wang et al., 2019). PRCoVs cause respiratory infections; TGEV, PEDV, SADS-CoVs, and PDCoVs cause acute gastroenteritis; and PHEV results in severe neurological and digestive disorders (Mora-Díaz et al., 2019; Wang et al., 2019). TGEV is a highly contagious, economically significant, and the oldest known swine CoV, mostly spread by feco–oral route affecting young pigs. TGEV virus targets the absorptive epithelial cells of villi causing its atrophy, leading to severe diarrhea, vomition, rapid weight loss, and death (Doyle and Hutchings, 1946; Weiss and Navas-Martin, 2005; Schwegmann-Weßels and Herrler, 2006). Currently, live attenuated and inactivated vaccines are available for the pregnant sow. PRCoV, an attenuated variant of TGEV, originated by the deletion of large 5’ region in the spike gene of the virus, is an example of the evolution of CoVs with altered virulence property and tropism (Saif, 2004). PEDV is an important enteric virus of swine which was introduced in a pig population in the early 1970s, mostly affecting growing and fattening pigs and is believed to be the consequence of spillover from bats, causing tremendous economic losses to the American, European and Asian pork industries (Lee, 2015; Banerjee et al., 2019). More than 8 million piglets died due to PEDV in 2013 in the USA, Canada, and Mexico (Lee, 2015). PEDV, mainly transmitted by the respiratory route, uses the same receptor as TGEV for entry in host cells and it is also closely related to Scotophilus bat coronavirus 512, human alphacoronaviruses HCoV-299E, and HCoV-NL63, which are likely to have common evolutionary precursors (Banerjee et al., 2019). Various vaccines are designed to cope with PEDV in swine populations (Lee, 2015; Hsueh et al., 2020). Digestive and nervous systems in pigs of all age groups are affected by PHEV, which was first isolated in 1962 and the virus was identified as a neurotropic virus, which showed subclinical presence worldwide (Mora-Díaz et al., 2019). SADS-CoV, having more than 86% and 96% sequence identity with bat alphacoronavirus HKU2-CoV and Rhinolophus spp., respectively, is another virulent swine alphacoronavirus causing enteritis in the small piglets, which indicates that SADS-CoV and HKU2-CoV descended from a common ancestor (Zhou et al., 2018). Recently, in 2012, PDCoV, which caused a mild infection to the pigs, was detected in Hong Kong in the porcine population, which is closely related with quail deltacoronavirus UAE-HKU30, and it was proposed that the virus originated from the recombination between sparrow CoV HKU15 and Bulbul CoV HKU11 by host switching activity between avian and mammalian CoVs (Jung et al., 2016). In the experimental infection, swines were found to be infected with Middle East respiratory syndrome coronavirus (MERS-CoV) and SARS-CoV, but not with SARS-CoV-2 (Wang et al., 2005; Vergara-Alert et al., 2017; Shi et al., 2020). Ruminants and wild animals are known to be infected by the number of CoVs. Bovine coronavirus (BCoV) is the oldest known ruminant CoV of betacoronavirus genus having the ability to cause severe clinical complications, including enteric diseases, like calf diarrhea in neonates and bloody diarrhea in adult cattle, and a respiratory form known as shipping fever in cattle of all age groups (Decaro et al., 2008; Suzuki et al., 2020). BCoVs are thought to be evolved from rodent CoVs and transmitted by the feco–oral route, mainly causing villous atrophy of small and large intestines leading to severe bloody diarrhea (Carman and Hazlett, 1992; Corman et al., 2018). Various bovine-like CoVs have been reported from domesticated animals, such as sheep and goats, water buffalo, llamas, alpacas, and some wild ruminants, such as sambar deer, white-tailed deer, sika deer, water deer, caribou, and elk (Jin et al., 2007; Kim et al., 2018). Similar type of viruses are also found in various species of antelopes, bisons, giraffes, and dromedary camels (Hasoksuz et al., 2007; Jin et al., 2007; Kim et al., 2018). Equine CoVs (ECoVs), first reported from the fecal samples in 1999 in North Carolina (USA), are the only CoVs that have been reported in horses, especially in foals of less than 2 weeks of age (Guy et al., 2000; Sanz et al., 2019). ECoVs are thought to be descendants of BCoVs, and are responsible for causing self-limiting enteritis in the horse population (Pusterla et al., 2018; Sanz et al., 2019). Horses are not naturally infected by MERS-CoVs; however, research in Egypt showed that 3 out of 42 donkeys were detected with MERS-CoV RNA in their respiratory specimen (Meyer et al., 2015; Kandeil et al., 2019). Canines are also infected by CoVs, two from alphacoronaviruses: canine enteric coronaviruses (CCoV-I and CCoV-II), and one from betacoronavirus genus: canine respiratory coronavirus (CRCoV) (Escutenaire et al., 2007; Licitra et al., 2014). Both CCoV-I and II affect the gastrointestinal tracts of dogs of all ages, but puppies are known to be affected more severely and simultaneous infection of both the virus may favor the recombination of the viral genome. TGEV is more analogous with CCoV and it is believed that TGEV originated from CCoV-II (Perlman and Netland, 2009). CRCoV, which causes severe respiratory infections, known as kennel cough, is more related to BCoVs (Erles et al., 2007). Feline populations can be infected with two CoVs from alphacoronavirus genus, viz. feline infectious peritonitis virus (FIPV) and feline enteric coronavirus (FECV), which mainly affect the respiratory tract, abdominal cavity, and central nervous system, leading to severe enteritis and infectious peritonitis mainly in very young and old animals with suppressed immunity for both domestic and wild felines (Tekes and Thiel, 2016). It is believed that FIPV originated from FECV due to mutation of the S gene (Rottier et al., 2005; Pradesh et al., 2020). Before the epidemic outbreak of SARS-CoV, bats were not considered as the natural host of CoVs. After that, many CoVs in bats have been identified and they are now considered as a natural reservoir of alphacoronavirus and betacoronavirus, and to date, more than 200 novel CoVs have been identified in bats (Poon et al., 2005; Chen et al., 2017). CoVs in bats have been reported from all continents, where the alphacoronavirus was found to be more widespread than betacoronavirus (Wong et al., 2019). Bats remain as an asymptomatic carrier and harbor CoVs persistently while searching for food and might shed virus in extensive areas close to humans by direct and indirect contacts (Fan et al., 2019). Also, in various countries, including China, bats are used as a source of food and in traditional Chinese medicine that increases the risk of transfer of novel zoonotic CoVs risking the human lives (Fan et al., 2019; Wong et al., 2019). Previous experiences with human CoVsBefore the outbreak of SARS in 2002 in Guangdong Province, China, CoVs were not considered as a highly pathogenic virus to the humans (Zhong et al., 2003). SARS-CoV shocked the entire world becoming the first epidemic of the 21st century with high virulence property and efficient transmissibility among human populations, infecting more than 8,000 peoples from 26 different countries and resulting in 774 deaths (WHO, 2004). The virus is mainly transferred through respiratory droplets and by the feco–oral route, and each positive person can infect two to four healthy individuals (Lee et al., 2003; Peiris et al., 2003a, 2003b). Although the incubation period is 4–7 days, viral load reaches peak on the 10th day, infecting all age groups and developing pneumonia with fever, myalgia, malaise, respiratory complications, pleurisy, and diarrhea as late symptoms (Peiris et al., 2003a; Scales et al., 2003). The outbreak of SARS not only affected human health but also heavily affected the global economy, with restrictions in travels and trade making a global loss of 40 billion dollars (To et al., 2013). After a decade of emergence of SARS-CoV, another highly pathogenic coronavirus, MERS-CoV, emerged in Saudi Arabia and other Middle Eastern countries (Zaki et al., 2012). MERS spread to 27 countries where a total of 2,494 positive cases had been reported with a case fatality of 34.4% globally, while Saudi Arabia alone had 2,102 cases with 780 deaths, 37.1% death rate, till the end of November 2019 (WHO, 2020d). The viral sequence obtained from infected humans and dromedary camels was found to be almost identical, which concludes that human infection with MERS originated from camels (Raj et al., 2014; Gautam et al., 2020). Majority of CoVs associated with humans cause many respiratory diseases, like nasal discharge, common cold, cough, pneumonia, and bronchitis and enteric and neurological problems, especially affecting the elderly, children, and immunocompromised patients (Pene et al., 2003; Walsh et al., 2013). Human CoVs (HCoVs) are recognized as one of the most rapidly evolving viruses, with the capacity to jump the species barrier and having found the new ecological niche with the novel epidemiologic phenomenon (Vijgen et al., 2005). Rapid urbanization and modern farming systems favor the emergence of novel HCoVs by frequent mixing of species, facilitating genomic recombination of the virus (Jones et al., 2013). Seven HCoVs have been identified till date which can infect humans, two belonging to the alphacoronavirus genus and five belonging to the gammacoronavirus genus (Table 2). Four HCoVs, namely HCoV-NL63, HCoV-229E, HCoV-OC43, and HCoV-HKU1, were known to infect humans before the emergence of SARS-CoV (2002), MERS-CoV (2012), and SARS-CoV-2 (2019). Including novel SARS-CoV-2, four HCoVs, HCoV-229E, HCoV-NL63, HCoV-OC43, and HCoV-HKU1, are globally circulating in human populations and are contributing to a majority of common cold infections in humans (Lorusso et al., 2020). It has been postulated that the natural hosts for HCoV-NL63 and HCoV-229E are bats and rodents for HCoV-OC43 and HKU1 CoVs. Some CoVs require intermediate hosts before infecting humans, like cattle for HCoV-OC43, alpacas for HCoV-229E, masked Palm civets for SARS-CoV, and dromedary camels for MERS-CoV (Hu et al., 2017; Cui et al., 2019). SARS-CoV and MERS-CoV have been transmitted to human by masked palm civets and dromedary camels, respectively (Zhong et al., 2003). Both of these viruses originated in bats, and this concludes the hypothesis that SARS-CoV-2 may also involve an intermediate animal host (Hu et al., 2017). Table 2. Coronaviruses in humans.
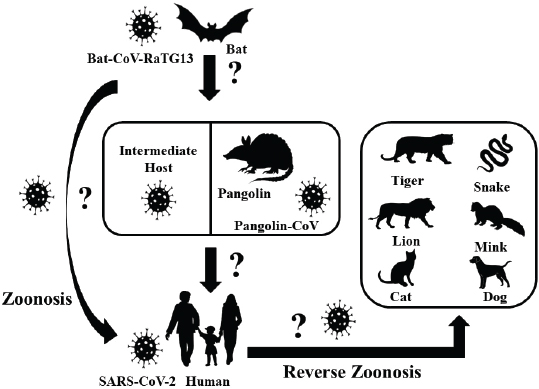
Coronavirus disease 2019 in animalsThe waves of COVID-19 from the Hubei province of Wuhan, China, have caused ripples all around the world and have created havoc with several thousand human deaths and infections across the world except Antarctica. Alongside humans, a number of animals, including dogs and cats (small and large), are also reported to be infected with the disease where transmissions might have occurred from pet owners and zookeepers (Rodriguez-Morales et al., 2020). SARS-CoV-2 has the ability to cross the species barrier, which makes it possible to infect humans, whereas COVID-19-positive humans facilitate the reverse zoonotic transmission of the virus from human to animals (Fig. 1) (Parry, 2020; Zhang et al., 2020b). Among the reported animals, an infected dog did not show any clinical signs, unlike cats. In March 2020, a Pomeranian and later a German Shepherd were found positive for COVID-19 infection in Hong Kong, whose owners were also the victims of the disease (Parry, 2020). The infected cats, wherein the transmission occurred from humans, were from Belgium, Hong Kong, and New York, that got infected in late March and April 2020 showed variations in the degrees of clinical signs, like mild respiratory illnesses (Parry, 2020). A tiger in a zoo in New York was also tested positive for the disease due to the association with a COVID-19-positive zoo employee (Parry, 2020). Positive cases of COVID-19 in domestic cats have also been reported in France, Spain, Germany, and Russia (Parry, 2020). COVID-19 has also been detected in two mink farms in The Netherlands where minks are supposed to transmit CoV to human (CDC, 2020; Yoo and Yoo, 2020). World Organization for Animal Health (OIE) has reported that, so far, there exists no relevant evidence of animal’s role in the spread of the SARS-CoV-2 in the human population (OIE – World Organization for Animal Health, 2020). One Health perspectiveThroughout history, including the current COVID-19 pandemic, human civilizations have been blighted with subtle and impervious emerging and reemerging zoonoses that cost millions of lives and a ruined global economy. The egress and growth of avian and swine influenza viruses, such as Zika virus, Ebola virus, SARS, MERS, and COVID-19, have caused panic around the world and demonstrated how difficult it is to counteract and respond to these changing dynamics of the outbreaks (Streinu-Cercel, 2014; Boeuf et al., 2016; Liu et al., 2020). The emerging diseases are shared between both animals and humans, where about 60% of the emerging pathogens have an animal origin (Jones et al., 2008). Emerging infectious diseases have always been considered as an economic threat, and to prevent the origin and spread of such diseases is of global importance (World Economic Forum, 2010; Morse et al., 2012). Scientists have been warning about the possibility of pandemics, but the world was not prepared in responding to them (Ogawa et al., 2009). Since 1940, more than two-third of the recorded 400 emerging infectious diseases have proved to be zoonotic in nature (Jones et al., 2008; Morse et al., 2012). The close interactions between the people, wildlife, livestock, and favorable environments are one of the major sources of recently emerging infectious diseases, and wildlife accounts for 71.8% of total zoonotic emerging infectious diseases (Caron et al., 2012). Even though the identification and realization of zoonotic diseases are of huge importance, the underlying causes of origin and spread of those diseases have not been given the attention they require (Murray and Daszak, 2013). Veterinarians and epidemiologists have often advocated about the holistic approach of One World One Health to shorten the knowledge gap on disease origin and transmission, but this has not been materialized yet to the extent needed. Animals and humans encounter each other on daily basis in similar ecological conditions for food, safety, companionships, and aesthetic purposes which create an opportunity for the spillover of viruses from their reservoirs (Taylor et al., 2001). It appears very important to constantly maintain the ecological and biological barrier to intervene in the zoonotic transmission of the disease, which is only possible with the ‘One Health’ concept. The recent COVID-19 pandemic is also thought to be the result of such close associations between humans and animals (Fig. 2) (Lorusso et al., 2020). In order to intervene in the transmission chain of the disease, it is extremely crucial to understand the nature of pathogen transmission and identify the range of reservoirs, hosts, and susceptible animals. During the infection, the receptor-binding domain (RBD) of the virus spike protein identifies the angiotensin-converting enzyme 2 (ACE2) of the host tissues mainly in the pneumocytes of lungs, heart, and kidneys (Tai et al., 2020). Therefore, the various species that have ACE2 receptors identical to humans can harbor the disease and get infected. After 96% resemblance of the full-length genome sequence of SARS-CoV-2, obtained from five infected patients, with bat CoV isolate RaTG13, bats were considered as the natural reservoirs (Zhou et al., 2020). It is still the matter of concern if the transmission occurs directly from the bats or there are some intermediate hosts involved (Jin et al., 2020). In the light of research on intermediate hosts, Ji et al. (2020) mentioned the probability of snakes as wildlife animal reservoir based on the relative synonymous codon usage bias. Similarly, Guo et al. (2020), through the introduction of virus host prediction using deep learning algorithm, predicted the infectivity pattern of SARS-CoV-2 and found closeness to minks. Malayan pangolins are the recent discoveries as the intermediate hosts. Pangolin CoVs are believed to be the second closest relative to SARS-CoV-2, after bat CoV RaTG13, with 91.02% identical genome and 97.1% RDB similarity (Zhang et al., 2020a).
Fig. 1. Possible origin of SARS-CoV-2, and zoonotic and reverse zoonotic transmission possibility. SARS-CoV-2=severe acute respiratory syndrome coronavirus-2; Bat-CoV=bat coronavirus; Pangolin-CoV=pangolin coronavirus. Apart from humans, different wild and domestic animals have been diagnosed with infections. The experimental inoculation of the virus on four rhesus macaques by Bao et al. (2020) showed weight loss by about 200–400 gm, reduced appetite, transient period increased respiration, and hunched posture without change in rectal temperature. In the necropsy of the same experiment, lung lesions, like interstitial pneumonia, alveolar epithelium degeneration, and inflammatory cells infiltration, were observed in HE staining along with opacity of lungs and interstitial markings observed in X-ray 7 days post-inoculation. Panthera tigris, Felis catus, and Canis lupus with ACE2 similarity of 85.70%, 85.22%, and 84.01%, respectively, also showed the infection and clinical signs (Canrong et al., 2020). Although many researches on non-human primates are yet to be carried out, it has been reported by Canrong et al. (2020) that the primates like Gorilla gorilla, Macaca nemestina, Papio Anubis, and Macaca fascicularis have ACE2 similarity of 99.01%, 95.34%, 95.34%, and 95.21%, respectively, when compared to humans. In the same experiment, they found that the free binding energy of SARS-CoV-2 to ACE2, which also defines the affinity of RBD to ACE2, in the above-listed animals is nearly −51.5 KJmol−1 which is very close to free binding energy of SARS-CoV-2 RBD in humans’ACE2 (−50.13 KJmol−1) (Canrong et al., 2020). Considering the very similar ACE2 receptor and close free binding energy of these primates when compared to humans, the possibility of the transmission of SARS-CoV-2 to those primates cannot be overlooked.
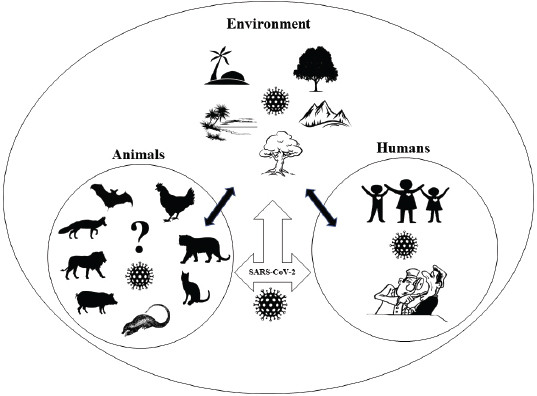
Fig. 2. One Health triad between animals, humans, and environment in the context of COVID-19. It is reported that the virus has been transmitted between several animals in nature (Lam et al., 2020). Therefore, veterinarians may be very important in identifying the disease dynamics of the CoVs because the virus is not novel for those veterinary virologists who have been working for a long time on the origin, infection, and transmission of CoVs on different animals, like cats, dogs, and pigs (Decaro and Lorusso, 2020). So far, there have been some shreds of evidence of COVID-19 transmission from humans to cats, as well where the veterinary perspective appears to be useful. One Health approach focuses on the interrelationship between human, animals, and environment for the benefits of all the associated factors (Fig. 2) (Zinsstag et al., 2011). Considering the recent global SARS-COV-2 pandemic, there are several areas where One Health approach must be taken into consideration. SARS-COV-2 is a coronavirus of bat origin, transmitted to humans either by spillover from the bats itself or from the intermediate hosts yet to be confirmed (El Zowalaty and Järhult, 2020). It seems very important to identify non-human hosts that may include wild and domestic animals in the concerned seafood market of the epicenter. Only after the identification of the possible intermediate hosts of COVID-19 by effective genetic analysis, a proper One Health approach can be applicable to break the chain of transmission from a reservoir to intermediate hosts, from intermediate hosts to humans and reverse spillover. If the intermediate hosts remain unrevealed, there is a possibility of new disease outbreaks in the near future. So, it is necessary to include representatives from all the disciplines, like epidemiologists, veterinarians, human health professionals, and environmentalists to break the transmission chain of the disease by a collaborative One Health approach (Kelly et al., 2017). Only through this approach, all the responsible factors, like humans, hosts animals, reservoirs, environmental situations, can be thought of as a single unit. By maintaining ecological barriers between the animals and humans in their respective habitats, controlling the unmanaged slaughter, and consumption of animals we can reduce the zoonotic risks. Furthermore, it is the role of the veterinarians to inspect the animals in their respective field to reduce the animal to animal transmission, and human health professionals should always be conscious about the possibility of reverse zoonosis. Only when all the concerned authorities work with a common vision and strong mentality, the world can progress as one for the development of effective vaccinations, control of current disease, and prevention of future pandemics. ConclusionIt is not a novel fact that different families of CoVs have affected different wild and domestic animals for several decades. But only after the first zoonotic incidence of SARS-CoV, at the beginning of this century, major attention was deviated toward CoVs. MERS-CoV in 2012 and the recent COVID-19 pandemic have once again proved how difficult it is to cope with the emergence of new diseases. Learning from the experiences of veterinary virologists dealing with several coronavirus infections in the past could be an important aspect for not only understanding the SARS-CoV-2 transmission dynamics but also for the development of vaccines and therapeutics. Likewise, environmental factors, like continuous deforestation and unmanaged modernization, have disturbed the ecological niche and broken the natural barriers of humans and animals creating more opportunity for pathogen spillover. Therefore, representatives from human, animal, and environmental sectors should come together to advance the One Health approach and to effectively manage the ongoing COVID-19 pandemic and any pandemics in the future. AcknowledgmentWe like to acknowledge the frontline workers in the fight against the COVID-19 pandemic and all the helping hands for completion of this article. Conflict of interestThe authors declare that there is no conflict of interest. Authors’ contributionAll authors have equal contributions. ReferencesArbour, N., Day, R., Newcombe, J. and Talbot, P.J. 2000. Neuroinvasion by human respiratory coronaviruses. J. Virol. 74, 8913–8921; doi:10.1128/jvi.74.19.8913-8921.2000. Arbour, N., Ekandé, S., Côté, G., Lachance, C., Chagnon, F., Tardieu, M., Cashman, N.R. and Talbot, P.J. 1999. Persistent infection of human oligodendrocytic and neuroglial cell lines by human coronavirus 229E. J. Virol. 73, 3326–3337; doi:10.1128/jvi.73.4.3326-3337.1999. Banerjee, A., Kulcsar, K., Misra, V., Frieman, M. and Mossman, K. 2019. Bats and coronaviruses. Viruses. 11(1), 41; doi:10.3390/v11010041. Bao , L., Deng, W., Gao, H., Xiao, C., Liu, J., Xue, J., Lv, Q., Liu, J., Yu, P., Xu, Y., Qi, F., Qu, Y., Li, F., Xiang, Z., Yu, H., Gong, S., Liu, M., Wang, G., Wang, S., Song, Z., Zhao, W., Han, Y., Zhao, L., Liu, X., Wei, Q. and Qin, C. 2020. Reinfection could not occur in SARS-CoV-2 infected rhesus macaques. bioRxiv; doi:10.1101/2020.03.13.990226. Boeuf, P., Drummer, H.E., Richards, J.S., Scoullar, M.J.L. and Beeson, J.G. 2016. The global threat of Zika virus to pregnancy: epidemiology, clinical perspectives, mechanisms, and impact. BMC Med. 14, 112; doi:10.1186/s12916-016-0660-0. Buonavoglia, C., Decaro, N., Martella, V., Elia, G., Campolo, M., Desario, C., Castagnaro, M. and Tempesta, M. 2006. Canine coronavirus highly pathogenic for dogs. Emerg. Infect. Dis. 12(3), 492–494; doi:10.3201%2Feid1203.050839.M Canrong, W., Mengzhu, Z., Yueying, Y., Mingxue, L., Yang, L., Peng, Z., Yali, W., Qiqi, W., Yang, X., Lixia, C. and Hua, L. 2020. In silico analysis of intermediate hosts and susceptible animals of SARS-CoV-2. chemRxiv; doi:10.26434/chemrxiv.12057996.v1. Carman, P.S. and Hazlett, M.J. 1992. Bovine coronavirus infection in Ontario 1990-1991. Can. Vet. J.=La Rev. Vet. Can. 33, 812–814. Caron, A., Morand, S. and de Garine-Wichatitsky, M. 2012. Epidemiological Interaction at the Wildlife/Livestock/Human Interface: Can We Anticipate Emerging Infectious Diseases in Their Hotspots? A Framework for Understanding Emerging Diseases Processes in Their Hot Spots. In: Morand, S., Beaudeau, F., Cabaret, J. (eds) New Frontiers of Molecular Epidemiology of Infectious Diseases. Springer, Dordrecht; https://doi.org/10.1007/978-94-007-2114-2_14. CDC. 2020. Coronavirus Disease 2019 (COVID-19). COVID-19 and animals. Available via https://www.cdc.gov/coronavirus/2019-ncov/daily-life-coping/animals.html (Accessed 20 April 2020). Chen, L., Liu, B., Wu, Z., Jin, Q. and Yang, J. 2017. DRodVir: a resource for exploring the virome diversity in rodents. J. Genet. Genomics. 44, 259–264; doi:10.1016/j.jgg.2017.04.004. Corman, V.M., Muth, D., Niemeyer, D. and Drosten, C. 2018. Hosts and sources of endemic human coronaviruses. Adv. Virus Res. 100, 163–188; doi:10.1016/bs.aivir.2018.01.001. Cui, J., Li, F. and Shi, Z.L. 2019. Origin and evolution of pathogenic coronaviruses. Nat. Rev. Microbiol. 17(3), 181–192; doi:10.1038%2Fs41579-018-0118-9. Decaro, N. and Lorusso, A. 2020. Novel human coronavirus (SARS-CoV-2): a lesson from animal coronaviruses. Vet. Microbiol. 244, 108693; doi:10.1016/j.vetmic.2020.108693. Decaro, N., Mari, V., Desario, C., Campolo, M., Elia, G., Martella, V., Greco, G., Cirone, F., Colaianni, M.L., Cordioli, P. and Buonavoglia, C. 2008. Severe outbreak of bovine coronavirus infection in dairy cattle during the warmer season. Vet. Microbiol. 126, 30–39; doi:10.1016/j.vetmic.2007.06.024. Dhama, K., Singh, S.D., Barathidasan, R., Desingu, P.A., Chakraborty, S., Tiwari, R. and Kumar, A.M. 2014. Emergence of avian infectious bronchitis virus and its variants need better diagnosis, prevention and control strategies: a global perspective” kuldeep. Pak. J. Biol. Sci. 17, 751–767; doi:10.3923/pjbs.2014.751.767. Doyle, L.P. and Hutchings, L.M. 1946. A transmissible gastroenteritis in pigs. J. Am. Vet. Med. Assoc. 108, 257–259. Du Toit, A. 2020. Outbreak of a novel coronavirus. Nat. Rev. Microbiol. 41, 579; doi:10.1038/s41579-020-0332-0. El Zowalaty, M.E. and Järhult, J.D. 2020. From SARS to COVID-19: a previously unknown SARS- related coronavirus (SARS-CoV-2) of pandemic potential infecting humans – call for a one health approach. One Health. 9, 100124; doi:10.1016/j.onehlt.2020.100124. Erles, K., Shiu, K.B. and Brownlie, J. 2007. Isolation and sequence analysis of canine respiratory coronavirus. Virus Res. 124, 78–87. doi:10.1016/j.virusres.2006.10.004. Escutenaire, S., Isaksson, M., Renström, L.H.M., Klingeborn, B., Buonavoglia, C., Berg, M., Belák, S. and Thorén, P. 2007. Characterization of divergent and atypical canine coronaviruses from Sweden. Arch. Virol. 152, 1507–1514; doi:10.1007/s00705-007-0986-1. Fan, W.S., Li, H.-M., He, Y.-N., Tang, N., Zhang, L.-H., Wang, H.-Y., Zhong, L., Chen, J.-C., Wei, T.-C., Huang, T., Mo, M.-L. and Wei, P. 2018. Immune protection conferred by three commonly used commercial live attenuated vaccines against the prevalent local strains of avian infectious bronchitis virus in southern China. J. Vet. Med. Sci. 80(9), 1438–1444; doi:10.1292/jvms.18-0249. Fan, Y., Zhao, K., Shi, Z.L. and Zhou, P. 2019. Bat coronaviruses in China. Viruses. 11, 210; doi:10.3390/v11030210. Gao, Z.C. 2020. Efficient management of novel coronavirus pneumonia by efficient prevention and control in scientific manner. Zhonghua Jie He He Hu Xi Za Zhi. 43(0), E001; doi:10.3760/cma.j.issn.1001-0939.2020.0001. Gautam, A., Kaphle, K., Shrestha, B. and Phuyal, S. 2020. Susceptibility to SARS, MERS, and COVID-19 from animal health perspective. Open Vet. J. 10(2), 164–177; doi:10.4314/ovj.v10i2.6. Gorbalenya, A.E., Baker, S.C., Baric, R.S., de Groot, R.J., Drosten, C., Gulyaeva, A.A., Haagmans, B.L., Lauber, C., Leontovich, A.M., Neuman, B.W., Penzar, D., Perlman, S., Poon, L.L., Samborskiy, D., Sidorov, I.A., Sola, I. and Ziebuhr, J. 2020. Severe acute respiratory syndrome-related coronavirus: the species and its viruses-a statement of the coronavirus study group. bioRxiv; https://doi.org/10.1101/2020.02.07.937862. Gorbalenya, A.E., Snijder, E.J. and Spaan, W.J.M. 2004. Severe acute respiratory syndrome coronavirus phylogeny: toward consensus. J. Virol. 78, 7863–7866; doi:10.1128/jvi.78.15.7863-7866.2004. Guo, Q., Li, M., Wang, C., Fang, Z., Wang, P., Tan, J., Wu, S., Xiao, Y. and Zhu, H. 2020. Host and infectivity prediction of Wuhan 2019 novel coronavirus using deep learning algorithm. bioRxiv; doi:10.1101/2020.01.21.914044. Guy, J.S., Breslin, J.J., Breuhaus, B., Vivrette, S. and Smith, L.G. 2000. Characterization of a coronavirus isolated from a diarrheic foal. J. Clin. Microbiol. 38, 4523–4526; doi:10.1128/jcm.38.12.4523-4526.2000. Hasoksuz, M., Alekseev, K., Vlasova, A., Zhang, X., Spiro, D., Halpin, R., Wang, S., Ghedin, E. and Saif, L.J. 2007. Biologic, antigenic, and full-length genomic characterization of a bovine-like coronavirus isolated from a giraffe. J. Virol. 81, 4981–4990; doi:10.1128/jvi.02361-06. Hsueh, F.C., Chang, Y.C., Kao, C.F., Hsu, C.W. and Chang, H.W. 2020. Intramuscular immunization with chemokine-adjuvanted inactive porcine epidemic diarrhea virus induces substantial protection in pigs. Vaccines. 8(1), 102; doi:10.3390/vaccines8010102. Hu, B., Zeng, L.P., Yang, X. L., Ge, X.Y., Zhang, W., Li, B., Xie, J.Z., Shen, X.R., Zhang, Y.Z., Wang, N., Luo, D.S., Zheng, X.S., Wang, M.N., Daszak, P., Wang, L.F., Cui, J. and Shi, Z.L. 2017. Discovery of a rich gene pool of bat SARS-related coronaviruses provides new insights into the origin of SARS coronavirus. PLoS Pathog. 13, e1006698; doi:10.1371/journal.ppat.1006698. Ji, W., Wang, W., Zhao, X., Zai, J. and Li, X. 2020. Cross-species transmission of the newly identified coronavirus 2019-nCoV. J. Med. Virol. 92, 433–440; doi:10.1002/jmv.25682. Jin, L., Cebra, C.K., Baker, R.J., Mattson, D.E., Cohen, S.A., Alvarado, D.E. and Rohrmann, G.F. 2007. Analysis of the genome sequence of an alpaca coronavirus. Virology. 365, 198–203; doi:10.1016/j.virol.2007.03.035. Jin, Y.H., Cai, L., Cheng, Z.S., Cheng, H., Deng, T., Fan, Y.P., Fang, C., Huang, D., Huang, L.Q., Huang, Q., Han, Y., Hu, B., Hu, F., Li, B.H., Li, Y.R., Liang, K., Lin, L.K., Luo, L.S., Ma, J., Ma, L.L., Peng, Z.Y., Pan, Y.B., Pan, Z.Y., Ren, X.Q., Sun, H.M., Wang, Y., Wang, Y.Y., Weng, H., Wei, C.J., Wu, D.F., Xia, J., Xiong, Y., Xu, H.B., Yao, X.M., Yuan, Y.F., Ye, T.S., Zhang, X.C., Zhang, Y.W., Zhang, Y.G., Zhang, H.M., Zhao, Y., Zhao, M.J., Zi, H., Zeng, X.T., Wang, Y.Y. and Wang, X.H. 2020. A rapid advice guideline for the diagnosis and treatment of 2019 novel coronavirus (2019-nCoV) infected pneumonia (standard version). Mil. Med. Res. 7, 16; doi:10.1186/s40779-020-0233-6. Jones, B.A., Grace, D., Kock, R., Alonso, S., Rushton, J., Said, M.Y., McKeever, D., Mutua, F., Young, J., McDermott, J. and Pfeiffer, D.U. 2013. Zoonosis emergence linked to agricultural intensification and environmental change. Proc. Natl. Acad. Sci. U. S. A. 110, 8399–8404; doi:10.1073/pnas.1208059110. Jones, K.E., Patel, N.G., Levy, M.A., Storeygard, A., Balk, D., Gittleman, J.L. and Daszak, P. 2008. Global trends in emerging infectious diseases. Nature. 451, 990–993; doi:10.1038/nature06536. Jung, K., Hu, H. and Saif, L.J. 2016. Porcine deltacoronavirus infection: Etiology, cell culture for virus isolation and propagation, molecular epidemiology and pathogenesis. Virus Res. 226, 50–59; doi:10.1016/j.virusres.2016.04.009. Kandeil, A., Gomaa, M., Shehata, M., El-Taweel, A., Kayed, A.E., Abiadh, A., Jrijer, J., Moatasim, Y., Kutkat, O., Bagato, O., Mahmoud, S., Mostafa, A., El-Shesheny, R., Perera, R.A.P.M., Ko, R.L.W., Hassan, N., Elsokary, B., Allal, L., Saad, A., Sobhy, H., McKenzie, P.P., Webby, R.J., Peiris, M., Ali, M.A. and Kayali, G. 2019. Middle East respiratory syndrome coronavirus infection in non-camelid domestic mammals. Emerg. Microbes Infect. 8, 103–108; doi:10.1080/22221751.2018.1560235. Kelly, T.R., Karesh, W.B., Johnson, C.K., Gilardi, K.V.K., Anthony, S.J., Goldstein, T., Olson, S.H., Machalaba, C. and Mazet, J.A.K. 2017. One health proof of concept: bringing a transdisciplinary approach to surveillance for zoonotic viruses at the human-wild animal interface. Prev. Vet. Med. 137, 112–118; doi:10.1016/j.prevetmed.2016.11.023. Kim, J.H., Jang, J.H., Yoon, S.W., Noh, J.Y., Ahn, M.J., Kim, Y., Jeong, D.G. and Kim, H.K. 2018. Detection of bovine coronavirus in nasal swab of non-captive wild water deer, Korea. Transbound. Emerg. Dis. 65, 627–631; doi:10.1111/tbed.12847. Kolesnikova, L., Slenczka, W., Brodt, H.R., Klenk, H.D. and Becker, S. 2003. Electron microscopy in diagnostics of SARS case. Microsc. Microanal. 9, 438–439; doi:10.1017/s1431927603035104. Lam, T.T.-Y., Shum, M.H.-H., Zhu, H.-C., Tong, Y.-G., Ni, X.-B., Liao, Y.-S., Wei, W., Cheung, W.Y.-M., Li, W.-J., Li, L.-F., Leung, G.M., Holmes, E.C., Hu, Y.-L. and Guan, Y. 2020. Identification of 2019-nCoV related coronaviruses in Malayan pangolins in southern China. bioRxiv; doi:10.1101/2020.02.13.945485. Lee, C. 2015. Porcine epidemic diarrhea virus: An emerging and re-emerging epizootic swine virus. Virol. J. 12(1), 193; doi:10.1186/s12985-015-0421-2. Lee, N., Hui, D., Wu, A., Chan, P., Cameron, P., Joynt, G.M., Ahuja, A., Yung, M.Y., Leung, C.B., To, K.F., Lui, S.F., Szeto, C.C., Chung, S. and Sung, J.J.Y. 2003. A major outbreak of severe acute respiratory syndrome in Hong Kong. N. Engl. J. Med. 348, 1986–1994; doi:10.1056/NEJMoa030685. Licitra, B.N., Duhamel, G.E. and Whittaker, G.R. 2014. Canine enteric coronaviruses: emerging viral pathogens with distinct recombinant spike proteins. Viruses. 6(8), 3363–3376; doi:10.3390/v6083363. Liu, C., Zhou, Q., Li, Y., Garner, L.V., Watkins, S.P., Carter, L.J., Smoot, J., Gregg, A.C., Daniels, A.D., Jervey, S. and Albaiu, D. 2020. Research and development on therapeutic agents and vaccines for COVID-19 and related human coronavirus diseases. ACS Cent. Sci. 6, 315–331; doi:10.1021/acscentsci.0c00272. Lorusso, A., Calistri, P., Petrini, A., Savini, G. and Decaro, N. 2020. Novel coronavirus (COVID-19) epidemic: a veterinary perspective. Vet. Ital. 56(1), 5–10; doi:10.12834/VetIt.2173.11599.1. Lu, H., Stratton, C.W. and Tang, Y.W. 2020. Outbreak of pneumonia of unknown etiology in Wuhan, China: the mystery and the miracle. J. Med. Virol. 92(4), 401–402; doi:10.1002/jmv.25678. Masters, P.S. 2006. The molecular biology of coronaviruses. Adv. Virus Res. 65, 193–292; doi:10.1016/S0065-3527(06)66005-3. McMahon, B.J., Morand, S. and Gray, J.S. 2018. Ecosystem change and zoonoses in the Anthropocene. Zoonoses Public Health 65, 755–765; doi:10.1111/zph.12489. Meyer, B., García-Bocanegra, I., Wernery, U., Wernery, R., Sieberg, A., Müller, M.A., Drexler, J.F., Drosten, C. and Eckerle, I. 2015. Serologic assessment of possibility for MERS-CoV infection in equids. Emerg. Infect. Dis. 21(1), 181–182; doi:10.3201/eid2101.141342. Mora-Díaz, J.C., Piñeyro, P.E., Houston, E., Zimmerman, J., Giménez-Lirola, L.G., 2019. Porcine hemagglutinating encephalomyelitis virus: a review. Front. Vet. Sci. 6, 53; doi:10.3389/fvets.2019.00053. Morse, S.S., Mazet, J.A.K., Woolhouse, M., Parrish, C.R., Carroll, D., Karesh, W.B., Zambrana-Torrelio, C., Lipkin, W.I. and Daszak, P. 2012. Prediction and prevention of the next pandemic zoonosis. Lancet. 9857, 1956–1965; doi:10.1016/S0140-6736(12)61684-5. Murray, K.A. and Daszak, P. 2013. Human ecology in pathogenic landscapes: two hypotheses on how land use change drives viral emergence. Curr. Opin. Virol. 3(1), 79–83; doi:10.1016/j.coviro.2013.01.006. Ogawa, V.A., Shah, C.A. and Nicholson, A. 2019. Exploring lessons learned from a century of outbreaks, National Academies of Sciences, Engineering, and Medicine; Health and Medicine Division; Board on Global Health; Forum on Microbial Threats. Washington, DC: National Academies Press. Available via https://doi.org/10.17226/25391. OIE – World Organisation for Animal Health. 2020. Questions and answers on COVID-19. Available via https://www.oie.int/scientific-expertise/specific-information-and-recommendations/questions-and-answers-on-2019novel-coronavirus/ (Accessed 22 April 2020). Paim, F.C., Bowman, A.S., Miller, L., Feehan, B.J., Marthaler, D., Saif, L.J. and Vlasova, A.N. 2019. Epidemiology of deltacoronaviruses (δ-CoV) and gammacoronaviruses (γ-CoV) in wild birds in the United States. Viruses. 11(10), 897; doi:10.3390/v11100897. Parry, N.M. 2020. COVID-19 and pets: when pandemic meets panic. Forn. Sci. Int. Rep. 2, 100090; doi:10.1016/j.fsir.2020.100090. Peiris, J.S.M., Lai, S.T., Poon, L.L.M., Guan, Y., Yam, L.Y.C., Lim, W., Nicholls, J., Yee, W.K.S., Yan, W.W., Cheung, M.T., Cheng, V.C.C., Chan, K.H., Tsang, D.N.C., Yung, R.W.H., Ng, T.K. and Yuen, K.Y. 2003a. Coronavirus as a possible cause of severe acute respiratory syndrome. Lancet. 361, 1319–1325; doi:10.1016/S0140-6736(03)13077-2. Peiris, J.S.M.P., Yuen, K.Y., Osterhaus, A.D.M.E. and Stöhr, K. 2003b. The severe acute respiratory syndrome. N. Engl. J. Med. 349, 2431–2441; doi:10.1056/NEJMra032498. Pene, F., Merlat, A., Vabret, A., Rozenberg, F., Buzyn, A., Dreyfus, F., Cariou, A., Freymuth, F. and Lebon, P. 2003. Coronavirus 229E-related pneumonia in immunocompromised patients. Clin. Infect. Dis. 37, 929–932; doi:10.1086/377612. Perlman, S. and Netland, J. 2009. Coronaviruses post-SARS: update on replication and pathogenesis. Nat. Rev. Microbiol. 7(6), 439–450; doi:10.1038/nrmicro2147. Poon, L.L.M., Chu, D.K.W., Chan, K.H., Wong, O.K., Ellis, T.M., Leung, Y.H.C., Lau, S.K.P., Woo, P.C.Y., Suen, K.Y., Yuen, K.Y., Guan, Y. and Peiris, J.S.M. 2005. Identification of a novel coronavirus in bats. J. Virol. 79, 2001–2009; doi:10.1128/jvi.79.4.2001-2009.2005. Pradesh, U., Pandit, P., Dayal, D., Pashu, U., Vigyan, C., Evam, V., Pradesh, U., Zoonosis, S. De, Pereira, S., Pereira, D., Malik, Y.S., Pradesh, U. and Rodriguez-morales, A.J. 2020. Coronavirus disease 2019 – COVID-19. Clin Microbiol Rev. 33(4), e00028–20; doi:10.1128/CMR.00028-20. Pusterla, N., Vin, R., Leutenegger, C.M., Mittel, L.D. and Divers, T.J. 2018. Enteric coronavirus infection in adult horses. Vet. J. 231, 13–18; doi:10.1016/j.tvjl.2017.11.004. Raj, V.S., Osterhaus, A.D.M.E., Fouchier, R.A.M. and Haagmans, B.L. 2014. MERS: emergence of a novel human coronavirus. Curr. Opin. Virol. 5, 58–62; doi:10.1016/j.coviro.2014.01.010. Ramadan, N. and Shaib, H. 2019. Middle east respiratory syndrome coronavirus (MERS-COV): a review. Germs. 9(1), 35; doi:10.18683/germs.2019.1155. Rodriguez-Morales, A.J., Dhama, K., Sharun, K., Tiwari, R. and Bonilla-Aldana, D.K. 2020. Susceptibility of felids to coronaviruses. Vet. Rec. 186, e21–e21; doi:10.1136/VR.M1671. Rottier, P.J.M., Nakamura, K., Schellen, P., Volders, H. and Haijema, B.J., 2005. Acquisition of macrophage tropism during the pathogenesis of feline infectious peritonitis is determined by mutations in the feline coronavirus spike protein. J. Virol. 79, 14122–14130; doi:10.1128/jvi.79.22.14122-14130.2005. Saif, L.J. 2004. Animal coronaviruses: what can they teach us about the severe acute respiratory syndrome? OIE Rev. Sci. Tech. 23(2), 643–660; doi:10.20506/rst.23.2.1513. Saif, L.J. 2020. Vaccines for Covid-19: perspectives, prospects, and challenges based on candidate SARS, MERS, and animal coronavirus vaccines. Eur. Med. J. doi:10.33590/emj/200324. Salata, C., Calistri, A., Parolin, C. and Palù, G. 2020. Coronaviruses: a paradigm of new emerging zoonotic diseases. Pathog. Dis. 77(9), ftaa006; doi:10.1093/femspd/ftaa006. Sanz, M.G., Kwon, S.Y., Pusterla, N., Gold, J.R., Bain, F. and Evermann, J. 2019. Evaluation of equine coronavirus fecal shedding among hospitalized horses. J. Vet. Intern. Med. 33, 918–922; doi:10.1111/jvim.15449. Scales, D.C., Green, K., Chan, A.K., Poutanen, S.M., Foster, D., Nowak, K., Raboud, J.M., Saskin, R., Lapinsky, S.E. and Stewart, T.E. 2003. Illness in intensive care staff after brief exposure to severe acute respiratory syndrome. Emerg. Infect. Dis. 9, 1205–1210; doi:10.3201/eid0910.030525. Schwegmann-Weßels, C. and Herrler, G. 2006. Transmissible gastroenteritis virus infection: a vanishing specter. Dtsch. Tierarztl. Wochenschr. 113(4), 157–159. Shi, J., Wen, Z., Zhong, G., Yang, H., Wang, C., Huang, B., Liu, R., He, X., Shuai, L., Sun, Z., Zhao, Y., Liu, P., Liang, L., Cui, P., Wang, J., Zhang, X., Guan, Y., Tan, W., Wu, G., Chen, H. and Bu, Z. 2020. Susceptibility of ferrets, cats, dogs, and other domesticated animals to SARS–coronavirus 2. Science. 368(6494), 1016–1020; doi:10.1126/science.abb7015. Smith, J., Sadeyen, J.R., Cavanagh, D., Kaiser, P. and Burt, D.W. 2015. The early immune response to infection of chickens with Infectious Bronchitis Virus (IBV) in susceptible and resistant birds. BMC Vet. Res. 11(1), 256; doi:10.1186/s12917-015-0575-6. Streinu-Cercel, A. 2014. Ebola virus disease – a global threat. Germs. 4(3), 58; doi:10.11599/germs.2014.1056. Suzuki, T., Otake, Y., Uchimoto, S., Hasebe, A. and Goto, Y. 2020. Genomic characterization and phylogenetic classification of bovine coronaviruses through whole genome sequence analysis. Viruses. 12(2), 123; doi:10.3390/v12020183. Tai, W., He, L., Zhang, X., Pu, J., Voronin, D., Jiang, S., Zhou, Y. and Du, L. 2020. Characterization of the receptor-binding domain (RBD) of 2019 novel coronavirus: implication for development of RBD protein as a viral attachment inhibitor and vaccine. Cell. Mol. Immunol. 17(6), 613–620; doi:10.1038/s41423-020-0400-4. Taylor, L.H., Latham, S.M. and Woolhouse, M.E.J. 2001. Risk factors for human disease emergence. Philos. Trans. R. Soc. B Biol. Sci. 356, 983–989; doi:10.1098/rstb.2001.0888. Tekes, G. and Thiel, H.J. 2016. Feline coronaviruses: pathogenesis of feline infectious peritonitis. In Advances in virus research. Cambridge MA: Academic Press Inc, pp: 193–218. Available via https://doi.org/10.1016/bs.aivir.2016.08.002. To, K.K.W., Hung, I.F.N., Chan, J.F.W. and Yuen, K.Y. 2013. From SARS coronavirus to novel animal and human coronaviruses. J. Thorac. Dis. 5(Suppl 2), S103; doi:10.3978/j.issn.2072-1439.2013.06.02. Tu, Y.F., Chien, C.S., Yarmishyn, A.A., Lin, Y.Y., Luo, Y.H., Lin, Y.T., Lai, W.Y., Yang, D.M., Chou, S.J., Yang, Y.P., Wang, M.L. and Chiou, S.H. 2020. A review of sars-cov-2 and the ongoing clinical trials. Int. J. Mol. Sci. 21(7), 2657; doi:10.3390/ijms21072657. Vergara-Alert, J., van den Brand, J.M.A., Widagdo, W., Muñoz, M., Raj, V.S., Schipper, D., Solanes, D., Cordón, I., Bensaid, A., Haagmans, B.L. and Segalés, J. 2017. Livestock susceptibility to infection with middle east respiratory syndrome coronavirus. Emerg. Infect. Dis. 23, 232–240; doi:10.3201/eid2302.161239. Vijgen, L., Keyaerts, E., Moës, E., Maes, P., Duson, G. and Van Ranst, M. 2005. Development of one-step, real-time, quantitative reverse transcriptase PCR assays for absolute quantitation of human coronaviruses OC43 and 229E. J. Clin. Microbiol. 43, 5452–5456; doi:10.1128/JCM.43.11.5452-5456.2005. Walsh, E.E., Shin, J.H. and Falsey, A.R. 2013. Clinical impact of human coronaviruses 229E and OC43 infection in diverse adult populations. J. Infect. Dis. 208, 1634–1642; doi:10.1093/infdis/jit393. Wang, M., Jing, H., Xu, H., Jiang, X., Kan, B., Liu, Q., Wan, K., Cui, B., Zheng, H., Cui, Z., Yan, M., Liang, W., Wang, H., Qi, X., Li, Z., Li, M., Chen, K., Zhang, E., Zhang, S., Hai, R., Yu, D., Xu, J., 2005. Surveillance on severe acute respiratory syndrome associated coronavirus in animals at a live animal market of Guangzhou in 2004. Zhonghua Liu Xing Bing Xue Za Zhi. 26, 84–87. Wang, Q., Vlasova, A.N., Kenney, S.P. and Saif, L.J. 2019. Emerging and re-emerging coronaviruses in pigs. Curr. Opin. Virol. 34, 1–42; doi:10.1016/j.coviro.2018.12.001. Weiss, S.R. and Navas-Martin, S. 2005. Coronavirus pathogenesis and the emerging pathogen severe acute respiratory syndrome coronavirus. Microbiol. Mol. Biol. Rev. 69, 635–664; doi:10.1128/MMBR.69.4.635-664.2005. Wong, A.C.P., Li, X., Lau, S.K.P. and Woo, P.C.Y. 2019. Global epidemiology of bat coronaviruses. Viruses. 11(2), 174; doi:10.3390/v11020174. World Economic Forum. 2010. Global risks 2010: a global risk network report, a global risk network report. Available via http://www3.weforum.org/docs/WEF_Global_Risks_Report_2010.pdf (Accessed 23 April 2020). World Health Organization (WHO). 2004. WHO guidelines for the global surveillance of severe acute respiratory syndrome (SARS), 2004. Available via https://www.who.int/csr/resources/publications/WHO_CDS_CSR_ARO_2004_1.pdf?ua=1 (Accessed 23 April 2020). World Health Organization (WHO). 2020a. Statement on the second meeting of the International Health Regulations (2005) Emergency Committee regarding the outbreak of novel coronavirus (2019-nCoV). Available via https://www.who.int/news-room/detail/30-01-2020-statement-on-the-second-meeting-of-the-international-health-regulations-(2005)-emergency-committee-regarding-the-outbreak-of-novel-coronavirus-(2019-NCoV) (Accessed 15 April 2020). World Health Organization (WHO). 2020b. WHO director-general’s opening remarks at the media briefing on COVID-19 - 11 March 2020. Available via https://www.who.int/dg/speeches/detail/who-director-general-s-opening-remarks-at-the-media-briefing-on-covid-19---11-march-2020 (Accessed 15 April 2020). World Health Organization (WHO). 2020c. Coronavirus disease (COVID-19) pandemic. Available via https://www.who.int/emergencies/diseases/novel-coronavirus-2019 (Accessed 15 April 2020). World Health Organization (WHO). 2020d. Middle East respiratory syndrome coronavirus (MERS-CoV). Available via https://www.who.int/health-topics/middle-east-respiratory-syndrome-coronavirus-mers#tab=tab_1 (Accessed 24 April 2020). Yoo, H.S. and Yoo, D. 2020. COVID-19 and veterinarians for one health, zoonotic- and reverse-zoonotic transmissions. J. Vet. Sci. 21(3), e51; doi:10.4142/jvs.2020.21.e51. Zaki, A.M., Van Boheemen, S., Bestebroer, T.M., Osterhaus, A.D.M.E. and Fouchier, R.A.M. 2012. Isolation of a novel coronavirus from a man with pneumonia in Saudi Arabia. N. Engl. J. Med. 367, 1814–1820; doi:10.1056/NEJMoa1211721. Zhang, T., Wu, Q. and Zhang, Z. 2020a. Probable pangolin origin of SARS-CoV-2 associated with the COVID-19 outbreak. Curr. Biol. 30, 1346–1351.e2; doi:10.1016/j.cub.2020.03.022. Zhang, X., Deng, T., Lu, J., Zhao, P., Chen, L., Qian, M., Guo, Y., Qiao, H., Xu, Y., Wang, Y., Li, X., Zhang, G., Wang, Z. and Bian, C. 2020b. Molecular characterization of variant infectious bronchitis virus in China, 2019: implications for control programmes. Transbound. Emerg. Dis. 67(3), 1349–1355; doi:10.1111/tbed.13477. Zhong, N.S., Zheng, B.J., Li, Y.M., Poon, L.L.M., Xie, Z.H., Chan, K.H., Li, P.H., Tan, S.Y., Chang, Q., Xie, J.P., Liu, X.Q., Xu, J., Li, D.X., Yuen, K.Y., Peiris, J.S.M. and Guan, Y. 2003. Epidemiology and cause of severe acute respiratory syndrome (SARS) in Guangdong, People’s Republic of China, in February, 2003. Lancet. 362, 1353–1358; doi:10.1016/S0140-6736(03)14630-2. Zhou, P., Fan, H., Lan, T., Yang, X.L., Shi, W.F., Zhang, W., Zhu, Y., Zhang, Y.W., Xie, Q.M., Mani, S., Zheng, X.S., Li, B., Li, J.M., Guo, H., Pei, G.Q., An, X.P., Chen, J.W., Zhou, L., Mai, K.J., Wu, Z.X., Li, D., Anderson, D.E., Zhang, L.B., Li, S.Y., Mi, Z.Q., He, T.T., Cong, F., Guo, P.J., Huang, R., Luo, Y., Liu, X.L., Chen, J., Huang, Y., Sun, Q., Zhang, X.L.L., Wang, Y.Y., Xing, S.Z., Chen, Y.S., Sun, Y., Li, J., Daszak, P., Wang, L.F., Shi, Z.L., Tong, Y.G. and Ma, J.Y., 2018. Fatal swine acute diarrhoea syndrome caused by an HKU2-related coronavirus of bat origin. Nature. 556, 255–259; doi:10.1038/s41586-018-0010-9. Zhou, P., Yang, X.L., Wang, X.G., Hu, B., Zhang, L., Zhang, W., Si, H.R., Zhu, Y., Li, B., Huang, C.L., Chen, H.D., Chen, J., Luo, Y., Guo, H., Jiang, R. Di, Liu, M.Q., Chen, Y., Shen, X.R., Wang, X., Zheng, X.S., Zhao, K., Chen, Q.J., Deng, F., Liu, L.L., Yan, B., Zhan, F.X., Wang, Y.Y., Xiao, G.F. and Shi, Z.L. 2020. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature. 579, 270–273; doi:10.1038/s41586-020-2012-7. Zinsstag, J., Schelling, E., Waltner-Toews, D. and Tanner, M. 2011. From “one medicine” to “one health” and systemic approaches to health and well-being. Prev. Vet. Med. 101, 148–156; doi:10.1016/j.prevetmed.2010.07.003. | ||
| How to Cite this Article |
| Pubmed Style Poudel U, Subedi D, Pantha S, Dhakal S. Animal coronaviruses and coronavirus disease 2019: Lesson for one health approach. Open Vet. J.. 2020; 10(3): 239-251. doi:10.4314/ovj.v10i3.1 Web Style Poudel U, Subedi D, Pantha S, Dhakal S. Animal coronaviruses and coronavirus disease 2019: Lesson for one health approach. https://www.openveterinaryjournal.com/?mno=113950 [Access: January 25, 2026]. doi:10.4314/ovj.v10i3.1 AMA (American Medical Association) Style Poudel U, Subedi D, Pantha S, Dhakal S. Animal coronaviruses and coronavirus disease 2019: Lesson for one health approach. Open Vet. J.. 2020; 10(3): 239-251. doi:10.4314/ovj.v10i3.1 Vancouver/ICMJE Style Poudel U, Subedi D, Pantha S, Dhakal S. Animal coronaviruses and coronavirus disease 2019: Lesson for one health approach. Open Vet. J.. (2020), [cited January 25, 2026]; 10(3): 239-251. doi:10.4314/ovj.v10i3.1 Harvard Style Poudel, U., Subedi, . D., Pantha, . S. & Dhakal, . S. (2020) Animal coronaviruses and coronavirus disease 2019: Lesson for one health approach. Open Vet. J., 10 (3), 239-251. doi:10.4314/ovj.v10i3.1 Turabian Style Poudel, Uddab, Deepak Subedi, Saurav Pantha, and Santosh Dhakal. 2020. Animal coronaviruses and coronavirus disease 2019: Lesson for one health approach. Open Veterinary Journal, 10 (3), 239-251. doi:10.4314/ovj.v10i3.1 Chicago Style Poudel, Uddab, Deepak Subedi, Saurav Pantha, and Santosh Dhakal. "Animal coronaviruses and coronavirus disease 2019: Lesson for one health approach." Open Veterinary Journal 10 (2020), 239-251. doi:10.4314/ovj.v10i3.1 MLA (The Modern Language Association) Style Poudel, Uddab, Deepak Subedi, Saurav Pantha, and Santosh Dhakal. "Animal coronaviruses and coronavirus disease 2019: Lesson for one health approach." Open Veterinary Journal 10.3 (2020), 239-251. Print. doi:10.4314/ovj.v10i3.1 APA (American Psychological Association) Style Poudel, U., Subedi, . D., Pantha, . S. & Dhakal, . S. (2020) Animal coronaviruses and coronavirus disease 2019: Lesson for one health approach. Open Veterinary Journal, 10 (3), 239-251. doi:10.4314/ovj.v10i3.1 |