| Research Article | ||
Open Vet. J.. 2024; 14(8): 1877-1895 Open Veterinary Journal, (2024), Vol. 14(8): 1877–1895 Research Article Characterization and molecular identification of the lumpy skin disease virus in cattle in the Mekong Delta of VietnamTran Ngoc Bich1,*, Le Quang Trung1, Truong Van Hieu2, Vo Tuan Khai Huyen3,4, Thai Quoc Hieu4, Huynh Truong Giang1 and Nguyen Tran Phuoc Chien11Faculty of Veterinary Medicine, College of Agriculture, Can Tho University, Can Tho, Vietnam 2Department of Animal Science and Veterinary Medicine, School of Agriculture and Aquaculture, Tra Vinh University, Tra Vinh, Vietnam 3Interdisciplinary Graduate Program in Animal Pathology and Disease Treatment, Faculty of Veterinary Medicine, College of Agriculture, Can Tho University, Can Tho, Vietnam 4Tien Giang Sub-Department of Animal Health, Ministry of Agriculture and Rural Development, Mỹ Tho, Vietnam *Corresponding Author: Tran Ngoc Bich. Faculty of Veterinary Medicine, College of Agriculture, Can Tho University, Can Tho City, Vietnam. Email: tnbich [at] ctu.edu.vn Submitted: 24/04/2024 Accepted: 23/07/2024 Published: 31/08/2024 © 2024 Open Veterinary Journal
ABSTRACTBackground: Lumpy skin disease (LSD) is caused by a virus belonging to the genus Capripoxvirus, exhibiting clinical symptoms ranging from mild signs to the development of nodules. LSD emerged in Asia and Southeast Asia, including Vietnam, in October 2020 and has since spread throughout the region, resulting in productivity and economic losses. Aim: This study aimed to investigate the virus-causing papular dermatitis in cattle from the Mekong Delta region of Vietnam by analyzing its GPCR gene and assessing its evolutionary relationship with sequences in the GenBank database. Methods: Blood samples (n=180) were collected from cattle farms in Ben Tre, Tien Giang, and Tra Vinh provinces. PCR targeting the P32 antigen gene was utilized to detect LSDV presence, and GPCR gene amplification was performed to assess genetic variability. Results: LSDV was detected in 8.33% (15/180) of the samples using PCR targeting the P32 antigen gene. Each sample that tested positive for LSDV demonstrated complete amplification of the GPCR gene. Sequence alignments and phylogenetic analyses of the GPCR gene revealed that Mekong Delta LSDV isolates shared genetic similarities and possessed a 12-nucleotide insertion comparable to strains from China in 2019 and Northern Vietnam in 2020. Conclusion: This study provides preliminary insights into the molecular characteristics of LSDV in cattle from the Mekong Delta region of Vietnam. The observed genetic relatedness to other LSDV sequences from Asia and Southeast Asia underscores the importance of regional surveillance and control measures. These findings contribute to the development of effective strategies for LSDV control and prevention. Keywords: GPCR, Lumpy skin disease virus, Characterization, Mekong Delta, Vietnam. IntroductionLumpy skin disease (LSD) is classified by the World Organisation for Animal Health (OIE) as a transboundary disease due to its rapid spread and the significant financial losses it incurs (Tuppurainen and Oura, 2012). LSD, caused by the Neethling strain of the Capripoxvirus from the Poxviridae family, was first identified in Zambia in 1929. It is prevalent across much of Africa, certain regions of the Middle East, and Turkey. According to data from 2020, the LSD epidemic has affected over 20 countries and territories, including Bangladesh, Bhutan, China, Taiwan, Djibouti, Hong Kong, India, Iraq, Mozambique, Myanmar, Namibia, Nepal, Russia, Saudi Arabia, Sri Lanka, Switzerland, Syria, Thailand, and Turkey (WAHIS, 2022). According to the online animal disease information system (WAHIS), LSD was first identified in Lang Son province of Vietnam in October 2020. Since then, LSD cases have been reported in 55 out of 63 provinces and cities across the country, spanning 457 districts and 4,280 communes. This has resulted in a total of 200,831 infected buffaloes and cows and the culling of 28,600 animals. Specifically, the provinces of Tien Giang, Long An, Tra Vinh, Vinh Long, and Ben Tre have reported 31 affected districts and 188 communes, with 4,558 infected cows and 803 culled cows (DAH, 2021). LSD is characterized by round, slightly elevated papules approximately 2–7 cm in diameter, which typically appear on the neck, legs, tail, and back shortly after the onset of fever (Şevik and Doğan, 2017). The disease leads to a significant decrease in milk production (ranging from 10% to 85%) due to elevated body temperature and the development of secondary mastitis. Additional consequences of the disease include irreversible skin damage, reduced growth rates in cattle, temporary or permanent sterility, miscarriage, and potential fatality (Şevik et al., 2016). Several specimens commonly used for viral genome detection include nodules, ulcers, secretions, semen, and blood from infected cattle (Bedeković et al., 2018; Wolf et al., 2021). PCR techniques focusing on P32 and GPCR genes are commonly used for identifying and analyzing LSDV (Seerintra et al., 2022; Koirala et al., 2022). Capripoxviruses can be genetically distinguished by the GPCR gene, found on the double-stranded DNA (dsDNA) of the LSDV genome and playing a role in immunomodulation in the host (Le Goff et al., 2009). Comparing the GPCR gene sequences of LSDV from wild field isolates and vaccine strains, it was shown that the wild type of LSDV genomes had a particular 12-bp deletion that was not present in vaccine strains (El-Tholoth and El-Kenawy, 2016; Lu et al., 2019). This discovery emphasizes the significance of the GPCR gene as a potential target for the creation of a “DIVA” (differentiation of infected from vaccinated animals) technique, which may accurately identify LSDV infections in herds that have been immunized with similar vaccinations (Agianniotaki et al., 2017; Bedeković et al., 2018). Although prior studies have examined the frequency of LSDV in Vietnam (Tran et al., 2021), there is less data available regarding the molecular attributes of the present LSDV strains found in the Mekong Delta region of Vietnam. Comprehending these molecular characteristics is crucial for the field of molecular epidemiology and the use of vaccines. Consequently, the purpose of this research was to examine the GPCR gene-based molecular features of LSDV strains in the blood of recently infected Vietnamese cows. Additionally, we looked at the phylogenetic link between the isolated Vietnamese LSDV strains and the sequences published in the database. Materials and MethodsSpecimen collection and DNA extractionThis study was conducted in three of Vietnam’s Mekong Delta provinces: Ben Tre, Tra Vinh, and Tien Giang (Fig. 1). Blood samples were collected from 180 cows suspected of being affected by LSD during the outbreak between January and December 2023. These samples were obtained from cows displaying LSD-like clinical signs and had not been vaccinated against LSDV previously. Approximately 1–3 ml of blood was collected and deposited in sterile tubes containing whole blood (EDTA tubes). The samples were collected aseptically following the guidelines outlined by the TCVN 8400:2010 (TCVN, 2010), TCCS 04:2020/TY–DT (MARD, 2020), and OIE (2017). Detailed information regarding suspected LSD-affected cattle, including age, gender, breed, and clinical signs, was also recorded. Each clinical sample was labeled with a unique sample ID upon collection. The samples were transported in a cooler box with ice packs to the Faculty of Veterinary Laboratory at Can Tho University and stored at −80°C for further molecular analysis. DNA extraction and PCR assayThe whole viral DNA was isolated from the acquired whole blood sample using the TopPURE® Serum Viral Extraction Kit (ABT, Vietnam) by the manufacturer’s instructions. The genomic DNA was extracted using the kit and then kept at a temperature of −20°C until it was needed for subsequent experiments. Next, the DNA extracts from LSDV-infected samples were used for conventional PCR amplification with a 192 bp region in the P32 (LSDV074) gene using pairs of primers: forward primer, 5′-TTTCCTGATTTTTCTTACTAT-3′, and reverse primer, 5′-AAATTATATACGTAAATAAC-3′, according to the PCR procedures, as described in a previous study (Ireland and Binepal, 1998). GPCR gene amplificationAll positive samples were processed to amplify the G-protein-coupled-chemokine-like receptor (GPCR) (LSDV011) gene using PCR according to primer forward 5′-GATGAGTATTGATAGATACCTAGCTGTAGTT-3′, and reverse primer, 5′-TGAGACAATCCAAACCACCAT-3′, were used for further DNA sequencing as previously described (Le Goff et al., 2009). The PCR amplification reactions were performed in a 25 μl volume containing 12 μl GoTaq® DNA Polymerase (Promega, USA), 3 μl template DNA, 1 μl of 10 μM each primer (forward and reverse), and 8 μl ultrapure water. The PCR was performed with the following thermal conditions: an initial denaturation step at 96°C for 5 minutes, followed by 35 cycles of denaturation at 95°C for 30 seconds, annealing at 50°C for 30 seconds, extension at 72°C for 30 seconds, and a final extension step at 72°C for 5 minutes. The PCR amplification results were analyzed using electrophoresis on a 1.5% agarose gel with ethidium bromide (1 μg/ml) in 1X Tris-acetate-EDTA (TAE) buffer. The visualization and imaging of the products were done using UV transillumination using a BIO-Rad instrument from the USA. Nucleotide sequencing and analysisThe amplicons of GPCR (1,158 bp) were purified from the PCR products using the TopPURE® PCR/Gel ADN purification kit (ABT, Vietnam). The products were sequenced using both forward and reverse strands at a commercial sequencing company (NamKhoa-Biotech, Vietnam). The Sanger sequencing technique, which used the ABI Prism BigDyeTM Terminator v1.1 cycle sequencing kit, was carried out with the assistance of an ABI PRISM 3,500 × l Genetic Analyzer that was utilized. The resulting LSDV GPCR sequences were repeatedly aligned using Geneious Prime® version 2024.0.2 and BioEdit® version 7.1.9, with final adjustments made manually, then evaluated for similarity to GenBank sequences using the BLAST program offered by NCBI (https://www.ncbi.nlm.nih.gov/). The Maximum Likelihood method in MEGA X software (Kumar et al., 2018) was employed for phylogenetic analysis to generate the phylogenetic tree for phylogenetic study. The confidence of the branching patterns of the trees was estimated using bootstrap analysis with 1,000 replications. The trees were visualized and annotated using the interactive tree of life (iTOL) (Letunic and Bork, 2024). The multiple sequence alignments of the partial GPCR genes were visualized using BioEdit® version 7.1.9. The GPCR sequences of the Mekong Delta Vietnam were submitted to the GenBank database and are available under accession numbers PP645759-PP645773.
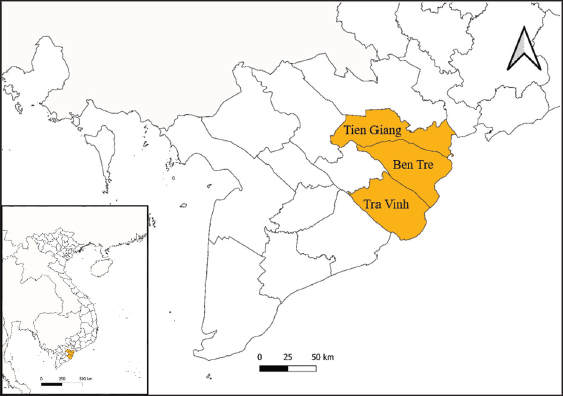
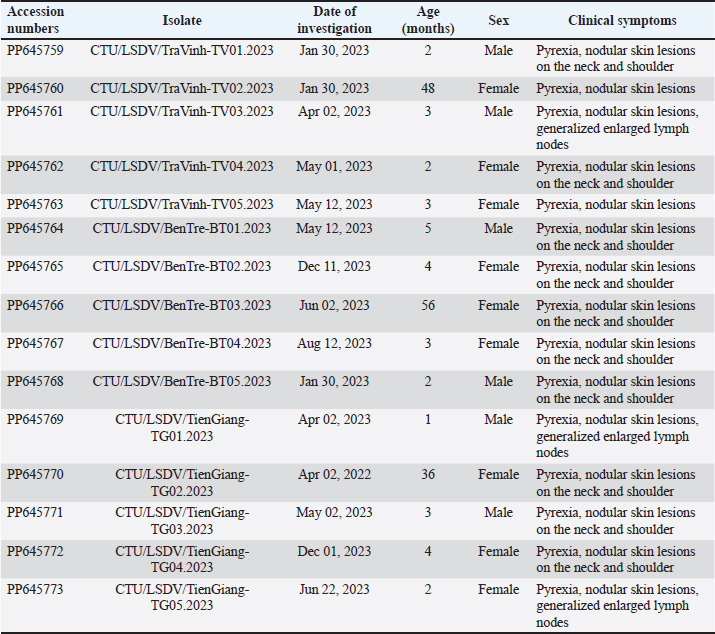
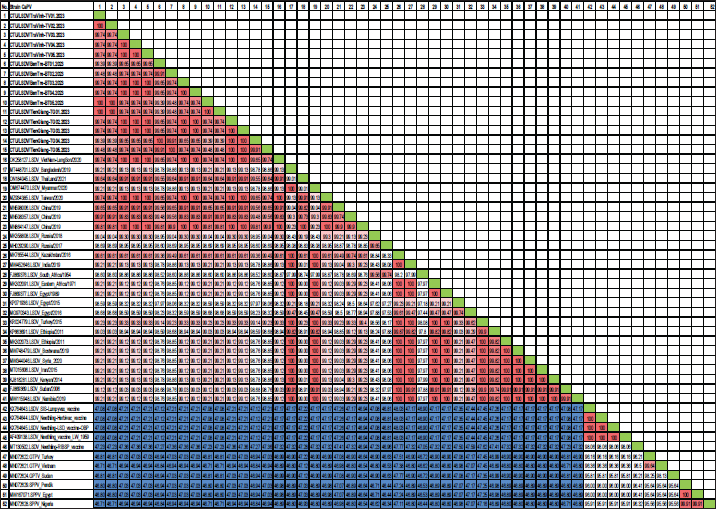
Fig. 1. The study area map shows three provinces in the Mekong Delta of Vietnam where the blood samples were collected. Ethical approvalThis study was carried out on naturally infected animals. The samples used for this study were diagnostic, and no experimental procedures were carried out in any animal. Written informed consent was obtained from the owners for the participation of their animals in this study. Results LSD outbreak investigation One hundred and eighty cattle suspected of being infected with LSDV from three provinces in the Mekong Delta of Vietnam were sampled: Tien Giang, Ben Tre, and Tra Vinh (Fig. 1). All sampled animals had not been previously vaccinated against LSDV. Cattle that were suspected of being infected with LSD had a wide range of clinical symptoms, such as depression, lack of appetite, fever, nasal and ocular discharges, enlarged superficial lymph nodes, circumscribed skin nodules on different regions of the body, and a fall in body condition score (Fig. 2). LSDV was isolated from fifteen cows and confirmed by PCR. Among these cows, eleven were calves between one and six months old. The remaining cows included two lactating cows and one dry cow (Table 1). Phylogenetic analysisLSDV was detected in fifteen out of 180 blood samples using PCR based on the P32 antigen gene. All fifteen positive samples exhibited clinical signs of LSD. In addition, partial GPCR sequences were amplified in approximately all positive samples. After quality checking and editing, the GPCR sequences of the Vietnam LSDV in beef cattle were deposited in GenBank under the accession numbers (Table 1). All fifteen LSDV samples from the Mekong Delta showed close genetic relatedness to LSDV strains from North Vietnam (accession number OK258127), Thailand (accession number ON184045), China (accession number MN598006, MN508357, and MN864147), and Taiwan (accession number MZ934385), with similarities ranging from 99.7% to 100%. Additionally, they exhibited similarities to LSDV strains from Russia (accession number MH029290 and MK358808), Myanmar (accession number OM674470), Bangladesh (accession number MT448701), and Egypt (accession number FJ869377, KP071936, and MG970343), ranging from 98.0% to 99.3% (Table S1).
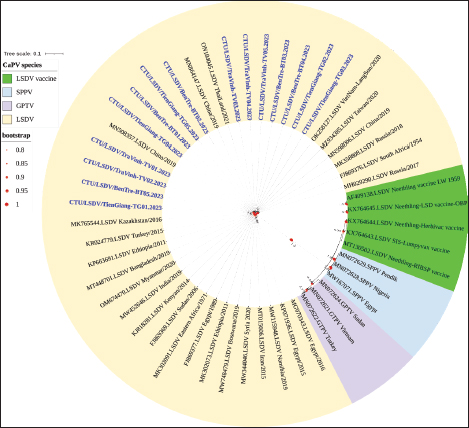
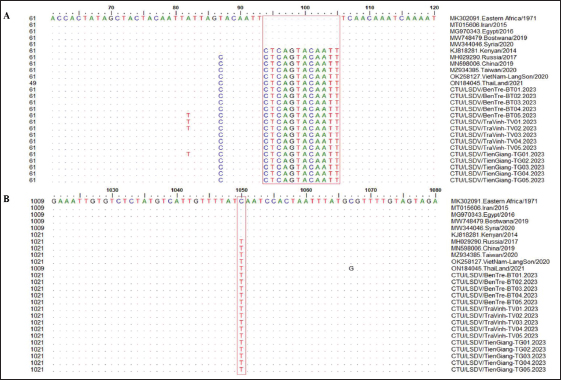
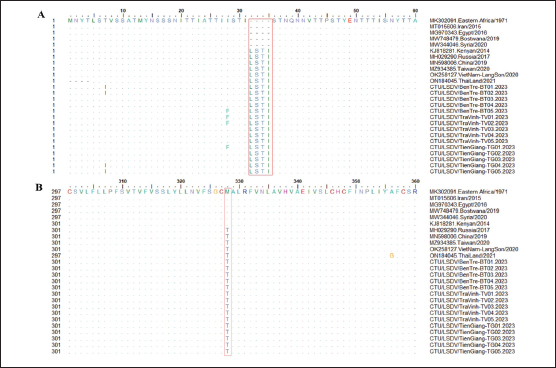
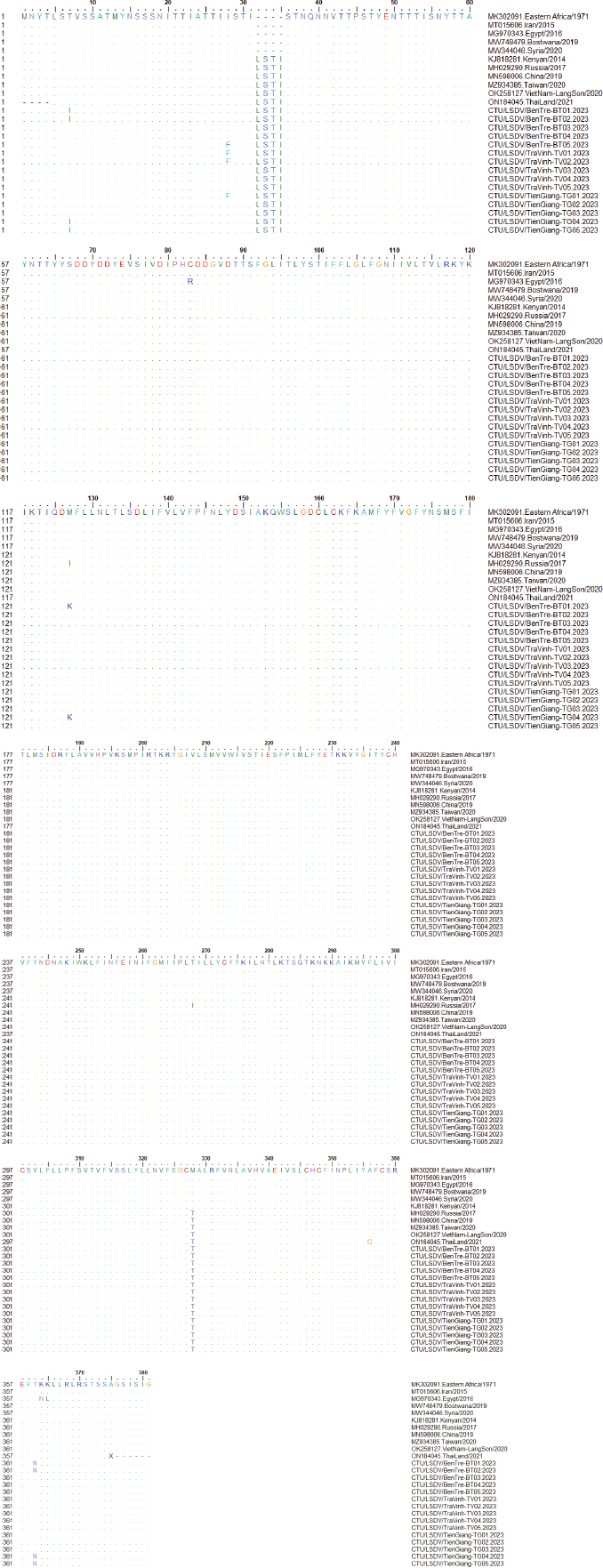
Fig. 2. Clinical signs of lumpy skin disease in cattle were observed in this study. The dairy calf with nodular skin (A)–(D) was infected with the lumpy skin disease virus and presented numerous circumscribed skin nodules throughout the body. Phylogenetic relationships among fifteen LSDV isolates from the Mekong Delta in Vietnam and all 26 LSDV GPCR genes from the GenBank database were analyzed. Additionally, five vaccine strains, three goatpox virus strains, and three sheeppox virus strains were included in the phylogeny (Fig. 3). The LSDV isolates from the Mekong Delta in Vietnam were discovered in the same group as other LSDV isolates from various countries, including North Vietnam (OK258127), China (MN598006), Russia (MH029290), Thailand (ON184045), and Taiwan (MZ934385) (Fig. 3). In addition, the study of the GPCR gene revealed an insertion of 12 base pairs (nucleotides 94 to 105) in the GPCR gene of cattle from the provinces of Tien Giang, Ben Tre, and Tra Vinh in the Mekong Delta region of Vietnam (Fig. 4; Fig. S1). The absence of specific amino acid residues (L32, S33, T34, and I35) and the presence of particular mutations (I28F and M318T) are identified as unique signatures among isolates from the Mekong Delta region and strains originating from North Vietnam, China, Thailand, Taiwan, and Russia. In comparison with the reference sequence MK302091.Eastern Africa/1971 (Fig. 5; Fig. S2). DiscussionThe current study examined and verified instances of LSD in the Mekong Delta region of Vietnam using PCR. The LSDVs obtained from the observed outbreaks were also analyzed utilizing GPCR gene sequencing. This research represents the first known instance of molecular characterization and phylogenetic analysis of the LSDV observed in this location in 2023. LSD is considered one of the most economically significant viral infections in cattle due to its detrimental effects on hides, milk production, mastitis, fertility, weight loss, and mortality. While the morbidity rate of LSD might exceed 100%, the fatality rate is below 10% (OIE, 2021). The initial occurrence of LSD in cattle was documented in Lang Son province, Vietnam, towards the conclusion of October 2020 (Tran et al., 2021). Following this, the LSDV subsequently disseminated to many additional regions within Vietnam. A prompt preventative strategy was implemented, utilizing the emergency vaccination Lumpyvac® (Vetal, Adiyaman, Turkey) sourced from Turkey. Furthermore, it was advised that farm owners in both infected and non-infested zones/provinces should also adopt biosecurity measures. Despite the implementation of a vaccination regimen for cattle and buffalo, inadequate vaccine coverage has resulted in ongoing LSD outbreaks that continue to inflict significant economic damage on the cattle industry. The current investigation used PCR targeting the P32 gene to identify the presence of LSDV in the blood of infected cattle. Using blood samples for early virus detection before cows show signs of lumps on the skin is a proactive approach to managing LSD in cattle, allowing for timely intervention and potentially reducing the spread of the disease (Tuppurainen et al., 2005; Aerts et al., 2021). The positive samples were then amplified, explicitly targeting the GPCR gene to detect and analyze LSDV outbreaks in some provinces in the Mekong Delta area of Vietnam. The presence of viral DNA was identified in 15 out of 180 samples (Table 1). All positive samples were obtained from cattle that had clinical symptoms of LSD and had not received prior vaccination against LSDV. Primarily, cows become infected with LSD when they are aged between one and six months. The positive rate of samples collected from suspected animals varied due to the occasional detection of LSDV viral DNA in blood during a short period of viremia from 4–11 days after infection (Aerts et al., 2021). There was no correlation between the duration of the viremia phase and the severity of clinical symptoms (Tuppurainen et al., 2005). Table 1. The summary of signalments, collected location, and clinical symptoms of 15 LSD-positive cattle.
The G protein-coupled chemokine receptor (GPCR) gene emerges as a potential candidate for elucidating the evolutionary connection between members of the Capripoxvirus genus and LSDV. This study, utilizing wild field and vaccine strains, employed primers from previous research to amplify LSDV, explicitly targeting the GPCR sequences (Le Goff et al., 2009). Multiple sequence alignments of the fifteen targeted LSDV genes within the GPCR revealed complete identity among LSDVs from three provinces in the Mekong Delta of Vietnam. Analysis of these sequences unveiled a 12-base pair nucleotide insertion between positions 94 and 105 (CTCAGTACAATT), leading to the alteration of four amino acids (LSTI) (aa 33–35) and a nucleotide change at position C1050T (aa M318T), thereby modifying the GPCR gene in LSDV in cattle. This characteristic was also observed in LSDV isolates from North Vietnam (OK258127), Thailand (ON184045), China (MN598006), Taiwan (MZ934385), Russia (MT129673), and Kenya (KJ818281) (Lu et al., 2019; Trinh et al., 2022; Singhla et al., 2022). Phylogenetic analysis indicated that LSDV strains found in 15 samples from three provinces in the Mekong Delta of Vietnam exhibited similarity to LSDV strains previously recovered from North Vietnam, China, Russia, and Thailand, suggesting a common progenitor. However, GPCR sequences obtained from strains in Eastern Africa (MK302081), Iran (MT015606), Egypt (MG970343), Botswana (MW748479), and Syria (MW818281) showed a distinct loss of 12 base pairs; these findings provide evidence that LSDV strains originating from Africa exhibit more significant genetic variation compared to Eurasian strains (Sameea Yousefi et al., 2018; Sprygin et al., 2020; Chibssa et al., 2021; Badhy et al., 2021). Strains originating from North Vietnam, China, Thailand, and Russia were classified as a distinct cluster due to their status as newly generated recombinant strains; these strains exhibited sequences similar to the Neethling vaccination strains, as previously described (Dao et al., 2020; Aleksandr et al., 2020; Ma et al., 2022; Zan et al., 2022). However, conducting a comprehensive genome sequence analysis is necessary to elucidate the discrepancy in the partial sequence alignment among the 15 strains and to track any genetic mutations in the LSDV. Table S1. Nucleotide sequence identity distances for the GPCR gene of local LSDV isolates compared to reference LSDV, sheep pox virus, and goat pox virus published in GenBank.
The molecular analysis of the gene sequences provided evidence that LSDV field strains were involved in the outbreak. LSDV has been documented in Southeast Asian and Asian countries, including Thailand, Laos, Cambodia, Myanmar, Malaysia, China, and Russia (Arjkumpa et al., 2021; Roche et al., 2021; Azeem et al., 2022). The disease may spread across borders by arthropod vectors, including stable flies, horse flies, and mosquitoes, facilitating the transmission of the virus from neighboring nations (Tuppurainen et al., 2021). A genetic comparison of LSDV strains from three provinces in the Mekong Delta of Vietnam showed that the 2023 strains were 100% identical to the strains collected in North Vietnam in 2020, suggesting that a single strain of the virus caused the outbreaks in Vietnam. Hence, to decrease losses to the farming community, LSD control methods must include immunization, biosecurity, vector management, and cow movement regulation. In addition, to effectively control and prevent the spread of LSDV, it is necessary to implement surveillance measures in various provinces to assess the prevalence of circulating LSDV strains. This will help identify the predominant LSDV candidates and facilitate the development of a suitable vaccine for the control of LSDV.
Fig. 3. Phylogenetic tree demonstrating the relationship between GPCR gene sequences obtained from the Mekong Delta in Vietnam (shown in blue and bold) and other Capripoxvirus GPCR gene sequences acquired from GenBank. LSDV-lumpy skin disease virus; GTPV-goat pox virus; SPPV-sheep pox virus.
Fig. 4. Multiple sequence alignments of the partial nucleotide sequences of the GPCR gene. The isolates from the Mekong Delta in Vietnam were aligned with representative LSDV sequences retrieved from GenBank. (A) Insertion of 12 nucleotides from positions 94 to 105 is evident in the sequences of the newly isolated LSDV when compared with strain MK302091. (B) The C to T change is highlighted in the red box. The dots indicate identical nucleotides in the alignment.
Fig. 5. Multiple sequence alignments of the partial amino acids sequences of the GPCR gene. The isolates from the Mekong Delta in Vietnam were aligned with representative LSDV sequences retrieved from GenBank. (A) Insertion of 4 amino acids from positions 32 to 35 is evident in the sequences of the newly isolated LSDV when compared with strain MK302091. (B) The M (Methionine) to T (Threonine) change is highlighted in the red box. The dots indicate identical amino acids in the alignment. ConclusionThis study utilizes GPCR gene sequencing to provide information on the molecular characteristics and phylogenetic analysis of LSDV circulating in the Mekong Delta of Vietnam. According to the findings, the LSDV strains obtained from this location have genetic resemblances to the strains discovered in China in 2019 and North Vietnam in 2020. This research has revealed a substantial amount of evidence that supports the extensive distribution of LSDVs throughout Asian nations and Southeast Asian countries. This data will be advantageous for researchers seeking a deeper understanding of LSDV molecular epidemiology. They will assist the government of Vietnam in enhancing its efforts to develop effective control measures against local LSDV outbreaks in the Mekong Delta and throughout the country. Furthermore, the results of this study are crucial for bolstering the national vaccination campaign against the LSD virus and for advancing the strategies for creating a potent vaccine against the LSD virus in Vietnam. AcknowledgmentThe authors thank the technical staff and students from the Faculty of Veterinary Medicine, College of Agriculture, Can Tho University, who helped with the sample collection and analysis. The authors would also like to acknowledge the cattle farmers of the area for their valuable cooperation. Conflict of interestThe authors declare that the research was conducted without any commercial or financial relationships that could be construed as a potential conflict of interest. Authors’ contributionsThis research was conducted with the contributions of all the authors. The authors all participated in the study design, analyzing the results, interpreting the results, and preparing the manuscript. All authors read and approved the final manuscript. FundingThis research is supported by the Vietnam Ministry-Level Scientific Project under Project Code No. B2023-TCT-13. Data availabilityThe following supporting information can be downloaded at: https://s.net.vn/KciJ. ReferencesAerts, L., Haegeman, A., De Leeuw, I., Philips, W., Van Campe, W., Behaeghel, I., Mostin, L. and De Clercq, K. 2021. Detection of clinical and subclinical lumpy skin disease using ear notch testing and skin biopsies. Microorganisms. 9(10), 2171. Agianniotaki, E.I., Tasioudi, K.E., Chaintoutis, S.C., Iliadou, P., Mangana-Vougiouka, O., Kirtzalidou, A., Alexandropoulos, T., Sachpatzidis, A., Plevraki, E., Dovas, C.I. and Chondrokouki, E. 2017. Lumpy skin disease outbreaks in Greece during 2015–2016, implementation of emergency immunization and genetic differentiation between field isolates and vaccine virus strains. Vet. Microbiol. 201, 78–84. Aleksandr, K., Pavel, P., Olga, B., Svetlana, K., Vladimir, R., Yana, P. and Alexander, S. 2020. Emergence of a new lumpy skin disease virus variant in Kurgan Oblast Russia in 2018. Arch. Virol. 165(6), 1343–1356. Arjkumpa, O., Suwannaboon, M., Boonrod, M., Punyawan, I., Liangchaisiri, S., Laobannue, P., Lapchareonwong, C., Sansri, C., Kuatako, N., Panyasomboonying, P. and Uttarak, P. 2022. The first lumpy skin disease outbreak in Thailand 2021: epidemiological features and spatio-temporal analysis. Fronti. Vet. Sci. 8, 799065. Azeem, S., Sharma, B., Shabir, S., Akbar, H. and Venter, E. 2022. Lumpy skin disease is expanding its geographic range: A challenge for Asian livestock management and food security. Vet. J. 279, 105785. Badhy, S.C., Chowdhury, M.G.A., Settypalli, T.B.K., Cattoli, G., Lamien, C.E., Fakir, M.A.U., Akter, S., Osmani, M.G., Talukdar, F., Begum, N. and Khan, I.A. 2021. Molecular characterization of lumpy skin disease virus (LSDV) emerged in Bangladesh reveals unique genetic features compared to contemporary field strains. BMC Vet. Res. 17, 01–11. Bedeković, T., Šimić, I., Krešić, N. and Lojkić, I. 2018. Detection of lumpy skin disease virus in skin lesions, blood, nasal swabs and milk following preventive vaccination. Transbound. Emerg. Dis. 65(2), 491–496. Chibssa, T.R., Sombo, M., Lichoti, J.K., Adam, T.I.B., Liu, Y., Elraouf, Y.A., Grabherr, R., Settypalli, T.B.K., Berguido, F.J., Loitsch, A. and Sahle, M. 2021. Molecular analysis of East African lumpy skin disease viruses reveals a mixed isolate with features of both vaccine and field isolates. Microorganisms. 9(6), 1142. DAH. Department of Animal Health. 2021. Report on the epidemic situation of papular dermatitis November 4, 2021. Ha Noi, Vietnam: Elsevier. Dao, T.D., Tran, L.H., Nguyen, H.D., Hoang, T.T., Nguyen, G.H., Tran, K.V.D., Nguyen, H.X., Van Dong, H., Bui, A.N. and Bui, V.N. 2022. Characterization of Lumpy skin disease virus isolated from a giraffe in Vietnam. Transbound. Emerg. Dis. 69(5), 3268–3272. El-Tholoth, M. and El-Kenawy, A.A. 2016. G-protein-coupled chemokine receptor gene in lumpy skin disease virus isolates from cattle and water buffalo (Bubalus bubalis) in Egypt. Transbound. Emerg. Dis. 63(6), 288–295. Ireland, D.C. and Binepal, Y.S. 1998. Improved detection of Capripoxvirus in biopsy samples by PCR. J. Virol. Met. 74(1), 1–7. Koirala, P., Meki, I.K., Maharjan, M., Settypalli, B.K., Manandhar, S. and Yadav, S.K. 2022. Molecular characterization of the 2020 outbreak of lumpy skin disease in Nepal. Microorganisms. 10(3), 539. Kumar, S., Stecher, G., Li, M., Knyaz, C. and Tamura, K. 2018. MEGA X: Molecular evolutionary genetics analysis across computing platforms. Mol. Biol. Evol. 35(6), 1547. Le Goff, C., Lamien, C.E., Fakhfakh, E., Chadeyras, A., Aba-Adulugba, E., Libeau, G., Tuppurainen, E., Wallace, D.B., Adam, T., Silber, R. and Gulyaz, V. 2009. Capripoxvirus G-protein-coupled chemokine receptor: a host-range gene suitable for virus animal origin discrimination. J. Gen. Virol. 90(8), 1967–1977. Letunic, I. and Bork, P. 2024. Interactive Tree of Life (iTOL) v6: recent updates to the phylogenetic tree display and annotation tool. Nucleic. Acids. Res. 52, 78–82. Lu, G., Xie, J., Luo, J., Shao, R., Jia, K. and Li, S. 2021. Lumpy skin disease outbreaks in China since 3 August 2019. Transbound. Emerg. Dis. 68(2), 216–219. Ma, J., Yuan, Y., Shao, J., Sun, M., He, W., Chen, J. and Liu, Q. 2022. Genomic characterization of lumpy skin disease virus in southern China. Transbound. Emerg. Dis. 69(5), 2788–2799. MARD. Ministry of Agriculture and Rural Development (TCVN 8402:2010). Animal disease -The institutional rules for the care and use of laboratory animals. National Academies Press: Ha Noi, Vietnam. MARD. Ministry of Agriculture and Rural Development. 2020. Facility standards (TCCS 04: 2020/TY–DT), including testing procedures to detect viruses that cause lympy skin disease. National Academies Press: Ha Noi, Vietnam. OIE. 2017. Chapter 2.4.13. Lumpy skin disease OIE Terrestrial Manual 2017, Paris, France. OIE. 2021. Lumpy skin disease: In Manual of diagnostic tests and vaccines for terrestrial animals. OIE: Paris, France. Roche, X., Rozstalnyy, A., TagoPacheco, D., Pittiglio, C., Kamata, A., Beltran Alcrudo, D., Bisht, K., Karki, S., Kayamori, J., Larfaoui, F., Raizman, E., VonDobschuetz, S., Dhingra, M.S. and Sumption, K. 2021. Introduction and spread of lumpy skin disease in South, East and Southeast Asia: Qualitative risk assessment and management. Rome, Italy: FAO, 183. Sameea Yousefi, P., Dalir-Naghadeh, B., Mardani, K. and Jalilzadeh-Amin, G. 2018. Phylogenetic analysis of the lumpy skin disease viruses in Northwest of Iran. Trop. Anim. Health. Prod. 50, 1851–1858. Seerintra, T., Saraphol, B., Wankaew, S. and Piratae, S. 2022. Molecular identification and characterization of Lumpy skin disease virus emergence from cattle in the Northeastern part of Thailand. J. Vet. Sci. 23(5), 73. Şevik, M. and Doğan, M. 2017. Epidemiological and molecular studies on lumpy skin disease outbreaks in Turkey during 2014–2015. Transbound. Emerg. Dis. 64(4), 1268–1279. Şevik, M., Avci, O., Doğan, M. and İnce, Ö. B. 2016. Serum biochemistry of lumpy skin disease virus-infected cattle. BioMed Res. Int. 2016(1), 6257984. Singhla, T., Boonsri, K., Kreausukon, K., Modethed, W., Pringproa, K., Sthitmatee, N., Punyapornwithaya, V. and Vinitchaikul, P. 2022. Molecular characterization and phylogenetic analysis of lumpy skin disease virus collected from outbreaks in Northern Thailand in 2021. Vet. Sci. 9(4), 194. Sprygin, A., Pestova, Y., Bjadovskaya, O., Prutnikov, P., Zinyakov, N., Kononova, S., Ruchnova, O., Lozovoy, D., Chvala, I. and Kononov, A. 2020. Evidence of recombination of vaccine strains of lumpy skin disease virus with field strains, causing disease. PLoS One. 15(5), 0232584. Tran, H.T.T., Truong, A.D., Dang, A.K., Ly, D.V., Nguyen, C.T., Chu, N.T., Hoang, T.V., Nguyen, H.T., Nguyen, V.T. and Dang, H.V. 2021. Lumpy skin disease outbreaks in Vietnam, 2020. Transbound. Emerg. Dis. 68(3), 977–980. Trinh, T.B.N., Nguyen, V.T., Nguyen, T.T.H., Mai, N.T.A., Le, P.N., Lai, T.N.H., Phan, T.H., Tran, D.H., Pham, N.T., Dam, V.P. and Nguyen, T.L. 2022. Molecular and histopathological characterization of lumpy skin disease in cattle in northern Vietnam during the 2020–2021 outbreaks. Arch. Virol. 167(11), 2143–2149. Tuppurainen, E., Dietze, K., Wolff, J., Bergmann, H., Beltran-Alcrudo, D., Fahrion, A., Lamien, C.E., Busch, F., Sauter-Louis, C., Conraths, F.J. and De Clercq, K. 2021. Vaccines and vaccination against lumpy skin disease. Vaccines. 9(10), 1136. Tuppurainen, E.S., Venter, E.H., and Coetzer, J.A.W. 2005. The detection of lumpy skin disease virus in samples of experimentally infected cattle using different diagnostic techniques. Onderstepoort. J. Vet. Res. 72(2), 153–164. Tuppurainen, E.S.M. and Oura, C.A.L. 2012. Lumpy skin disease: an emerging threat to Europe, the Middle East and Asia. Transbound. Emerg. Dis. 59(1), 40–48. WAHIS. World Animal Health Information System. 2022. Lumpy Skin Disease (LSD)—OIE—Asia. Available via https://rr-asia.oie.int/en/projects/lumpy-skin-disease-lsd/ (Accessed 22 April 2024). Wolff, J., Beer, M. and Hoffmann, B. 2021. Probe-based real-time qPCR assays for a reliable differentiation of Capripoxvirus species. Microorganisms. 9(4), 765. Zan, X., Huang, H., Guo, Y., Di, D., Fu, C., Wang, S., Wu, Y., Wang, J., Wang, Y., Ma, Y. and Chai, C. 2022. Molecular characterization of a novel subgenotype of lumpy skin disease virus strain isolated in Inner Mongolia of China. BMC Vet. Res. 18(1), 295.
Fig. S1. Multiple sequence alignments of the partial nucleotide sequences of the GPCR gene.
Fig. S2. Multiple sequence alignments of the partial amino acids sequences of the GPCR gene. | ||
| How to Cite this Article |
| Pubmed Style Bich TN, Trung LQ, Hieu TV, Huyen VTK, Hieu TQ, Giang HT, Chien NTP. Characterization and molecular identification of the lumpy skin disease virus in cattle in the Mekong Delta of Vietnam. Open Vet. J.. 2024; 14(8): 1877-1895. doi:10.5455/OVJ.2024.v14.i8.16 Web Style Bich TN, Trung LQ, Hieu TV, Huyen VTK, Hieu TQ, Giang HT, Chien NTP. Characterization and molecular identification of the lumpy skin disease virus in cattle in the Mekong Delta of Vietnam. https://www.openveterinaryjournal.com/?mno=198939 [Access: January 25, 2026]. doi:10.5455/OVJ.2024.v14.i8.16 AMA (American Medical Association) Style Bich TN, Trung LQ, Hieu TV, Huyen VTK, Hieu TQ, Giang HT, Chien NTP. Characterization and molecular identification of the lumpy skin disease virus in cattle in the Mekong Delta of Vietnam. Open Vet. J.. 2024; 14(8): 1877-1895. doi:10.5455/OVJ.2024.v14.i8.16 Vancouver/ICMJE Style Bich TN, Trung LQ, Hieu TV, Huyen VTK, Hieu TQ, Giang HT, Chien NTP. Characterization and molecular identification of the lumpy skin disease virus in cattle in the Mekong Delta of Vietnam. Open Vet. J.. (2024), [cited January 25, 2026]; 14(8): 1877-1895. doi:10.5455/OVJ.2024.v14.i8.16 Harvard Style Bich, T. N., Trung, . L. Q., Hieu, . T. V., Huyen, . V. T. K., Hieu, . T. Q., Giang, . H. T. & Chien, . N. T. P. (2024) Characterization and molecular identification of the lumpy skin disease virus in cattle in the Mekong Delta of Vietnam. Open Vet. J., 14 (8), 1877-1895. doi:10.5455/OVJ.2024.v14.i8.16 Turabian Style Bich, Tran Ngoc, Le Quang Trung, Truong Van Hieu, Vo Tuan Khai Huyen, Thai Quoc Hieu, Huynh Truong Giang, and Nguyen Tran Phuoc Chien. 2024. Characterization and molecular identification of the lumpy skin disease virus in cattle in the Mekong Delta of Vietnam. Open Veterinary Journal, 14 (8), 1877-1895. doi:10.5455/OVJ.2024.v14.i8.16 Chicago Style Bich, Tran Ngoc, Le Quang Trung, Truong Van Hieu, Vo Tuan Khai Huyen, Thai Quoc Hieu, Huynh Truong Giang, and Nguyen Tran Phuoc Chien. "Characterization and molecular identification of the lumpy skin disease virus in cattle in the Mekong Delta of Vietnam." Open Veterinary Journal 14 (2024), 1877-1895. doi:10.5455/OVJ.2024.v14.i8.16 MLA (The Modern Language Association) Style Bich, Tran Ngoc, Le Quang Trung, Truong Van Hieu, Vo Tuan Khai Huyen, Thai Quoc Hieu, Huynh Truong Giang, and Nguyen Tran Phuoc Chien. "Characterization and molecular identification of the lumpy skin disease virus in cattle in the Mekong Delta of Vietnam." Open Veterinary Journal 14.8 (2024), 1877-1895. Print. doi:10.5455/OVJ.2024.v14.i8.16 APA (American Psychological Association) Style Bich, T. N., Trung, . L. Q., Hieu, . T. V., Huyen, . V. T. K., Hieu, . T. Q., Giang, . H. T. & Chien, . N. T. P. (2024) Characterization and molecular identification of the lumpy skin disease virus in cattle in the Mekong Delta of Vietnam. Open Veterinary Journal, 14 (8), 1877-1895. doi:10.5455/OVJ.2024.v14.i8.16 |