| Research Article | ||
Open Vet J. 2024; 14(7): 1644-1657 Open Veterinary Journal, (2024), Vol. 14(7): 1644–1657 Research Article Phylogenetic analysis of Staphylococcus aureus enterotoxin A gene in Iraqi breed cows with bovine mastitis: Implications for disease managementAlaa Jawad1, Sabreen Noori Dagman1, Mohammed Mahdi Yaseen2 and Hassan Al-Karagoly3*1Department of Microbiology, College of Veterinary Medicine, University of Al-Qadisiyah, Al-Diwaniyah, Iraq 2Department of Public Health, College of Veterinary Medicine, University of Al-Qadisiyah, Al-Diwaniyah, Iraq 3Department of Internal and Preventive Medicine, College of Veterinary Medicine, University of Al-Qadisiyah, Al-Diwaniyah, Iraq *Corresponding Author: Hassan Al-Karagoly. Department of Internal and Preventive Medicine, College of Veterinary Medicine, University of Al-Qadisiyah, Al-Diwaniyah, Iraq. Email: hassan.aliwee [at] qu.edu.iq Submitted: 14/05/2024 Accepted: 26/06/2024 Published: 31/07/2024 © 2024 Open Veterinary Journal
AbstractBackground: Although milk is nutritionally valuable, it also serves as a significant medium for the transmission of pathogens and their toxins. Aim: This study aimed to investigate the role of enterotoxin gene A (SEA) in the development of bovine mastitis. We accomplished this by examining milk through polymerase chain reaction (PCR) testing, amino acid substitution analysis, and phylogenetic analysis. Methods: A total of fifty milk samples were collected from locally bred dairy cows in Al-Diwaniyah, located in southern Iraq. We employed the VITEK-2 platform to validate the diagnosis of Staphylococcus aureus and confirm the results of routine tests (culturing and biochemical tests). Subsequently, the genetic mutation and phylogeny analysis were achieved utilizing DNA sequencing to 16S rRNA and enterotoxin A genes. Results: 66% (33/50) of the milk samples found to be contain S. aureus by the VITEK-2 system. Furthermore, 25/33 of the samples were positive by the PCR test. While 60% (15 out of 25) tested positive for the SEA gene. After genomic analysis, we identified amino acid substitutions of serine, glutamine with arginine, tyrosine with cysteine, and aspartic acid with glycine at positions 9, 101, 119, 187, and 191. The phylogenetic investigation demonstrated a genetic relationship between our isolates (Iraqi isolates) and isolates from Indian and the United States. Conclusion: Our study indicated the widespread distribution of the enterotoxin gene A (SEA) of S. aureus among dairy cows. The molecular study revealed significant changes in key amino acids that could play an important role in the bacterium's pathogenesis. The phylogenetic similarities among S. aureus samples from various countries suggest that the bacteria has spread globally. Keywords: S. aureus, 16S rRNA gene, Amino acid substitutions, Phylogenetic analysis, Mastitis. IntroductionMastitis has a huge impact on dairy production, negatively impacting livestock health, well-being, and the quality of milk while incurring substantial damage to the dairy industry (Le Maréchal et al., 2011). Staphylococcus aureus, which is a widely prevalent and extremely contagious bacteria, is known to be responsible for both clinical and subclinical mastitis (Guimarães et al., 2017). Different genes encode several virulence factors in S. aureus, which significantly contribute to the induction of inflammation, degeneration, and necrosis. The ClfA, ClfB, FnbA, and FnbB genes encode proteins that bind fibrinogen and promote bacterial aggregation, clot formation, and tissue colonization. Additionally, sspA and sspB genes encode proteases (particularly V8 protease) that degrade host proteins and expedite tissue invasion. While hysA encodes hyaluronidase, which dissolves connective tissue hyaluronic acid to disseminate pathogens, The Geh gene encodes lipases, which breakdown lipids and feed microorganisms. Also, alpha-hemolysin (α-toxin) is produced by the HLA gene and is responsible for damaging host cell membranes. Moreover, the lukS-PV and lukF-PV genes encode pantan-valentine leukocidin, which destroys leukocytes and suppresses the immune response of the host (Ullah et al., 2022). Additionally, tst encodes toxic shock syndrome toxin-1 (TSST-1), a superantigen that promotes systemic inflammation. SEA and SEB enterotoxins cause gastrointestinal symptoms and act as superantigens. The icaADBC gene encodes polysaccharide intercellular adhesin, which is crucial for the development of biofilm matrix and bacterial adherence. Cap genes produce capsular polysaccharides, which protect the bacterium. The Spa gene encodes protein A, which suppresses opsonization and phagocytosis by binding to antibodies' Fc domains. By inhibiting complement activation, SCIN protects bacteria against complement-mediated lysis. The infection of mammary glands with Staphylococci (particularly S. aureus) is thought to be a multi-stage process involving multiple phases and complicated biological interactions (Dego et al., 2002). During milking, minor wounds, or environmental contamination, bacteria primarily enter the udder through the teat canal. Staphylococcus bacteria can adhere tightly to the extracellular matrix of the epithelial cells of the teat using surface proteins known as cell wall-anchored proteins that are covalently connected to peptidoglycan (Foster et al., 2014; Foster, 2020). This helps the bacteria grow in that area. The bacteria then begin to create biofilms, which are clusters of bacterial cells surrounded by an extracellular matrix, increasing their resistance to antibiotics and the host's defense mechanisms making it more difficult to eliminate bacteria (Kiran et al., 2022). Because the bacteria produce several digestive enzymes, they penetrate the teat duct and disperse to the inter lobes of the mammary glands, causing damage to the alveolar and ductal cells. After that, leukocytes (neutrophils) diapedesis to the infection site and then release cytokines to recruit more immune cells and cause inflammation. Additionally, Staphylococci produce toxins such as α-toxin and β-toxin, which damage and destroy mammary gland tissue (Anderson et al., 2012). This causes inflammation, swelling, and redness in the udder, causing pain and discomfort for the cow. The collection of dead cells and germs causes pus to develop, complicating the disease even further. The development of mastitis has a negative impact on milk quality and production. It is also possible for certain cases to progress to a chronic disease in which pathogens remain in the udder for extended periods of time. Chronic mastitis infections can cause irreversible harm to the mammary glands and dramatically limit milk output. Mastitis causes a negative economic impact by reducing both the amount and quality of produce, resulting in severe financial losses for breeders (Ibrahim, 2017). The presence of staphylococci in milk can cause contamination of dairy products. Furthermore, failure to appropriately remove some of the toxins produced by S. aureus during the dairy product manufacturing process might be harmful to human health. Infected cows might transfer the infection to the other cows on the property. The use of intensive treatment is necessary to prevent and control the spread of Staphylococcal disease, intensive treatment is required. Unfortunately, frequent and extensive use of antibiotics to treat mastitis can lead to the development of antibiotic-resistant bacterial strains (Sipahi et al., 2023). Resistance to antibiotics represents a global problem facing public health. Recent studies (Salam et al., 2023; Alara and Alara, 2024) reported that antimicrobial resistance (AMR) contributes to 1.27 million deaths and 4.95 million cases of illness worldwide. Furthermore, several researchers have focused on the emergence of multidrug-resistant bacteria. To find the appropriate antibiotic, as well as to find new strains that are resistant to multiple drugs and study their resistance genes, it is important to do regular antimicrobial susceptibility testing (Algammal et al., 2020; Shafiq et al., 2022; Worku et al., 2022). Staphylococci are responsible for a variety of infectious diseases in both humans and animals (Guo et al., 2020). In animals, the most recorded illnesses include mastitis and dermatitis in cattle, sheep, goats, and horses. It also causes botryomycosis in pigs and suppurative infections in pets (Peton and Le Loir, 2014). While it is mostly responsible for food poisoning in humans, the most notable symptoms associated with gastrointestinal tract problems include cramps, nausea, vomiting, and diarrhea. These symptoms usually start to show up 2 to 4 hours after having spoiled food (Kimberlin et al., 2022). Individual differences in the intensity of these symptoms and the amount of SE consumed also play a role. The current study aims to investigate the role of the enterotoxin A gene of S. aureus in the pathogenesis of mastitis in dairy cows, using routine and confirmatory tests such as the VITEK-2 and polymerase chain reaction (PCR) tests, as well as genetic analysis related to mutations and phylogenetic. Materials and MethodsSample collectionWe collected 50 milk samples under sterile conditions (Vacheyrou et al., 2011) from dairy Iraqi breed cows, sourced from several farms in the Al-Diwaniyah province in southern Iraq. We then transported the collected samples to the College of Veterinary Medicine at the University of Al-Qadisiyah Laboratory for culturing, biochemical testing, and molecular testing. Isolation and identification of S. aureusWith minor adjustments, we achieved the isolating and identification of S. aureus in accordance with Singh and Prakash (2008) methodology. For the enrichment process, we used the peptone water enrichment broth provided by HiMedia Private Ltd. After that, we thoroughly combined a 10 ml or 10 g specimen with 90 ml of sterile enrichment broth and subjected it to incubation at 37°C for a duration of 24 hours. The specific medium used for S. aureus isolation was Baird Parker Agar (BPA) (HiMedia Private Ltd., India). We spread a small amount of culture from the enrichment onto BP agar and then incubated it at 37°C for 48 hours. The presence of jet-black colonies encircled by a white halo was indicative of the likely presence of S. aureus. After that, the pure cultures were inoculated onto nutrient agar (HiMedia Pvt. Ltd., India) and then placed them in an incubator at a temperature of 37°C for 24 hours. We then subjected the cultures to biochemical assays for further characterization. Morphological characteristicsWe made the smear by applying the isolated culture to a clean, grease-free glass slide and then staining it using Gram's staining procedure (Habib et al., 2015). We then examined the stained specimens under a microscope. Gram-positive, coccus-shaped cells, organized in irregular clusters resembling a bunch of grapes, were present in the smear examination. Biochemical analysisWe conducted biochemical assays using the catalase test, coagulase test, DNase test, acetoin production, oxidase test, and D-mannitol fermentation to verify the presence of S. aureus. Identification of S. aureus using VITEK2 systemBacterial identification were achieved by using cultural and biochemical testes VITEK2 cards (Biomerieux, USA) injected with fluids directly obtained from culture bottles are excellent for rapid identification and susceptibility testing of S.aureus (Nimer et al., 2016). A sufficient number of colonies were transferred from a pure culture using a sterile brush (Albispro.com, Kalisz, Poland) or applicator stick (Key Scientific Products, USA) into a 1275 mm transparent plastic (polystyrene) test tube (Falcon®, Deutschland) containing 3.0 ml of 0.45% NaCl (pH 4.5–7.0) (Baxter Ltd., USA), which is a sterile saline solution. A Densi Chek turbidity meter was used to measure the turbidity, which was set to 0.50–0.6 Mf (Nimer et al., 2016). The findings were evaluated the next day. Staphylococcus aureus detection via molecular methodsTo detect S. aureus, special primers (Bioneer, Daejeon, South Korea) were designed to amplify the 16s rRNA and the Enterotoxin A genes. Responsible for the encoding SEA gene. The sequences of these primers are presented in Table 1. Bacterial genome DNA extractionIn this study, the genomic DNA of S. aureus was extracted from newly grown bacterial cultures, following the standard protocol described in previous works (Sitthisak, 2011; Vremerã et al., 2011). The PrestoTM Small Genomic DNA Bacteria Kit (Geneaid, China) was employed for this purpose. To ensure the quality and quantity of the extracted DNA, validation was performed using a Nanodrop spectrophotometer®TM 2000 (ThermoFisher, USA). Subsequently, the validated DNA samples were stored at −20°C in a refrigerator until further analysis. The PCRThe targeted 16s rRNA gene (500 bp) was detected by PCR test using the AccuPowerPCR PreMix Kit (Bioneer, Daejeon, South Korea) (Psifidi et al., 2015). For that purpose, 20 µl of the PCR reaction solution was prepared based on the instructions of the kit, including 5 µl of DNA, 2 µl of 10 pmol of F and R primers (Table 1), 9 µl of green®-master mix PCR, and 4 µl of nuclease-free water. The mixture was vortexed quickly before being placed in the thermocycler (MJ-Mini BioRad, USA) to commence the amplifying procedure, which involved denaturation for 1 cycle at 94°C/1 minute. Then 30 cycles of denaturation and annealing at 940°C and 57/30 seconds, respectively, followed by 40 seconds at 720°C. Then, a 7-minutes cycle of the final extension at 720°C. Ten microliters of PCR product were then loaded onto an agarose gel (GeneOn GmbH, Germany) containing three microliters of ethidium bromide (HA Life Science, India). Five ml were drawn from a 100–2000 bp marker (Bioneer, Daejeon, South Korea) at one well. The gel was run for one hour at 100 V and 80 amps. The gel and bands were then looked at under a UV-light-based gel documentary (Alpha Laboratories Ltd., Germany). The positive 16s rRNA samples were presented for the investigation of the virulence gene enterotoxin-A using the specific primers for that purpose (Table 1). The same thermocycler condition were applied for the virulence gene enterotoxin A gene (550 bp) except for an annealing time of 55 seconds utilizing (Monistero et al., 2018). Sequencing of DNAStaphylococcus aureus was detected through DNA sequencing (Morse et al., 2002; Ghebremedhin et al., 2008). After confirming the presence of the 16s rRNA gene, the necessary PCR product was isolated from an agarose gel. Bioneer Ltd. Company (Daejeon, South Korea) received purified gene products for sequencing. The bioEdit tool (Tamura et al., 2021) was employed to analyze mutations and amino acid variations. Subsequently, the sequence files were analyzed using MEGA software version 11 to conduct phylogenetic analysis based on the isolates from the current study and those obtained from GenBank (Tamura et al., 2021). Table 1. Presents the specific primers designed for the 16S rRNA gene and the S. aureus enterotoxin A gene, along with their respective product sizes.
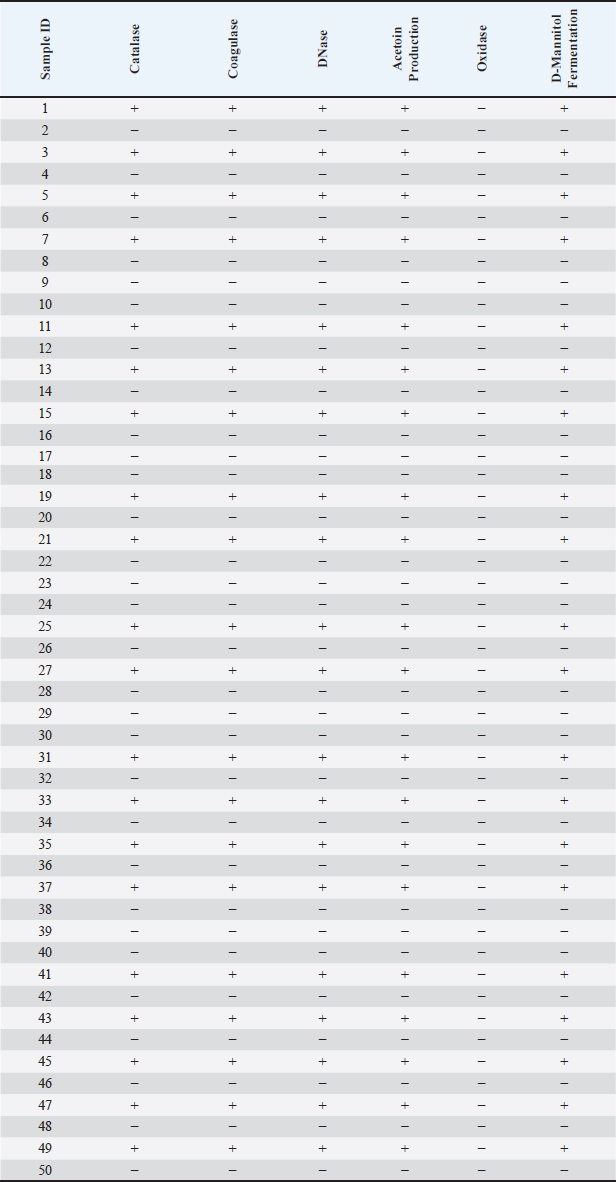
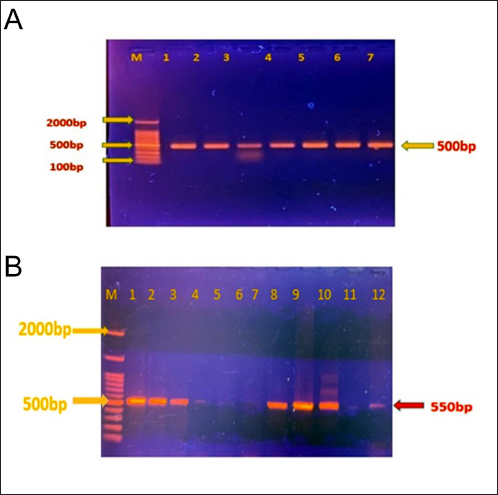
Statistical analysisWe used statistical analysis to implement our study's strategies. We computed sample S. aureus prevalence, investigated the association between biochemical parameters and S. aureus using descriptive statistics, and compared it using the Chi-Square test. We compared VITEK2's S. aureus identification accuracy to PCR using the Kappa statistic. We reported extracted DNA yield and purity using descriptive statistics and compared results among procedures or samples using an ANOVA. Ethical approvalThe techniques employed in this investigation, which included collecting milk samples from animals, were approved by the College of Veterinary Medicine's ethical committee (ref no. 122/2023). All participants agreed to the collection of samples and requested that their identities be kept private. ResultsThe purpose of this study was to examine the genetic makeup and prevalence of S. aureus in milk samples from dairy cows, with an emphasis on the gene A enterotoxin (SEA). Our hypothesis posited that a substantial number of S. aureus isolates would include the SEA gene, and that genetic differences in these isolates might impact their pathogenicity and resistance characteristics. To fulfill our objectives, we obtained Fifty milk specimens from dairy cows, then isolated and identified S. aureus using conventional culture and biochemical testing. Phenotypic features of retrieved isolatesDuring the smear analysis, Gram-stained S. aureus specimens revealed Gram-positive, coccus-shaped cells clustered in irregular clusters, similar to a bunch of grapes. On BPA, the presence of jet-black colonies surrounded by a white halo indicated S. aureus. The results of the biochemical test revealed that the catalase, coagulase, DNase, and acetoin production tests produced a total of 43 positive results out of 50 (Table 2). Nonetheless, the oxidase test continues to show negative findings. As a result, 86% of the S. aureus isolates tested positive for the biochemical tests. VITEK2 system and PCR analysisWe utilized the VITEK2 system to verify and deploy molecular techniques, such as PCR and DNA sequencing, to identify and analyze the bacterial genome. Regarding the results recorded by VITEK2 system, S. aureus was found to be present in 33/50 (66%) of milk samples. Similarly, the 16s rRNA gene, regarded to be relevant for diagnosis, was detected in 25/33 (75%) of the specimens. After finding out what percentage of S. aureus samples were positive for the 16S rRNA gene, we did more tests to see if the enterotoxin A gene was present. Out of the total number of samples, we discovered that 15, which accounts for 60%, tested positive for this gene (Fig. 1A and B). Table 2. Biochemical test criteria of S. aureus Isolates.
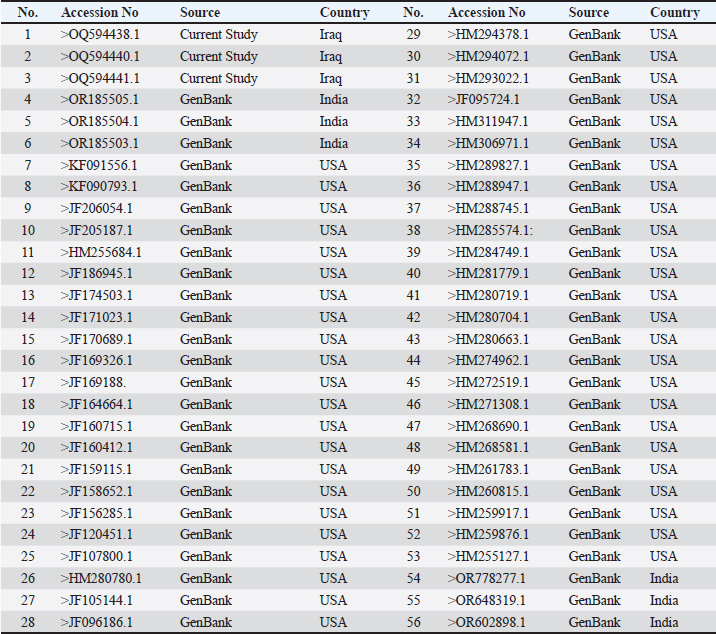
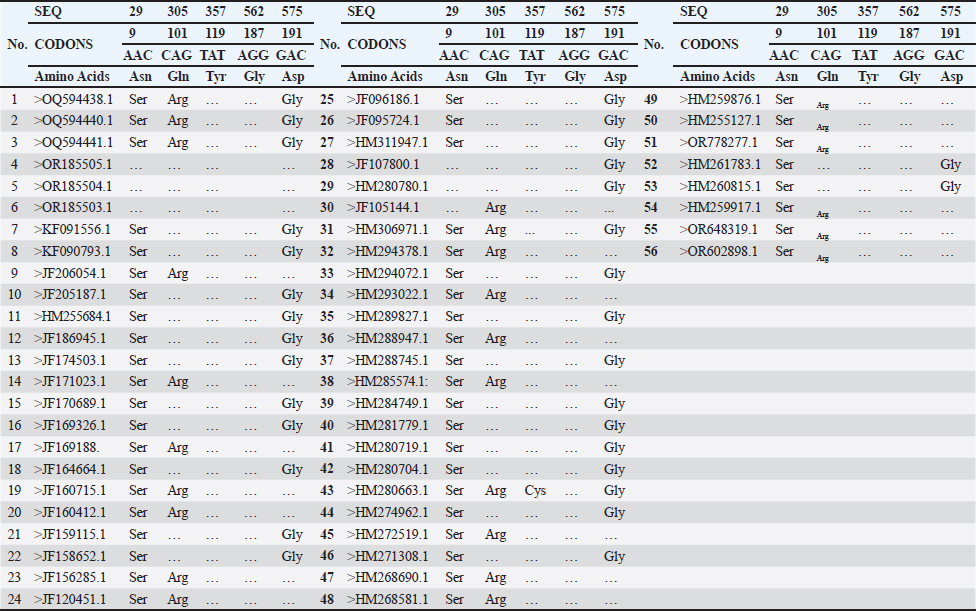
DNA sequencing analysisThe partial sequences of the targeted 16s rRNA genes were submitted to GenBank with the accession codes OQ594438, OQ594440, and OQ594441. Table 3 illustrates the correlation between the strains from the current study isolates and 53 other accession numbers from GenBank.
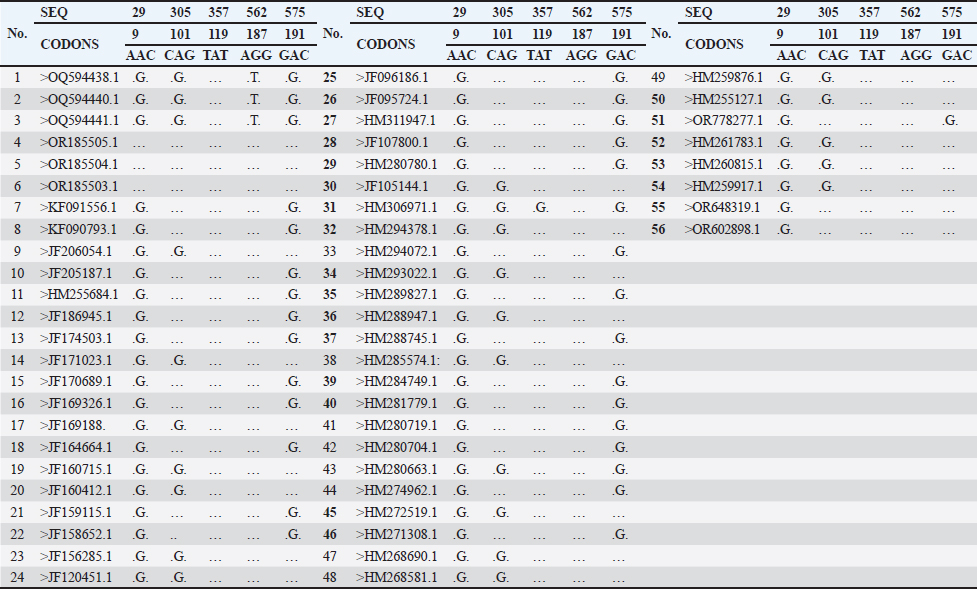
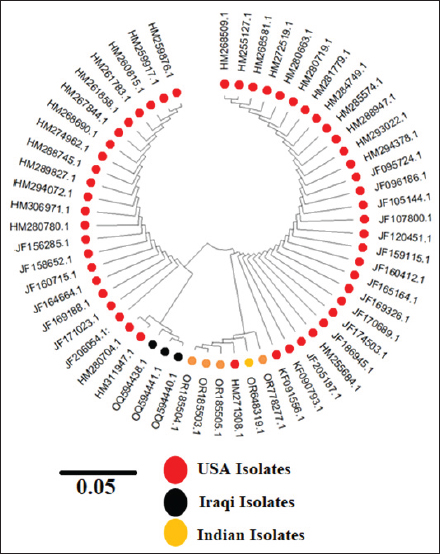
Fig. 1. (A) S. aureus 16S rRNA gene PCR product analysis on an agarose gel electrophoresis. Where M is a marker (2000–100 bp), and lanes (1–7) denote positive dairy cow samples at 500 bp PCR product. (B) S. aureus PCR product analysis of Enterotoxin A gene (SEA), where M: marker (2000–100 bp), lane (1–3), (8–10) positive samples dairy cow samples at 550 bp PCR product. The partial sequencing analysis in our current study identified a mutation within the S. aureus 16S rRNA gene (Tables 4 and 5). We recorded a switch between asparagine (Asn) and serine (Ser). A single nucleotide substitution in the DNA sequence led to this alteration, replacing a specific nucleotide with another, resulting in a change in the encoded amino acid during translation. For example, a change from the DNA sequence AAC (coding for asparagine) to AGC (coding for serine) at the mRNA level would result in this amino acid change. Point mutations in the DNA sequence typically cause substitutions between glutamine (Gln) and arginine (Arg) or tyrosine (Tyr) and cysteine (Cys) in the genetic code of S. aureus. These mutations substituted a single nucleotide, leading to the incorporation of a different amino acid into the protein during translation. These changes could have different effects on different parts of S. aureus' biology depending on where they happened and what roles the proteins they changed played. To figure out what these mutations did, we had to do a lot of genetic testing to see how they changed the functions of bacterial proteins and how they affected S. aureus's body and how it interacted with its surroundings. Figure 2 revealed the genetic characteristics and phylogenetic relationship between Iraqi S. aureus isolates and other genbank isolates (especially Indian and USA isolates). This genetic similarity, which resulted from a common ancestor or genetic exchange events, can explain the global spread of S. aureus through human travel and trade. If the isolates shared antibiotic resistance genes or mutations, this could have indicated a shared evolutionary pressure due to the widespread use of antibiotics in healthcare settings around the world. It was possible that human movement, such as travel, immigration, or animal transport, had facilitated the transmission of S. aureus strains between Iraq, India, and the USA. It is believed that cooperation between research or clinical institutions in different countries will lead to the sharing of bacterial isolates for study or treatment, which contributes to understanding the observed association between isolates from different geographical regions. Table 3. Accession codes employed in this study to conduct the phylogenetic analysis.
DiscussionRaw (unpasteurized) milk poses a significant risk to public health because it may contain S. aureus and associated enterotoxins. Our research focused mainly on the suspected role of S. aureus enterotoxins (SEA) in causing mastitis in Iraqi dairy cows. To achieve this goal, the use of the PCR test is necessary and decisive (Sharma et al., 2000). In this study, 86% (43/50) of the milk samples tested positive for catalase, coagulase, DNase, and acetoin. This showed that S. aureus was present. The oxidase test was negative, which is consistent with the specific features of this bacteria (Abbas et al., 2014). Iraq conducted several research studies on cow's milk samples to investigate the presence of various pathogens, including staphylococci (Khudaier et al., 2013; Hasan and Hoshyar, 2019; Sheet, 2022). In their 2019 investigation, Hasan and Hoshyar discovered that 70% of the samples contained S. aureus isolates that possessed the SEA gene. They emphasized that SEA gene isolation is critical for investigating S. aureus' development. The research specifically links this gene to a higher occurrence of food poisoning. Also, Tong et al. (2015) emphasize the importance of studying and understanding bacterial population control (Tong et al., 2015). While, 33 out of 50 milk samples, or 66%, tested positive for S. aureus using the Vitek-2 technology. Subsequent investigations employing the identical methodology also noted this proportion, or a similar approximation. Table 4. A total of 24 places variations found in multiple sequence alignment of field S. aureus isolates from the current investigation [>OQ594438.1, > OQ594440.1, and > OQ594441.1) and 53 GenBank nucleotide sequences.
Table 5. Amino acids substitution mutations recorded in the study isolates [>OQ594438.1, > OQ594440.1, and > OQ594441.1] compared to the 53 genbank isolates of S. aureus isolated from different sources.
Fig. 2. The phylogenetic tree of S. aureus based on partial sequences of the 16s rRNA gene that classifies strains by host and geographic location; our isolates are colored black. Molecular identification using 16s rRNA revealed a percentage of 25/33 (75%) of the total 33 positive that diagnostic by VITEK-2 (Kim et al., 2008). In this study, PCR testing showed that some S. aureus isolates do not have enterotoxin genes. Several factors contribute to the absence of enterotoxin genes in some S. aureus isolates (Bianchi et al., 2014; Muş et al., 2023). First, according to various studies, S. aureus shows a high level of genetic variation, which allows it to adapt to its surrounding environment (Adame-Gómez et al., 2020). According to recent studies, S. aureus has unique gene sets, like enterotoxin genes, which are crucial as they are associated with inflammation and necrosis (Varshney et al., 2009). Second, these genes can be moved by mobile genetic elements like plasmids, transposons, or prophages (Malachowa and DeLeo, 2010). This causes heterogeneity in the presence of enterotoxin genes, depending on whether an isolate received these mobile elements (Malachowa and DeLeo, 2010). Moreover, selective pressure plays a role in promoting bacterial proliferation during food poisoning, as enterotoxin genes continue to increase in some cases while gradually disappearing in others (Medved’ová et al., 2017). Finally, environmental factors influence enterotoxin gene expression, and some isolates may have silent or non-functional genes due to mutations or regulatory mechanisms. These variables all contribute to differences in the presence of enterotoxin genes across S. aureus strains (Krismer et al., 2014). The sequencing and phylogenetic tree revealed that four isolates received accession numbers for OQ594438, OQ594439, OQ594440, and OQ594441. Our phylogenetic analysis of S. aureus isolates from bovine mastitis cases exist in numerous clades with clear relationships between them. This results disagree with Pizauro et al. (2021) who indicated that the phylogenetic analysis of S. aureus from bovine mastitis cases showed that the clinical and subclinical isolates exist in numerous clades with no clear relationships between them. The phylogeny based on the 16S RNA gene showed that Staphylococcus spp are all closely related to one another and have a common ancestor that causes mastitis. In most cases, subclinical and clinical strains of the same species belonged to the same lineage. Changes to certain amino acid sequences in S. aureus could have a big effect on the formation of galaxies and, by extension, on their ability to control bacterial activity (Canfield et al., 2013). Amino acid substitutions in S. aureus can have a major impact on bacterial pathogenicity (Herron et al., 2002). Amino acids are essential for protein function, and any alteration in the amino acid sequence can cause major changes in the structure and function of bacterial proteins, impacting various aspects of bacterial pathogenesis. These characteristics include bacteria's ability to create toxins, resistance to antibiotics, and ability to cling to and enter host cells (Ng and Henikoff, 2006). It is believed that the amino acids found in enzyme and toxin-active sites are some of the most significant (Pons et al., 2019). Replacing it can have a significant impact on the protein's function. Mutations in toxin-encoding genes, for example, can alter bacteria's ability to create toxins. Also, the amino acids in areas where antibiotics interact are very important. Changes in antibiotic-targeted proteins, like the changes that happen in PBP2a in methicillin-resistant S. aureus, can make bacteria resistant to antibiotics (Goeders and Van Melderen, 2014). Furthermore, amino acids that help bacteria adhere to host cells have a significant impact. Mutations in the proteins that allow bacteria to connect to and enter host cells can impair the bacteria's capacity to cause infection. Changes in surface proteins, such as protein A, can influence bacterial adherence to host cells (Ozma et al., 2022). Biochemical analysis and functional investigations of impacted proteins can help to understand how these mutations alter bacterial pathogenicity (Price et al., 2018). Various factors may contribute to the evolutionary similarities between Iraqi, Indian, and American strains of S. aureus that cause mastitis in cows. For instance, genetic interchange among various lineages might happen due to human migration and the flow of culture and trade between nations (Lakhundi and Zhang, 2018). Furthermore, the presence of common environmental pressures, such as antibiotic usage and comparable climatic conditions, could result in the dissemination and development of resistant strains in many geographical areas (Nübel et al., 2008). It is plausible that the adoption of comparable farming methods and the sharing of genetic resources across various populations might potentially enhance the genetic resemblance among breeds. Therefore, the impact of environmental and anthropogenic variables across different geographic regions may be responsible for the similarities in the evolutionary and transmission patterns of these strains (Feng et al., 2008). After establishing the percentage of S. aureus samples that tested positive for the 16 gene, we conducted additional testing to detect the presence of the enterotoxin A gene. Out of the total number of samples, we discovered that 15, which accounts for 60%, tested positive for this gene. ConclusionThe current study found that 66% of milk samples were positive for S. aureus when detected by the VITEK-2 platform, suggesting its extensive distribution between dairy cows. Despite the PCR test revealing only 75% of the samples as positive, the enterotoxin gene diagnosis identified S. aureus. Among these samples, the study showed that 60% of the Staphylococcus isolates contained this enterotoxin gene, which means that this gene is common in S. aureus, which strengthens the hypothesis that this gene is involved in causing mastitis in dairy cows. The phylogenetic results in this study show a genetic similarity between the strains isolated in Iraq and those isolated in India and the United States, indicating common origins or genetic exchange between these geographically different strains. This genetic similarity could be a result of the transmission of bacteria facilitated by travel, trade, and the transportation of animals and animal products. Understanding these genetic links and identifying factors that influence the distribution and evolution of S. aureus strains is important for guiding public health policies and taking necessary preventive actions to limit the spread of infections and antibiotic resistance. AcknowledgmentWe would like to convey our gratitude and appreciation to the proprietors of dairy cattle farms for supplying the essential samples that facilitated the completion of our research. Additionally, we extend our thanks and appreciation to the College of Veterinary Medicine, Al-Qadisiyah University, for their technical assistance, including the provision of laboratories and scientific support, which were crucial for the research. Conflict of interestThere are no conflicts of interest between the authors and the subject matter of the paper. Author ContributionsMohammed Mahdi Yaseen1 and Hassan Al-Karagoly were concerned with conceptualization and methodology; Mohammed Mahdi Yaseen and Alaa Jawad conducted formal analysis; Alaa Jawad, Hassan Al-Karagoly and Sabreen Noori Dagman were responsible for investigation, data curation, and study validation; Mohammed Mahdi Yaseen and Sabreen Noori Dagman were involved in the visualization and original draft preparation; Hassan Al-Karagoly worked on writing review and editing and assumed supervisory responsibilities; Alaa Jawad and Hassan Al-Karagoly were followed project administration. All authors gave approval to the final version of the manuscript. FundingThis research did not receive specific funding. All authors were contributing to supporting this work in a self-supporting manner. Authors gave the authority to the author (Alaa Jawad) for covering the costs of publication. Data availabilityData employed for verifying the outcomes of this investigation are accessible upon request from the corresponding author. ReferencesAbbas, B.A., Khudor, M.H. and Hanoon, B.M. 2014. Isolation and identification of Staphylococcus aureus from bovine and the detection of its coagulase gene (coa) using polymerase chain reaction (PCR). Sci. Res. Ess. 9(20), 864868. Adame-Gómez, R., Castro-Alarcón, N., Vences-Velázquez, A., Toribio-Jiménez, J., Pérez-Valdespino, A., Leyva-Vázquez, M.A. and Ramírez-Peralta, A. 2020. Genetic diversity and virulence factors of S. aureus isolated from food, humans, and animals. Int. J. Microbiol. 2020(1), 1048097. Alara, J.A. and Alara, O.R. 2024. An overview of the global alarming increase of multiple drug resistant: a major challenge in clinical diagnosis. Infect Disord Drug Targets 24(3), 2642. Algammal, A.M., Enany, M.E., ElTarabili, R.M., Ghobashy, M.O. and Helmy, Y.A. 2020. Prevalence, antimicrobial resistance profiles, virulence and enterotoxinsdeterminant genes of MRSA isolated from subclinical bovine mastitis in Egypt. Pathogens 9(5), 362. Anderson, M.J., Lin, Y.C., Gillman, A.N., Parks, P.J., Schlievert, P.M. and Peterson, M.L. 2012. Alphatoxin promotes Staphylococcus aureus mucosal biofilm formation. Front. Cell. Infect. Microbiol. 2, 64. Bianchi, D., Gallina, S., Bellio, A., Chiesa, F., Civera, T. and Decastelli, L. 2014. Enterotoxin gene profiles of Staphylococcus aureus isolated from milk and dairy products in Italy. Lett. Appl. Microbiol. 58(2), 190196. Canfield, G.S., Schwingel, J.M., Foley, M.H., Vore, K.L., Boonanantanasarn, K., Gill, A.L., Sutton, M.D. and Gill, E.R. 2013. Evolution in fast forward: a potential role for mutators in accelerating Staphylococcus aureus pathoadaptation. J. Bacteriol. 195(3), 615628. Dego, O.K., Van Dijk, J. and Nederbragt, H. 2002. Factors involved in the early pathogenesis of bovine Staphylococcus aureus mastitis with emphasis on bacterial adhesion and invasion. Rev. Vet. Q. 24(4), 181198. Feng, Y., Chen, C.J., Su, L.H., Hu, S., Yu, J. and Chiu, C.H. 2008. Evolution and pathogenesis of Staphylococcus aureus : lessons learned from genotyping and comparative genomics. FEMS Microbiol. Rev. 32(1), 2337. Foster, T.J. 2020. Surface proteins of Staphylococcus epidermidis. Front. Microbiol. 11, 566153. Foster, T.J., Geoghegan, J.A., Ganesh, V.K. and Höök, M. 2014. Adhesion, invasion and evasion: the many functions of the surface proteins of Staphylococcus aureus . Nature Rev. Microbiol. 12(1), 4962. Ghebremedhin, B., Layer, F., Konig, W. and Konig, B. 2008. Genetic classification and distinguishing of Staphylococcus species based on different partial gap, 16S rRNA, hsp60, rpoB, sodA, and tuf gene sequences. J. Clin. Microbiol. 46(3), 10191025. Goeders, N. and Van Melderen, L. 2014. Toxinantitoxin systems as multilevel interaction systems. Toxins (Basel) 6(1), 304324. Guimarães, F., Manzi, M., Joaquim, S., RichiniPereira, V. and Langoni, H. 2017. Outbreak of methicillinresistant Staphylococcus aureus (MRSA)associated mastitis in a closed dairy herd. J. Dairy Sci. 100(1), 726730. Guo, Y., Song, G., Sun, M., Wang, J. and Wang, Y. 2020. Prevalence and therapies of antibioticresistance in Staphylococcus aureus . Front. Cell. Infect. Microbiol. 10, 107. Habib, F., Rind, R., Durani, N., Bhutto, A.L., Buriro, R.S., Tunio, A., Aijaz, N., Lakho, S.A., Bugti, A.G. and Shoaib, M. 2015. Morphological and cultural characterization of Staphylococcus aureus isolated from different animal species. J. Appl. Environ. Biol. Sci. 5(2), 1526. Hasan, N.S. and Hoshyar, D.F. 2019. Detection of enterotoxigenic Staphylococcus aureus strains in raw milk of cows reared in Erbil province, Iraq. Zanco. J. Pure Appl. Sci. 31(4), 5060. Herron, L.L., Chakravarty, R., Dwan, C., Fitzgerald, J.R., Musser, J.M., Retzel, E. and Kapur, V. 2002. Genome sequence survey identifies unique sequences and key virulence genes with unusual rates of amino acid substitution in bovine Staphylococcus aureus . Infect. Immu. 70(7), 39783981. Ibrahim, N. 2017. Review on mastitis and its economic effect. Can. J. Sci. Res. 6(1), 1322. Khudaier, B.Y., Abbas, B.A. and Khudaier, A.M. 2013. Detection of methicillin resistant Staphylococcus aureus isolated from human and animals in Basrah province, Iraq. MRVSA 2, 1221. Kim, M., Heo, S.R., Choi, S.H., Kwon, H., Park, J.S., Seong, M.W., Lee, D.H., Park, K.U., Song, J. and Kim, E.C. 2008. Comparison of the microscan, VITEK 2, and crystal GP with 16S rRNA sequencing and MicroSeq 500 v2.0 analysis for coagulasenegative staphylococci. BMC Microbiol. 8, 17. Kimberlin, D., Fischer, M., Long, S.S. and Prober, C.G. 2022. Principles and practice of pediatric infectious diseases ebook. Elsevier Health Sciences. Kiran, F., Karaca, B. and Erdoğan, A.F. 2022. Microbial biofilms in veterinary medicine. Ankara Üniversitesi Veteriner Fakültesi Dergisi 70(1), 107114. Krismer, B., Liebeke, M., Janek, D., Nega, M., Rautenberg, M., Hornig, G., Unger, C., Weidenmaier, C., Lalk, M. and Peschel, A. 2014. Nutrient limitation governs Staphylococcus aureus metabolism and niche adaptation in the human nose. PLoS Pathogens 10(1), e1003862. Lakhundi, S. and Zhang, K. 2018. Methicillinresistant Staphylococcus aureus : molecular characterization, evolution, and epidemiology. Clin. Microbiol. Rev. 31(4), 10–1128; doi:10.1128/cmr.0002000018. Le Maréchal, C., Seyffert, N., Jardin, J., Hernandez, D., Jan, G., Rault, L., Azevedo, V., François, P., Schrenzel, J. and Van De Guchte, M. 2011. Molecular basis of virulence in Staphylococcus aureus mastitis. PLoS One 6(11), e27354. Malachowa, N. and DeLeo, F.R. 2010. Mobile genetic elements of Staphylococcus aureus . Cell. Mol. Life Sci. 67, 30573071. Medved’ová, A., Havlíková, A. and Valík, L. 2017. Staphylococcus aureus enterotoxin production in relation to environmental factors. The rise of virulence and antibiotic resistance in Staphylococcus aureus. London, UK; IntechOpen, pp: 145167. Monistero, V., Graber, H.U., Pollera, C., Cremonesi, P., Castiglioni, B., Bottini, E., CeballosMarquez, A., LassoRojas, L., Kroemker, V. and Wente, N. 2018. Staphylococcus aureus isolates from bovine mastitis in eight countries: genotypes, detection of genes encoding different toxins and other virulence genes. Toxins (Basel) 10(6), 247. Morse, R., O'Hanlon, K. and Collins, M.D. 2002. Phylogenetic, amino acid content and indel analyses of the beta subunit of DNAdependent RNA polymerase of grampositive and gramnegative bacteria. Int. J. Syst. Evol. Microbiol. 52(5), 14771484. Muş, T.E., Cetinkaya, F., Soyutemiz, G.E. and Erten, B. 2023. Toxigenic genes of coagulasenegative staphylococci and Staphylococcus aureus from milk and dairy. J. Agricul. Sci. 29(4), 924932. Ng, P.C. and Henikoff, S. 2006. Predicting the effects of amino acid substitutions on protein function. Ann. Rev. Genom. Human Genet. 7, 6180. Nimer, N., AlSaa'da, R. and Abuelaish, O. 2016. Accuracy of the VITEK 2 system for a rapid and direct identification and susceptibility testing of gramnegative rods and grampositive cocci in blood samples. Eastern Mediterranean Health J. 22(3), 193200. Nübel, U., Roumagnac, P., Feldkamp, A., Song, J.H., Ko, K.S., Huang, Y.C., Coombs, G., Ip, M., Westh, H. and Skov, R. 2008. Frequent emergence and limited geographic dispersal of methicillinresistant Staphylococcus aureus . Proc. Nat. Acad. Sci. 105(37), 1413014135. Ozma, M.A., Khodadadi, E., Rezaee, M.A., Asgharzadeh, M., Aghazadeh, M., Zeinalzadeh, E., Ganbarov, K. and Kafil, H.S. 2022. Bacterial proteomics and its application in pathogenesis studies. Curr. Pharma. Biotechnol. 23(10), 12451256. Peton, V. and Le Loir, Y. 2014. Staphylococcus aureus in veterinary medicine. Infect. Genet. Evol. 21, 602615. Pizauro, L.J.L., de Almeida, C.C., Silva, S.R., MacInnes, J.I., Kropinski, A.M., Zafalon, L.F., de Avila, F.A. and de Mello Varani, A. 2021. Genomic comparisons and phylogenetic analysis of mastitisrelated Staphylococci with a focus on adhesion, biofilm, and related regulatory genes. Sci. Rep. 11(1), 17392. Pons, B.J., Vignard, J. and Mirey, G. 2019. Cytolethal distending toxin subunit B: a review of structure–function relationship. Toxins (Basel), 11(10), 595. Price, M.N., Wetmore, K.M., Waters, R.J., Callaghan, M., Ray, J., Liu, H., Kuehl, J.V., Melnyk, R.A., Lamson, J.S. and Suh, Y. 2018. Mutant phenotypes for thousands of bacterial genes of unknown function. Nature 557, 503–509. Psifidi, A., Dovas, C.I., Bramis, G., Lazou, T., Russel, C.L., Arsenos G. and Banos, G. 2015. Comparison of eleven methods for genomic DNA extraction suitable for largescale wholegenome genotyping and longterm DNA banking using blood samples. PLoS One 10(1), e0115960. Salam, M.A., AlAmin, M.Y., Salam, M.T., Pawar, J.S., Akhter, N., Rabaan, A.A. and Alqumber, M.A. 2023. Antimicrobial resistance: a growing serious threat for global public health. In: Healthcare. MDPI: pp: 1946. Shafiq, M., Zeng, M., Permana, B., Bilal, H., Huang, J., Yao, F., Algammal, A.M., Li, X., Yuan, Y. and Jiao, X. 2022. Coexistence of bla ndm–5 and tet (x4) in international highrisk Escherichia coli clone st648 of human origin in China. Front. Microbiol. 13, 1031688. Sharma, N.K., Rees C.E. and Dodd, C.E. 2000. Development of a singlereaction multiplex PCR toxin typing assay for Staphylococcus aureus strains. Appl. Environ. Microbiol. 66(4), 13471353. Sheet, O.H. 2022. Molecular detection of mecA gene in methicillinresistant Staphylococcus aureus isolated from dairy mastitis in Nineveh Governorate, Iraq. Iraqi J. Vet. Sci. 36(4), 939943. Singh, P. and Prakash, A. 2008. Isolation of Escherichia coli, Staphylococcus aureus and Listeria monocytogenes from milk products sold under market conditions at Agra region. Acta Agriculturae Slovenica 92(1), 8388. Sipahi, N., Kaya, E., Çelik C. and Pınar, O. 2023. The characterization and betalactam resistance of Staphylococcal community recovered from raw bovine milk. Antibiotics 12(3), 556. Sitthisak, S. 2011. Epidemiology of community associated methicillin resistance Staphylococcus aureus . TMJ, 11(1), 6273. Tamura, K., Stecher, G. and Kumar, S. 2021. MEGA11: molecular evolutionary genetics analysis version 11. Mol. Biol. Evol. 38(7), 30223027. Tong, S.Y., Davis, J.S., Eichenberger, E., Holland, T.L. and Fowler Jr, G.R. 2015. Staphylococcus aureus infections: epidemiology, pathophysiology, clinical manifestations, and management. Clin. Microbiol. Rev. 28(3), 603661 Ullah, N., Nasir, S., Ishaq, Z., Anwer, F., Raza, T., Rahman, M., Alshammari, A., Alharbi, M., Bae, T. and Rahman, A. 2022. Comparative genomic analysis of a Panton–Valentine leukocidinpositive ST22 communityacquired methicillinresistant Staphylococcus aureus from Pakistan. Antibiotics 11(4), 496. Vacheyrou, M., Normand, A.C., Guyot, P., Cassagne, C., Piarroux, R. and Bouton, Y. 2011. Cultivable microbial communities in raw cow milk and potential transfers from stables of sixteen French farms. Int. J. Food Microbiol. 146(3), 253262. Varshney, A.K., Mediavilla, J.R., Robiou, N., Guh, A., Wang, X., Gialanella, P., Levi, M.H., Kreiswirth, B.N. and Fries, B.C. 2009. Diverse enterotoxin gene profiles among clonal complexes of Staphylococcus aureus isolates from the Bronx, New York. Appl. Environ. Microbiol. 75(21), 68396849. Vremerã, T., Iancu, L.S., Logigan, C., Nãstase, E., Miftode, E., Luncã, C. and Dorneanu, O. 2011. Optimization of triplex realtime PCR for detecting Staphylococcus aureus mecA, pvl, and nuc genes. Roum. Arch. Microbiol. Immunol. 70(2), 6973. Worku, M., Belay, G. and Tigabu, A. 2022. Bacterial profile and antimicrobial susceptibility patterns in cancer patients. PLoS One 17(4), e0266919 | ||
| How to Cite this Article |
| Pubmed Style Jawad A, Dagman SN, Yaseen MM, Al-karagoly H. Phylogenetic analysis of Staphylococcus aureus enterotoxin A gene in Iraqi breed cows with bovine mastitis: Implications for disease management. Open Vet J. 2024; 14(7): 1644-1657. doi:10.5455/OVJ.2024.v14.i7.14 Web Style Jawad A, Dagman SN, Yaseen MM, Al-karagoly H. Phylogenetic analysis of Staphylococcus aureus enterotoxin A gene in Iraqi breed cows with bovine mastitis: Implications for disease management. https://www.openveterinaryjournal.com/?mno=201625 [Access: June 02, 2025]. doi:10.5455/OVJ.2024.v14.i7.14 AMA (American Medical Association) Style Jawad A, Dagman SN, Yaseen MM, Al-karagoly H. Phylogenetic analysis of Staphylococcus aureus enterotoxin A gene in Iraqi breed cows with bovine mastitis: Implications for disease management. Open Vet J. 2024; 14(7): 1644-1657. doi:10.5455/OVJ.2024.v14.i7.14 Vancouver/ICMJE Style Jawad A, Dagman SN, Yaseen MM, Al-karagoly H. Phylogenetic analysis of Staphylococcus aureus enterotoxin A gene in Iraqi breed cows with bovine mastitis: Implications for disease management. Open Vet J. (2024), [cited June 02, 2025]; 14(7): 1644-1657. doi:10.5455/OVJ.2024.v14.i7.14 Harvard Style Jawad, A., Dagman, . S. N., Yaseen, . M. M. & Al-karagoly, . H. (2024) Phylogenetic analysis of Staphylococcus aureus enterotoxin A gene in Iraqi breed cows with bovine mastitis: Implications for disease management. Open Vet J, 14 (7), 1644-1657. doi:10.5455/OVJ.2024.v14.i7.14 Turabian Style Jawad, Alaa, Sabreen Noori Dagman, Mohammed Mahdi Yaseen, and Hassan Al-karagoly. 2024. Phylogenetic analysis of Staphylococcus aureus enterotoxin A gene in Iraqi breed cows with bovine mastitis: Implications for disease management. Open Veterinary Journal, 14 (7), 1644-1657. doi:10.5455/OVJ.2024.v14.i7.14 Chicago Style Jawad, Alaa, Sabreen Noori Dagman, Mohammed Mahdi Yaseen, and Hassan Al-karagoly. "Phylogenetic analysis of Staphylococcus aureus enterotoxin A gene in Iraqi breed cows with bovine mastitis: Implications for disease management." Open Veterinary Journal 14 (2024), 1644-1657. doi:10.5455/OVJ.2024.v14.i7.14 MLA (The Modern Language Association) Style Jawad, Alaa, Sabreen Noori Dagman, Mohammed Mahdi Yaseen, and Hassan Al-karagoly. "Phylogenetic analysis of Staphylococcus aureus enterotoxin A gene in Iraqi breed cows with bovine mastitis: Implications for disease management." Open Veterinary Journal 14.7 (2024), 1644-1657. Print. doi:10.5455/OVJ.2024.v14.i7.14 APA (American Psychological Association) Style Jawad, A., Dagman, . S. N., Yaseen, . M. M. & Al-karagoly, . H. (2024) Phylogenetic analysis of Staphylococcus aureus enterotoxin A gene in Iraqi breed cows with bovine mastitis: Implications for disease management. Open Veterinary Journal, 14 (7), 1644-1657. doi:10.5455/OVJ.2024.v14.i7.14 |