| Research Article | ||
Open Vet. J.. 2024; 14(8): 2073-2078 Open Veterinary Journal, (2024), Vol. 14(8): 2073–2078 Research Article Molecular identification of Staphylococcus aureus-related enterotoxin genes in cheese samplesZahira Abdljabbar Al-Zuhairi1, Esraa Taher Muslim1, Orooba Meteab Faja1*, Ziad M. Alkhozai2 and Basima Jasim Mohammed11Collage of Veterinary Medicine, University of Al-Qadisiyah, Al-Diwaniyah City, Iraq 2Collage of Biotechnology, University of Al-Qadisiyah, Al-Diwaniyah City, Iraq *Corresponding Author: Orooba Meteab Faja. Department of Public Health, College of Veterinary Medicine, University of Al-Qadisiyah, Al-Diwaniyah City, Iraq. Email: orooba.faja [at] qu.edu.iq Submitted: 26/06/2024 Accepted: 20/07/2024 Published: 31/08/2024 © 2024 Open Veterinary Journal
ABSTRACTBackground: Dairy products are considered some important sources of various nutritional compounds; however, pathogenic bacterial growth is a critical destructive factor to these products leading to consumer health and system financial crises. Aim: The current study was carried out to identify if there is any presence of Staphylococcus aureus-related enterotoxin genes in cheese samples. Methods: The research included the collection of 35 samples. The samples passed through conventional cultivation processes and a PCR method to detect the presence of icaA, sea, hla, and fnbA enterotoxin genes in these samples. Results: The conventional identification revealed the growth of S. aureus from the cheese samples. The PCR findings recorded the presence of the icaA, sea, hla, and fnbA in 31 (88.5%), 27 (77%), 19 (54%), and 12 (34%), respectively, of cheese samples. The sequencing revealed close similarities with global isolates, which reached up to 98.5% of identity. Conclusion: The current results indicate the presence of enterotoxin genes of S. aureus in high rates in the dairy products examined, which reveals critical problems of food safety due to the possible presence of enterotoxins in consumer dairy products. Keywords: Enterotoxin genes, Food-borne diseases, Food safety, Staphylococcus aureus. IntroductionThe incidence of foodborne diseases has increased over time, posing a significant global human health concern, with an estimated annual occurrence of over 600 million individuals. Foodborne microorganisms, such as Staphylococcus aureus, cause numerous diseases and pose a danger to people health and economic stability due to their detrimental impact on health. Such microorganisms sometimes are present in dairy products and are considered important sources of food-borne diseases. Various pathogenic bacteria have the ability to infect food items during production, processing, preservation, and transportation before they are consumed. Foodborne infections are responsible for causing extensive gastrointestinal diseases, which have been predicted to result in a substantial economic and health impact (Martinović et al., 2016; Carstens et al., 2019; Ling et al., 2019; Faour-Klingbeil and Todd, 2020). Children under the age of five account for over 30% of deaths caused by foodborne illnesses, as reported by the WHO. Foodborne bacteria can lead to a range of symptoms, ranging from mild to severe, including diarrhea and potentially debilitating illnesses including meningitis. Young children are especially susceptible to foodborne infections due to their less robust immune systems. Additionally, they may have an increased propensity to engage in behaviors such as placing their hands in their mouths or consuming food that has been compromised with germs (Scallan Walter et al., 2020; Qiu et al., 2021; Van Puyvelde et al., 2021; Belina et al., 2021; Prata et al., 2021; Akter et al., 2021). Approximately 25% of the US population suffers from foodborne diseases annually, despite the fact that American cuisine is considered one of the cleanest providers of food globally. Several studies have discovered that the occurrence and importance of various foodborne illnesses are influenced by the interactions between disease-causing microbes, humans, food, and the surrounding environment. Foodborne disorders are caused by harmful microbes, including bacteria, viruses, fungi, and parasites. Among these, bacterial microbes are the most frequent causes and can exhibit various traits and activities (Malachowa and DeLeo, 2010; Hoffmann et al., 2021; Ishaq et al., 2021; Jahan et al., 2021; Saravanan et al., 2021; Zarkani and Schikora, 2021; Ge et al., 2022). Several bacteria, including Clostridium botulinum and C. perfringens, have the ability to generate spores and have exceptional heat resistance. For instance, S. aureus and C. botulinum are microorganisms that may generate heat-resistant toxins. Typically, they exhibit mesophilic characteristics, indicating that their optimal temperature range for growth is between 20°C and 45°C. In addition, certain bacteria like Listeria monocytogenes, which are responsible for causing foodborne illnesses, have the ability to last under refrigerated conditions or temperatures below 10°C. These infections can still be transferred to humans (Elbehiry et al., 2017; Söderqvist et al., 2017; Schirone et al., 2017). Staphylococcus aureus enterotoxins exhibit resistance to high temperatures, degrading enzymes, destroying agents, and a broad spectrum of pH levels. Therefore, it remains intact in the gastrointestinal tract, where it can bypass the stomach, and target the intestinal cells. As S. aureus thrives in many harsh environments, it has the potential to contaminate livestock products throughout processing and preparation (Argudín et al., 2010). Dairy products are considered some important sources of various nutritional compounds; however, pathogenic bacterial growth is a critical destructive factor to these products leading to consumer health and system financial crises. The current study was carried out to identify if there is any presence of S. aureus-related enterotoxin genes in cheese samples. Materials and MethodsSamplesThe research included the collection of 35 samples, which were divided equally into cheese samples. Staphylococcus aureus was isolated from cow milk cheese purchased from a local market located in Al-Diwaniyah City, Iraq, using the microbiological procedures outlined in the Ministry of Food and Drug Safety’s published guidelines (2018). The samples from cheese were diluted by a factor of ten in a solution of 0.1% peptone water prior to their inoculation onto agar plates nutrient agar, blood agar (BA), and brain heart infusion agar utilizing the pour plate technique. The plates were then incubated at a temperature of 37°C for a duration of 24 hours. Subsequently, the colonies were sub-cultivated to get a culture that was free from impurities. The isolates were preserved in the event that the microorganisms were required to be identified. The strains were determined based on their Gram staining, morphological features of colonies, biochemical features (including sugar fermentation), as well as catalase and coagulase analyses. The isolates were confirmed using a 16S rRNA gene-originated PCR test. This investigation involved the testing of 70 isolates of S. aureus recovered from 35 cheese samples. Detection of enterotoxin-based genesDNA extraction The isolation of genomic DNA from bacterial isolates (SAD6, SAD7, SAD8, SAD9, and SAD10) was performed using the methodology outlined in the Wizard Genomic DNA Purification Kit, Promega. To perform the RNA lysis, 3 μl of RNase solution was used. The mixture was subjected to incubation at a temperature of 37°C for 15 minutes. In the case of protein precipitation, 200 μl of Protein Precipitation Solution was introduced into the cell lysate. Subsequently thoroughly blended by the vortex. Subsequently, the sample was subjected to incubation at a low temperature of –30°C. Subsequently, the sample was subjected to centrifugation at a speed of 13,000 revolutions per minute for a duration of 5 minutes. The DNA was NanoDrop-estimated. PCR for bacterial genotyping The PCR depended on the 16S rRNA gene as a target by utilizing the primer set; 27F: AGAGTTTGATCATGGCTCAG and 1492R: GGTTACCTTGTTACGACTT (Jarraud et al., 2002). The PCR reaction of 50 µl included 10–100 ng DNA template, 20 mM Tris-HCl (pH >8), 50 mM KCl, 1.5 mM MgCl2, 2 μl each DNTP, 0.4 μM per primer, and 2.5 U DNA polymerase. The PCR thermal was: 94°C elongated for 120 seconds for a primary denaturation, 35 repeats of a main denaturing step, an annealing process, and a main-extension program at 94°C elongated for 60 seconds, 55°C elongated for 90 seconds, and 72°C elongated for the 60 seconds, respectively, and 72°C elongated for 180 seconds of a final-extension step. Then, the purified PCR products were sent to Sanger-method-based sequencing in Korea. The resulting data were processed and analyzed using NCBI websites and MEGA X software for the alignment and phylogenetic tree building. Table 1. Primers used in the study.
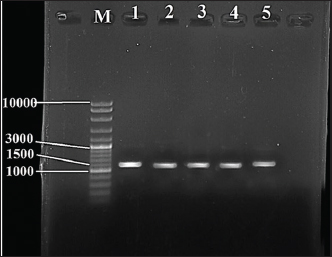
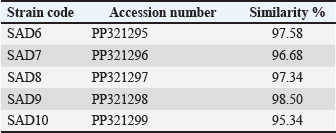
Fig. 1. Image of 16S rRNA-based PCR-1%-agarose gel electrophoresis of nine isolates of Staphylococcus aureus of cheese samples. M: ladder; Lane 1 to 5: SAD6, SAD7, SAD8, SAD9, and SAD10, respectively. PCR for the enterotoxin genes The samples were passed through a conventional PCR method to detect the presence of icaA, sea, hla, and fnbA enterotoxin genes in the samples. The Accupower PCR PreMix kit used was purchased from (Bioneer, Daejeon, Korea). The thermal conditions were 95°C for 3–5 minutes for the first step. In 30–35 cycles, 95°C for 30 seconds for the denaturation, 52°C and 57°C for 30–45 seconds for the annealing, and 72°C for 1 minute for the extension, and 72°C for 5–10 minutes for the final extension. The primers are listed in Table 1. Ethical approval The study has been approved by the ethics committee of The College of Veterinary Medicine, University of Al-Qadisiyah, Al-Diwaniyah City, Iraq. ResultsThe cultivation and conventional identification revealed the growth of S. aureus from the cheese samples. For the genotyping, the PCR amplified at 1,500 bp (Fig. 1). The PCR findings recorded the presence of the icaA, sea, hla, and fnbA in 31 (88.5%), 27 (77%), 19 (54%), and 12 (34%), respectively, of cheese samples (Fig. 2). The amplification of these genes was successfully recorded in cheese samples. The sequencing of the isolates revealed close relationships of the current isolates with isolates deposited previously in the GeneBank by researchers from around the world (Table 2 and Fig. 3). DiscussionOver 20 enterotoxin and enterotoxin-like coding genes have been identified as playing a substantial role in various phases of host colonization, gastrointestinal infections, the invasion of skin and mucus, and host defense systems. Food has a crucial role in the transmission and dissemination of antibiotic resistance genes. Staphylococcus aureus harbors pathogenicity genes on movable chromosomal components, including plasmids and staphylococcal pathogenic islands. These components have the ability to transmit horizontally across different strains. Enterotoxins from S. aureus in cheese can overcome body defense mechanisms, such as resistance to breakdown by digestion enzymes and acids, heat resistance, and encouraging the production of high amounts of cytokines (Malachowa and DeLeo, 2010; Wu et al., 2011; Alibayov et al., 2014).
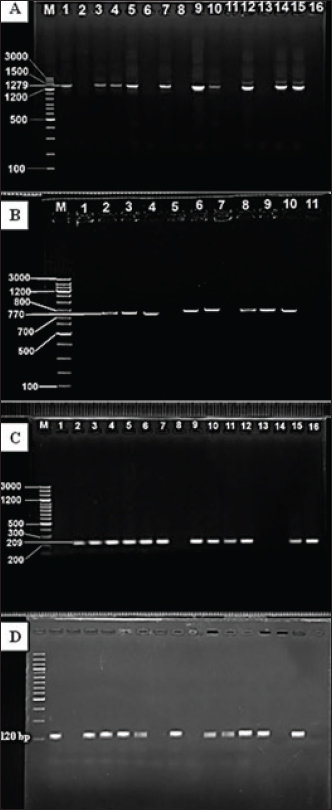
Fig. 2. Image of agarose gel electrophoresis based on the PCR-directed amplification of Staphylococcus aureus enterotoxin genes from cheese. (A) fnbA gene; (1,3,4,5,7,9,10,12,14,15): Positive PCR samples. (B) icaA gene; (2,3,4,6,8,9,10): Positive PCR samples (C) A hla; (2,3,4,5,6,7,9,10,11,12,15,16): Positive PCR samples. (D) sea gene; (1,3,4,5,8,10,11,12,13,15): Positive PCR samples. M: 100bp ladder. Table 2. Current and GeneBank-based comparison of Staphylococcus aureus isolates sequences of cheese samples.
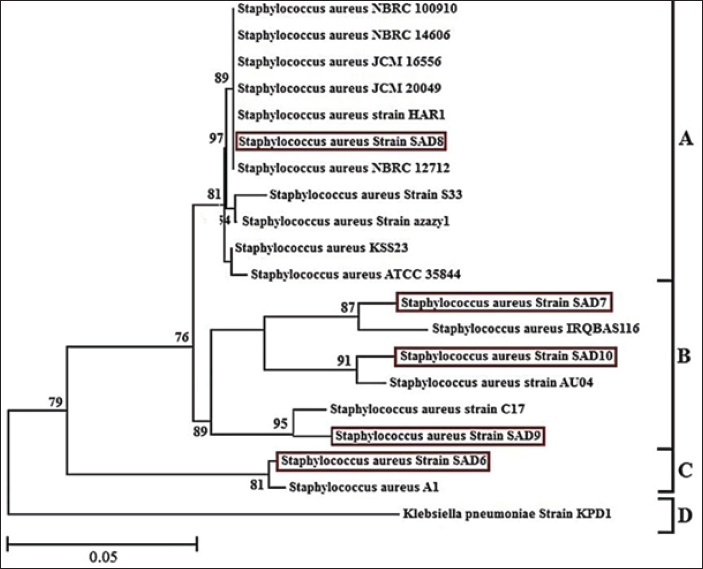
Fig. 3. Phylogenetic tree based on the 16S rRNA gene for cheese-sample-isolated Staphylococcus aureus. Mashouf et al (2015) found that S. aureus strains with enterotoxigenic properties most commonly exhibited Staphylococcal Enterotoxin A. Furthermore, the seg gene was the prevailing enterotoxin among their newly discovered enterotoxin genes. The sea gene was identified in 25.5% (n=25), and it was also found to exist alongside with the seg gene in 14.3% (n=14) of cases. Our analysis identified the presence of the strain exclusively in one sample isolated from cheese. This outcome was congruent with the study conducted by Alibayov et al. (2014) In the same way, Cramton et al. (1999) noted that even if S. aureus strains include the ica locus, they may still be unable to generate biofilm in a laboratory setting due to the fact that biofilm development on non-living surfaces is greatly influenced by the specific circumstances under which the bacteria are grown. A study conducted by Arciola et al. (2001 a,b) revealed that red forms of slime-positive S. aureus were shown to be deficient in the icaA and icaD. They proposed that the altered physical characteristics may be linked to the complete removal of the ica locus. The study conducted by Fooladi et al. (2010) revealed that around 32% of the dairy products examined were found to be contaminated. Unconventional milking practices, such as utilizing unclean milking equipment or bare hands, can also transmit S. aureus. The majority of the isolates examined in this investigation included the sea gene, with the sed gene being present in subsequent frequency. Additional researchers have proposed the prevalence of the sea (Imani Fooladi et al., 2010; Kamarehei et al., 2013; Seyoum et al., 2016). The findings obtained from the current study revealed that many enterotoxin genes can coexist inside a single isolate. This is necessary to reduce the hazard of S. aureus contamination in foods and improve health services, applying other tracks of controlling at all costumes of food processing. Pelisser et al. (2009) informed the needed number of live bacteria at least 106 colony-forming units per gram (CFU/g) of S. aureus for producing of enterotoxins (Argudín et al., 2010). We have to bear this issue in mind while we are facing with food hygiene issues and we have to use many food preservative techniques in order to control bacterial growth in foods. To investigate the grouping of the isolates of the present study, they were compared with the results of other global and Iraqi isolates which published in PubMed. Travelling efforts of humans transport many bacteria from one country to another and import of animals and migrating birds which many all the time to different areas around the world influences the evolution process for many bacteria physiology and leads to different genomic differences of isolates (Jarraud et al., 2002; Aguiar et al., 2024; Aleksic et al., 2024). The current findings tell us that the genomes of enterotoxins genes of S. aureus found in a high rate of the dairy products samples which tested in the current study, showing some critical situations about food safety and surely consumers are threatened with the enterotoxins possible presence in the dairy products which they are consuming. AcknowledgmentsNone. Conflict of interestNo conflict of interest is presented in the current study. FundingThis work was self-funded Data availabilityThe datasets used and/ or analyzed during the current study are available from the corresponding author (Orooba Meteab Faja) on reasonable request. Authors’ contributionsAll authors participated equally in this work. ReferencesAguiar, R. A. C., Ferreira, F. A., Rubio Cieza, M. Y., Silva, N. C. C., Miotto, M., Carvalho, M. M., Bazzo, B. R., Botelho, L. A. B., Dias, R. S. and De Dea Lindner, J. 2024. Staphylococcus aureus Isolated From Traditional Artisanal Raw Milk Cheese from Southern Brazil: Diversity, Virulence, and Antimicrobial Resistance Profile. J. Food Prot. 87(6), 100285. Akter, R., Rahman, M. H., Bhattacharya, T., Kaushik, D., Mittal, V. and Parashar, J. 2021. Novel coronavirus pathogen in humans and animals: an overview on its social impact, economic impact, and potential treatments. Environ. Sci. Pollut. Res. Int. 28(48), 68071–68089. Aleksic, B., Udovicki, B., Kovacevic, J., Miloradovic, Z., Djekic, I., Miocinovic, J., Tomic, N. and Smigic, N. 2024. Microbiological assessment of dairy products produced by small-scale dairy producers in Serbia. Foods (Basel, Switzerland) 13(10), 1456. Alibayov, B., Zdeňková, K., Purkrtová, S., Demnerová, K., and Karpíšková, R. 2014. Detection of some phenotypic and genotypic characteristics of Staphylococcus aureus isolated from food items in the Czech Republic. Ann. Microbiol. 64(4), 1587–1596. Arciola, C. R., Baldassarri, L., and Montanaro, L. 2001. Presence of icaA and icaD genes and slime production in a collection of Staphylococcal strains from Catheter-Associated Infections. J. Clin. Microbiol. 39(6), 2151–2156. Arciola, C. R., Collamati, S., Donati, E., and Montanaro, L. 2001a. A rapid PCR method for the detection of slime-producing strains of Staphylococcus epidermidis and S. aureus in periprosthesis infections. Diagn. Mol. Pathol. 10(2), 130–137. Argudín, M. Á., Mendoza, M. C., and Rodicio, M. R. 2010b. Food poisoning and Staphylococcus aureus Enterotoxins. Toxins (Basel). 2(7), 1751–1773. Belina, D., Hailu, Y., Gobena, T., Hald, T., and Njage, P. M. K. 2021. Prevalence and epidemiological distribution of selected foodborne pathogens in human and different environmental samples in Ethiopia: a systematic review and meta-analysis. One Health Outlook. 3, 19. Carstens, C. K., Salazar, J. K., and Darkoh, C. 2019. Multistate outbreaks of foodborne illness in the United States Associated with fresh produce From 2010 to 2017. Front. Microbiol. 22(10), 2667. Cramton, S. E., Gerke, C., Schnell, N. F., Nichols, W. W., and Götz, F. 1999. The intercellular adhesion (ica) Locus is present in Staphylococcus aureus and is required for biofilm formation. Infect. Immun. 67(10), 5427–5433. Elbehiry, A., Marzouk, E., Hamada, M., Al-Dubaib, M., Alyamani, E. and Moussa, I.M. 2017. Application of MALDI-TOF MS fingerprinting as a quick tool for identification and clustering of foodborne pathogens isolated from food products. New Microbiol. 40(4), 269–278. Faour-Klingbeil, D., and Todd, E. C. D. 2020. Prevention and control of foodborne diseases in Middle-East North African Countries: review of national control systems. Int. J. Environ. Res. Public Health. 17(1), 70. Ge, H., Wang, Y., and Zhao, X. 2022. Research on the drug resistance mechanism of foodborne pathogens. Microb. Pathog. 162, 105306. Hoffmann, S., Ashton, L., and Ahn, J. W. 2021. Food safety: a policy history and introduction to avenues for economic research. Appl. Econ. Perspect. Policy. 43(2), 680–700. Imani Fooladi, A., Tavakoli, H., and Naderi, A. 2010. Detection of enterotoxigenic Staphylococcus aureus isolates in domestic dairy products. Iran J. Microbiol. 2(3), 137–142. Ishaq, A. R., Manzoor, M., Hussain, A., Altaf, J., Rehman, S. U. and Javed, Z. 2021. Prospect of microbial food borne diseases in Pakistan: a review. Braz. J. Biol. 81(4), 940–953. Jahan, N. A., Lindsey, L. L., and Larsen, P. A. 2021. The role of peridomestic rodents as reservoirs for zoonotic foodborne pathogens. Vector Borne Zoonotic Dis. 21(3), 133–148. Jarraud, S., Mougel, C., Thioulouse, J., Lina, G., Meugnier, H. and Forey, F. 2002. Relationships between Staphylococcus aureus genetic background, virulence factors, agr groups (Alleles), and human disease. Infect. Immun. 70(2), 631–641. Kamarehei, F., Ghaemi, E. A., and Dadgar, T. 2013. Prevalence of enterotoxin A and B genes in Staphylococcus aureus isolated from clinical samples and healthy carriers in Gorgan City, North of Iran. Indian J. Pathol. Microbiol. 56(3), 265–268. Ling, Y., Zhu, Y., Fan, H., Zha, H., Yang, M. and Wu, L. 2019. Rapid method for detection of Staphylococcus aureus in feces. J. Biomed. Nanotechnol. 15(6), 1290–1298. Malachowa, N., and DeLeo, F. R. 2010. Mobile genetic elements of Staphylococcus aureus. Cell Mol. Life Sci. 67(18), 3057–3071. Martinović, T., Andjelković, U., Gajdošik, M. Š., Rešetar, D., and Josić, D. 2016. Foodborne pathogens and their toxins. J. Proteomics. 147, 226–235. Mashouf, R. Y., Hosseini, S. M., Mousavi, S. M., and Arabestani, M. R. 2015. Prevalence of enterotoxin genes and antibacterial susceptibility pattern of Staphylococcus aureus strains isolated from Animal Originated Foods in West of Iran. Oman Med. J. 30(4), 283–290. Pelisser, M.R., Klein, C.S., Ascoli, K.R. and Zotti, T.R. 2009. Ocurrence of Staphylococcus aureus and multiplex pcr detection of classic enterotoxin genes in cheese and meat products. Braz. J. Microbiol. 40(1), 145–148. Prata, J. C., da Costa, J. P., Lopes, I., Andrady, A. L., Duarte, A. C., and Rocha-Santos, T. 2021. A one health perspective of the impacts of microplastics on animal, human and environmental health. Sci. Total Environ. 777, 146094. Qiu, H., Zhang, Y., Li, Z., Jiang, P., Guo, S. and He, Y. 2021. Donepezil Ameliorates Pulmonary Arterial Hypertension by Inhibiting M2-Macrophage Activation. Front. Cardiovasc. Med. 8, 639541. Saravanan, A., Kumar, P. S., Hemavathy, R. V., Jeevanantham, S., Kamalesh, R. and Sneha, S. 2021. Methods of detection of food-borne pathogens: a review. Environ. Chem. Lett. 19(1), 189–207. Scallan Walter, E. J., McLean, H. Q., and Griffin, P. M. 2020. Hospital discharge data underascertain enteric bacterial infections among children. Foodborne Pathog. Dis. 17(9), 530–532. Schirone, M., Visciano, P., Tofalo, R., and Suzzi, G. 2017. Editorial: Biological Hazards in Food. Front. Microbiol. 7, 2154. Seyoum, E. T., Mekonene, T. K., Woldetsadik, D. A., and Zewudie, B. M. 2016. Enterotoxin gene profile of Staphylococcus aureus isolates recovered from bovine milk produced in central Ethiopia. J. Infect. Dev. Ctries. 10(2), 138–142. Söderqvist, K., Lambertz, S. T., Vågsholm, I., Fernström, L. L., Alsanius, B. and Mogren, L. 2017. Fate of Listeria monocytogenes, pathogenic Yersinia enterocolitica, and Escherichia coli O157:H7 gfp+ in ready-to-eat salad during cold storage: what is the risk to consumers? J. Food Prot. 80(2), 204–212. Van Puyvelde, L., Aissa, A., Panda, S. K., De Borggraeve, W. M., Mukazayire, M. J., and Luyten, W. 2021. Bioassay-guided isolation of antibacterial compounds from the leaves of Tetradenia riparia with potential bactericidal effects on food-borne pathogens. J. Ethnopharmacol. 273, 113956. Vasudevan, P., Nair, M. K. M., Annamalai, T., and Venkitanarayanan, K. S. 2003. Phenotypic and genotypic characterization of bovine mastitis isolates of Staphylococcus aureus for biofilm formation. Vet. Microbiol. 92(1), 179–185. Wu, D., Li, X., Yang, Y., Zheng, Y., Wang, C. and Deng, L. 2011. Superantigen gene profiles and presence of exfoliative toxin genes in community-acquired meticillin-resistant Staphylococcus aureus isolated from Chinese children. J. Med. Microbiol. 60(Pt 1), 35–45. Xu, J., Tan, X., Zhang, X., Xia, X., and Sun, H. 2015. The diversities of staphylococcal species, virulence and antibiotic resistance genes in the subclinical mastitis milk from a single Chinese cow herd. Microb. Pathog. 88, 29–38. Zarkani, A. A., and Schikora, A. 2021. Mechanisms adopted by Salmonella to colonize plant hosts. Food Microbiol. 99, 103833. | ||
| How to Cite this Article |
| Pubmed Style Al-zuhairi ZA, Muslim ET, Faja OM, Alkhozai ZM, Mohammed BJ. Molecular identification of Staphylococcus aureus related enterotoxin genes in cheese samples. Open Vet. J.. 2024; 14(8): 2073-2078. doi:10.5455/OVJ.2024.v14.i8.36 Web Style Al-zuhairi ZA, Muslim ET, Faja OM, Alkhozai ZM, Mohammed BJ. Molecular identification of Staphylococcus aureus related enterotoxin genes in cheese samples. https://www.openveterinaryjournal.com/?mno=207145 [Access: January 25, 2026]. doi:10.5455/OVJ.2024.v14.i8.36 AMA (American Medical Association) Style Al-zuhairi ZA, Muslim ET, Faja OM, Alkhozai ZM, Mohammed BJ. Molecular identification of Staphylococcus aureus related enterotoxin genes in cheese samples. Open Vet. J.. 2024; 14(8): 2073-2078. doi:10.5455/OVJ.2024.v14.i8.36 Vancouver/ICMJE Style Al-zuhairi ZA, Muslim ET, Faja OM, Alkhozai ZM, Mohammed BJ. Molecular identification of Staphylococcus aureus related enterotoxin genes in cheese samples. Open Vet. J.. (2024), [cited January 25, 2026]; 14(8): 2073-2078. doi:10.5455/OVJ.2024.v14.i8.36 Harvard Style Al-zuhairi, Z. A., Muslim, . E. T., Faja, . O. M., Alkhozai, . Z. M. & Mohammed, . B. J. (2024) Molecular identification of Staphylococcus aureus related enterotoxin genes in cheese samples. Open Vet. J., 14 (8), 2073-2078. doi:10.5455/OVJ.2024.v14.i8.36 Turabian Style Al-zuhairi, Zahira Abdljabbar, Esraa Taher Muslim, Orooba Meteab Faja, Ziad M. Alkhozai, and Basima Jasim Mohammed. 2024. Molecular identification of Staphylococcus aureus related enterotoxin genes in cheese samples. Open Veterinary Journal, 14 (8), 2073-2078. doi:10.5455/OVJ.2024.v14.i8.36 Chicago Style Al-zuhairi, Zahira Abdljabbar, Esraa Taher Muslim, Orooba Meteab Faja, Ziad M. Alkhozai, and Basima Jasim Mohammed. "Molecular identification of Staphylococcus aureus related enterotoxin genes in cheese samples." Open Veterinary Journal 14 (2024), 2073-2078. doi:10.5455/OVJ.2024.v14.i8.36 MLA (The Modern Language Association) Style Al-zuhairi, Zahira Abdljabbar, Esraa Taher Muslim, Orooba Meteab Faja, Ziad M. Alkhozai, and Basima Jasim Mohammed. "Molecular identification of Staphylococcus aureus related enterotoxin genes in cheese samples." Open Veterinary Journal 14.8 (2024), 2073-2078. Print. doi:10.5455/OVJ.2024.v14.i8.36 APA (American Psychological Association) Style Al-zuhairi, Z. A., Muslim, . E. T., Faja, . O. M., Alkhozai, . Z. M. & Mohammed, . B. J. (2024) Molecular identification of Staphylococcus aureus related enterotoxin genes in cheese samples. Open Veterinary Journal, 14 (8), 2073-2078. doi:10.5455/OVJ.2024.v14.i8.36 |