| Research Article | ||
Open Vet. J.. 2024; 14(11): 2980-2988 Open Veterinary Journal, (2024), Vol. 14(11): 2980-2988 Research Article Molecular identification of Klebsiella species from pneumonic goats, IraqHanaa Khaleel Ibraheim1, Khadeeja S. Madhi2, Alyaa Sabti Jasim1 and Hasanain A. J. Gharban3*1Department of Microbiology, College of Veterinary Medicine, University of Basrah, Basra, Iraq 2Department of Microbiology, College of Medicine, University of Basrah, Basra, Iraq 3Department of Internal and Preventive Veterinary Medicine, College of Veterinary Medicine, University of Wasit, Wasit, Iraq *Corresponding Author: Hasanain A. J. Gharban. Department of Internal and Preventive Veterinary Medicine, College of Veterinary Medicine, University of Wasit, Wasit, Iraq. Email: hghirban [at] uowasit.edu.iq Submitted: 20/08/2024 Accepted: 16/10/2024 Published: 30/11/2024 © 2024 Open Veterinary Journal
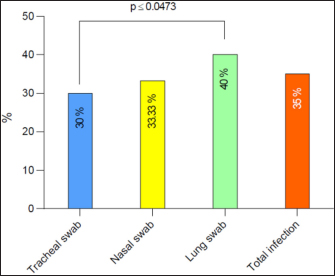
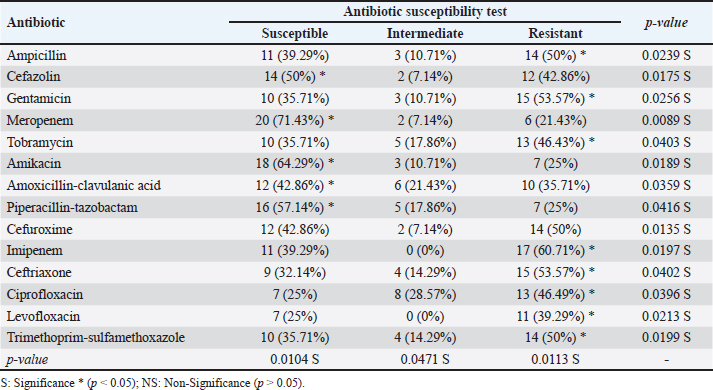
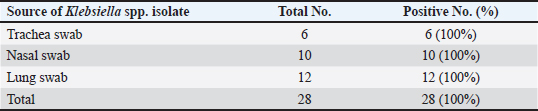
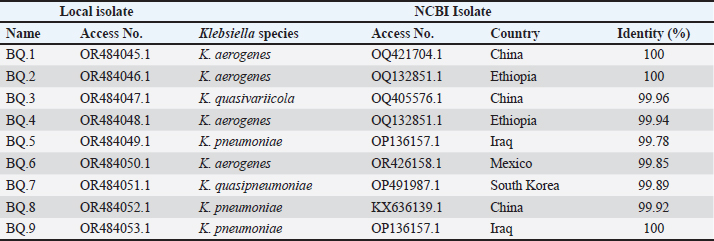
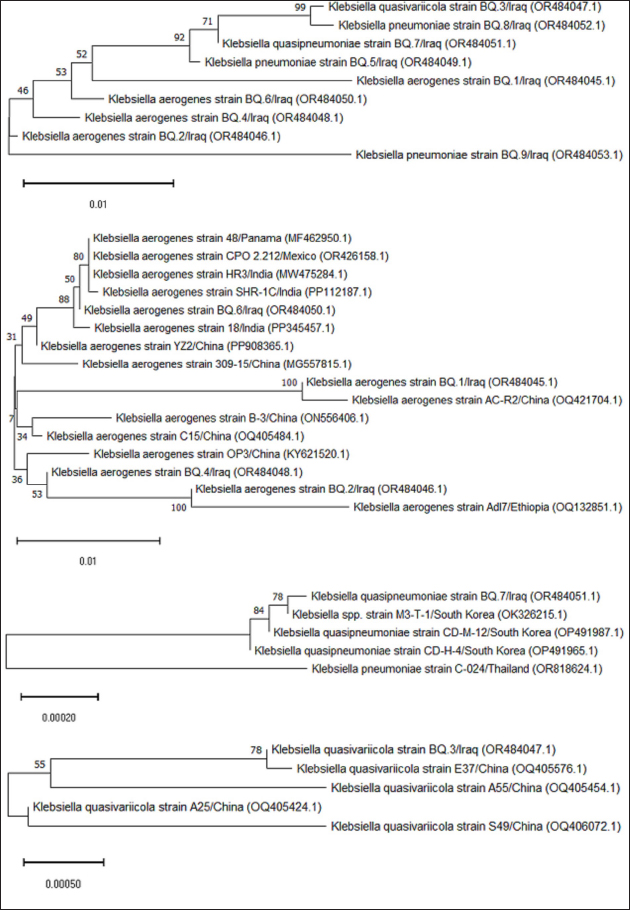
AbstractBackground: In goats, acute and chronic respiratory infections are often characterized by a rapidly progressing clinical course with little opportunity to develop an effective antibiotic therapy. Aim: This study aimed to identify Klebsiella spp. in pneumonic goats, assess its antibiotic susceptibility, and confirm the molecular phylogenetics of Klebsiella spp. Methods: A total of 80 pneumonic goats were selected from the slaughterhouses located in Basra province (Iraq) from June to November 2023, and each animal was subjected to obtaining only one sample. The studied samples were included 30 nasal swabs obtained from the lived goats, while 30 lung samples in addition to 20 tracheal swabs were collected from slaughtered goats. All study samples were inoculated onto MacConkey agar and tested biochemically. Eleven types of antibiotics were served in the Kirby-Bauer disc diffusion method to identify the susceptibility of Klebsiella spp. Positive culture isolates were tested molecularly using the polymerase chain reaction (PCR) and then sequenced for phylogenetic analysis of study isolates. Results: The findings indicated that 35% of samples were positive to Klebsiella spp. including 30% in trachea, 33.33% in nasal swabs, and 40% in lungs. Klebsiella colonies appeared on MacConkey agar as bright pink mucoid texture; while on blood agar, they were large, glossy, mucoid, whitish-grey, spherical, and free of hemolysis. Biochemically, all isolates were exhibited a negative reactivity to motility, oxidase, indole, and methyl red, but positives to urease, citrate utilization, catalase, and Voges-Proskauer, acid and gas production. Antibiotic susceptibility testing revealed the high susceptibility of Klebsiella isolates to meropenem (71.43%), and intermediate susceptibility to ciprofloxacin (28.57%), but high resistance to imipenem (60.71%). Targeting the 16S rRNA gene, PCR results confirmed all tested isolates as Klebsiella spp. Finally, phylogenetic analysis of 9 positive isolates demonstrated the identity of local Klebsiella isolates to Klebsiella aerogenes (no=4), Klebsiella pneumoniae (no=3), Klebsiella quasivariicola (no=1), and Klebsiella quasipneumoniae (no=1). Conclusion: Our study confirms the presence of K. aerogenes, K. quasivariicola, and K. quasipneumoniae in pneumonic goats, highlighting the importance of molecular phylogeny in the detection of new Klebsiella species. However, furthermore studies are necessary to investigate various Klebsiella species/strains in goats and other domestic animals. Keywords: Caprine pneumonia, K. aerogenes, K. quasipneumoniae, K. quasivariicola, K. pneumoniae. IntroductionKlebsiella is a Gram-negative, rod-shaped, capsulated bacterium belonging to the Enterobacteriaceae family. This bacterium causes various infections in both animals and humans with different outcomes that range from mild to moderate and severe even death (Sathyavathy and Madhusudhan 2020; Wu et al., 2021). Respiratory infections due to Klebsiella spp. are often characterized clinically by a rapidly progressing course and complicated by the occurrence of lung abscesses and multi-lobular involvement, which leave little chance of effective antibiotic therapy. Moreover, the development of antibiotic resistance among nosocomial isolates of Klebsiella pneumoniae has limited the therapeutic activity in the treatment of these infections (Khalid et al., 2022). Pneumonia is an inflammation of lung tissues that infects additionally either bronchioles to result in bronchopneumonia or pleura leading to pleuropneumonia (Emikpe et al., 2019). However, almost all pneumonic infections have been attributed to multifactorial etiologies such as parasites, bacteria, and viruses, alongside with environmental factors like extreme weather (cold or hot), and management practices that increase the strength of infection (Al-Ammiri et al., 2016). In goats, acute respiratory infections lead to poor weight gain, mortality, and great economic losses due to morbidities and mortalities, medical cares, and preventive measures (Saleh and Allam 2014). Whilst, chronic infections may cause endotoxemia, septicemia, and even death due to insufficiency and severe systemic illness (Mekibib et al., 2019). During respiratory outbreaks, the selection of effective antibiotics is of great importance due to the rapid progression of lung injury and endotoxin release (Nejibanand Al-Amery 2018). Therefore, this study was performed to isolate Klebsiella spp. from different samples of pneumonic goats, with the assessment of antibiotic susceptibility, and confirmation of different Klebsiella species by the molecular phylogenetic analysis. Materials and MethodsPreparation of culture media and reagentsFollowing the manufacturer’s instructions (HiMedia, India), media (MacConkey and blood agars), brain-heart infusion broth, reagents of chemical tests (oxidase, catalase, and Kovac’s), and Gram stain were prepared. SamplesA total of 80 pneumonic goats were selected from slaughterhouses located in Basra province (Iraq) from June to November 2023, and each animal was subjected to obtaining only one sample. The studied samples were included 30 nasal swabs obtained from the lived goats, while 30 lung samples in addition to 20 tracheal swabs were collected from slaughtered goats. The collected samples were transported to the Microbiology Lab (College of Veterinary Medicine, University of Basrah) using the brain-heart infusion broth. Bacterial isolationDifferent samples were inoculated aseptically on MacConkey and blood agars at 37°C for 24 hours, and subsequently, the suspected colonies were purified to identify their morphological characteristics (Quinn et al., 2011). Additional biochemical tests including oxidase test, catalase test, urease activity test, coagulase test, sugar fermentation, indole production test, and gas production test were done. Antibiotic susceptibility testThe Kirby-Bauer disc diffusion method was performed on Muller Hinton agar (Oxoid, UK) to identify the susceptibility of Klebsiella isolates toward 11 types of ready-to-use antibiotic discs based on the inhibition zone developed around each antibiotic. Molecular examinationPrestoTM Mini gDNA Bacteria Kit (Geneaid, Taiwan) was used to extract the genomic DNAs from purified Klebsiella colonies. Nanodrop spectrophotometer (Thermo Scientific, UK) was served to measurement the purity and concentration of DNAs. Targeting the 16S rRNA gene, a set of primers was designed [(F: 5’ AGA GTT TGA TCC TGG C-3’) and (R: 5’- GGT TAC CTT GTT ACG ACT T-3’)] to preparing the MasterMix tubes (Bioneer/South Korea) at a final volume of 50 µl (Munaff and Chmagh 2014). Thermocycler for Mastermix tubes was done as follows: 1 cycle for initial denaturation (92°C/2 minutes); 30 cycles for denaturation (94°C/30 seconds), annealing (52°C/45 seconds), and extension (72°C/1 minute); and 1 cycle final extension (72°C/5 minutes). Electrophoresis of PCR products was performed in 3% agarose gel stained with 3 μl Ethidium Bromide at 100V and 80A for 1 hour. Positive samples were indicated under the UV transilluminator (Clinx Science, China) at approximately 1,500 bp. PhylogenyThe DNAs of 9 molecularly positive samples were sent for sequencing in the Macrogen Company (South Korea) following the Modified Sanger dideoxynucleotide sequencing method. The sequence data were received via private email, and the local Klebsiella isolates were named, submitted to NCBI-GenBank database, and analyzed phylogenetically through the MEGA-11 Software. Statistical analysisThe t-test and One-Way Analysis of Variance in the GraphPad Prism Software (version 6.0.1) to estimate significant differences between study results at p ≤ 0.05 (Wahab et al., 2024). Ethical approvalThis study follows the ethics guidelines of the College of Veterinary Medicine (University of Wasit, Iraq) under the awarded access number (WU/CVM: 138-9-2023). ResultsCulture and biochemical testingThe total findings revealed that 35% (28/80) of samples were positive Klebsiella spp., including 30% (6/20) in trachea, 33.33% (10/30) in nasal swabs, and 40% (12/30) in lung (Fig. 1). Klebsiella grown on MacConkey agar appeared as bright pink and mucoid colonies; while on blood agar, the colonies shown large, glossy, mucoid, whitish-grey, spherical and free-hemolysis appearance. Biochemically, all isolates exhibited a negative reaction to motility, oxidase, indole, and methyl red. A positive reaction was observed to urease, citrate utilization, catalase, and Voges-Proskauer with the generation of acid and gas in the glucose fermentation test. Antibiotic susceptibility testingKlebsiella spp. isolates were shown a high susceptibility to meropenem (71.43%), and intermediate susceptibility to ciprofloxacin (28.57%) but a high resistance to imipenem (60.71%). Other findings revealed variable rates of susceptibility as following: cefazolin (50%), amikacin (64.29%), amoxicillin-clavulanic acid (42.86%), and piperacillin-tazobactam (57.14%). However, resistance to ampicillin (50%), gentamicin (53.57%), tobramycin (46.43%), ceftriaxone (53.57%), ciprofloxacin (46.49%), levofloxacin (39.29%), and trimethoprim-sulfamethoxazole (50%) was detected (Table 1). Molecular examination and phylogenyTargeting the 16S rRNA gene, PCR assay confirmed that all tested isolates were Klebsiella (Table 2, Fig. 2). Phylogenetic analysis of 9 positive isolates demonstrated significant identity between the local Klebsiella spp. isolates and various Klebsiella species existed in the NCBI-GenBank as follows: 4 local isolates identical to Klebsiella aerogenes of Chinese (BQ.1), Ethiopian (BQ.2 and BQ4), and Mexican (BQ6) origin; 3 isolates identical to K. pneumoniae of Iraqi (BQ.5 and BQ9) and Chinese (BQ.8) origins; 1 isolate identical to Klebsiella quasipneumoniae of South Korean origin (BQ.7); and 1 isolate identical to Klebsiella quasivariicola of Chinese (BQ.3) origin. Analysis of homology sequence identity revealed a range of similarity (*) and substitution mutation between the local and the NCBI-BLAST Klebsiella species at 99.78%–100% and 0.0002%–0.01%, respectively (Table 3, Figs. 3 and 4).
Fig. 1. Total positive isolates of Klebsiella spp. by MacConkey agar, blood agar and biochemical tests. DiscussionGoat is an important domestic animal which known which adaptable to diverse environmental conditions ranging from arid deserts to lush mountainous regions; however, several respiratory infections remain of serious concern (Zhou et al., 2017). In the current study, 35% of study animals were infected with Klebsiella spp. In comparison to other national studies, the prevalence rate of Klebsiella infection was 22% in Saudi Arabia (Mansour et al., 2014), 17% (Yaseen et al., 2019), 13.83% (Ahmed and Abdullah, 2022), and 6% (Mohammed, 2023); whereas internationally, it was 5.6% in India (Aher et al., 2012), 36% in Egypt (Ali and Abu-Zaid, 2019), and 51.72% in Nigeria (Adam et al., 2023). However, variations in the prevalence of respiratory bacteria may be reflected by the sampling scheme, sample size, diagnostic methods serve, and pathogenicity of Klebsiella strains. Common opportunistic Klebsiella strains significantly affect animals that undergo a compromised or weakened immunity (Hu et al., 2021). In the last decade, many researchers have claimed the increasing rate of Klebsiella infection due to to the pathogenic characteristics and virulence factors of this bacterium (Younis et al., 2016; Wang et al., 2020; Zhu et al., 2021). These characteristics could enhance the growth of a colony in a host. Moreover, highly invasive and virulent Klebsiella strains could infect healthy animals leading to severe acquired infections such as pneumonia, necrotizing fasciitis, meningitis, and pyogenic liver abscess (Marques et al., 2019; Russo and Marr, 2019). In the respiratory tract, the colonized pathogenic Klebsiella in pulmonary tissues establish a prominent lesion and causes severe pneumonia through modification resistance to phagocytosis by immune cells (Bengoechea and Sa Pessoa, 2019; Priyanka et al., 2020). Furthermore, Klebsiella tends to cause chronic infections due to two main factors: the first is the secretion of various enzymes that inhibit certain antibiotics to rendering pathogen resistance, and the second is the development of immune-evading biofilms in vivo (Padmini et al., 2017; Abbas et al., 2024). Table 1. Antibiotic susceptibility testing of Klebsiella isolates using the Kirby-Bauer disc diffusion method (Total No: 28)
Table 2. Molecular PCR results for testing a total of 28 suspected Klebsiella isolates.
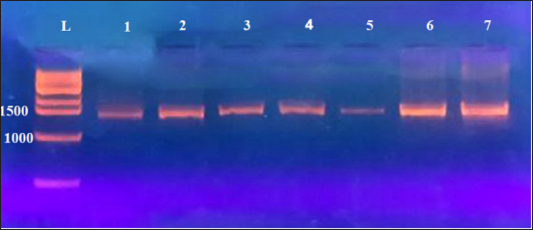
Fig. 2. Electrophoresis of 3% agarose gel stained with Ethidium Bromide to amplify PCR products. Lane L: Ladder marker at 3,000 bp; Lanes 1–7: Some positive PCR products to Klebsiella spp. at 1,500 bp. Table 3. NCBI-BLAST Homology sequence identity of local BQ Klebsiella strains in goats comparing the NCBI-GenBank Klebsiella species targeting the 16S rRNA gene.
Fig. 3. Multiple sequence alignment analysis of local BQ Klebsiella strains in goats comparing with the NCBI-GenBank Klebsiella species targeting the 16S rRNA gene. Analysis conducted using the Clustal W alignment tool in MEGA 11 Software showed the presence of nucleotide alignment similarity as (*) and substitution mutations.
Fig. 4. Phylogenetic tree analysis of the 16S rRNA gene partial sequence of the local BQ Klebsiella strains in goats and identity with the NCBI-GenBank Klebsiella species. The analysis was built using the Unweighted Pair Group technique with Arithmetic Mean (UPGMA tree method), and the evolutionary distances were calculated using the Maximum Composite Likelihood method in MEGA 11. In this study, results indicated that Klebsiella isolates were significantly resistant to several classes of antibiotics, likely due to their frequent use in treating patients (Gao et al., 2020a, b). Conversely, the apparent resistance rate of Klebsiella isolates to imipenem was higher than reported by others (Hu et al., 2021; Su et al., 2022; Liza et al., 2024). Dapgh et al. (2019) mentioned that irregular and excessive administration of antibiotics has increased the prevalence of bacterial resistance and transmission of infections between animals as well as from animals to humans with complicating the treatment strategy. Mukuna et al. (2023) studied the antimicrobial susceptibility profile of pathogenic and commensal bacteria recovered from cattle and goats suggesting that both animals could act as reservoirs of multi-drug resistance bacteria. Liza et al. (2024) concluded that the presence of extended-spectrum β-lactamase-producing multidrug resistance K. pneumoniae isolates that posing a substantial public health threat. Klebsiella is difficult to detect with frequent misclassification in clinical microbiology laboratories (Shankar et al., 2018). Several investigations have been conducted to determine the efficacy of different strategies to identifying various species of Klebsiella (Mukherjee et al., 2020). Molecular PCR assay has proven as more valuable and highly sensitive and specific technique in the diagnosis of Klebsiella species when compared to other traditional procedures (Hansen et al., 2020). Phylogenetic analysis of study isolates indicates presence of new Klebsiella species in goats including K. aerogenes, K. quasivariicola, and K. quasipneumoniae. Worldwide, several previous and recent studies have shown some of these species in patients with epidemic resistance to antibiotics and phagocytosis (Curie et al., 1978; Williams et al., 1983; Potter et al., 2018; Zhang et al., 2022; Delik et al., 2024). This might explain the prevalence of antibiotic resistance among the study isolates. ConclusionThis study lies in the first determination of three new species of Klebsiella in pneumonic goats in Iraq (K. aerogenes, K. quasivariicola, and K. quasipneumoniae), along with the identification of their distinct species based on NCBI-GenBank database. In addition, the current study clearly identifies the susceptibility and resistance of Klebsiella isolates to various antibiotics. However, moreover, studies are necessary to investigate various Klebsiella species/strains in goats and other domestic animals based on molecular phylogeny. AcknowledgmentsThe authors thank all workers and veterinarians who contributed to completing the current work. FundingNo funds were received to complete this work. Authors’ contributionHKI: Molecular examination of bacterial isolate. KSM: Bacterial isolation. ASJ: Antimicrobial susceptibility testing. HAJG: Samples collection, phylogeny, and statistical analysis of study results. All authors were approved the final copy of the manuscript. Conflict of interestThe authors have declared no conflict of interest. Data availabilityAll obtained data were included in this manuscript. ReferencesAbbas, R., Chakkour M., Zein, El Dine, H., Obaseki, EF., Obeid, S.T., Jezzini, A. and Ezzeddine, Z. 2024. General overview of Klebsiella pneumonia: epidemiology and the role of siderophores in its pathogenicity. Biology 13(2), 1–19. Adam, M., Akeem, A.O., Barka, S.A., Olu, S.S.V., Abiodun, A.A. and David, O.F.S. (2023). Pathological and molecular investigation of some bacterial in the lungs and livers of red sokoto goats slaughtered at Ilorin, Kwara State, Nigeria. Med. Kedokteran Hewan, 34(2), 87–101. Aher, T., Roy, A. and Kumar, P. 2012. Molecular detection of virulence genes associated with pathogenicity of Klebsiella spp. isolated from the respiratory tract of apparently healthy as well as sick goats. Israel J. Vet. Med. 67(4), 249–252. Ahmed, B.A. and Abdullah, M.A. 2022. Isolation and molecular diagnosis of the main bacterial species causing Pneumonia in small ruminants in the Duhok Abattoir-Kurdistan region of Iraq. Microb. Biosys. 7(2), 66–73. Al-Ammiri, H.H., Abd-Allh, A.H. and Hamid, M. 2016. Isolation and identification of aerobic bacteria detected from sheep infected with pneumonia. Adv. Environ. Biol. 10(5), 214–220. Ali, A. and Abu-Zaid, K. 2019. Study on Klebsiella pneumoniae causing respiratory infection in small ruminants. Anim. Health Res. J. 7, 57–67. Bengoechea, J.A. and Sa Pessoa, J. 2019. Klebsiella pneumoniae infection biology: living to counteract host defences. FEMS Microbiol. Rev. 43(2), 123–144. Curie, K., Speller, D.C.E., Simpson, R.A., Stephens, M. and Cooke P.I. 1978. A hospital epidemic caused by a gentamicin-resistant Klebsiella aerogenes. Epidemiol. Infect. 80(1), 115–123. Dapgh, A.N., Hakim, A.S., Abouelhag, H.A., Abdou, A.M. and Elgabry EA 2019. Detection of virulence and multidrug resistance operons in Pseudomonas aeruginosa isolated from Egyptian Baladi sheep and goat. Vet. World. 12(10), 1524–1528. Delik, E., Eroğlu, B. and Tefon-Öztürk, B.E. 2024. Evaluation of the in vitro effects of concentrations of antibiotics on three Enterobacteriaceae isolates. World J. Microbiol. Biotechnol. 40(2), 1–16. Emikpe, B.O., Jarikre, T.A., Akpavie, S.O., Opoku-Agyemang, T., Asare, D. and Folitse, R.D. 2019. Histological and immunohistochemical assessments of pneumonia in sheep slaughtered at Ibadan, Nigeria and Kumasi, Ghana. J. Immunoassay Immunochem. 40(3), 300–313. Gao, L., Lv, Y. and Li, Y. 2020a. Analysis of the drug resistance of carbapenem-resistant Klebsiella pneumoniae in the China antimicrobial resistance surveillance trial program, 2007–2018. Microb. Drug Resist. 26(8), 944–950. Gao, Q., Shen, Z., Qin, J., Liu, Y. and Li, M. 2020b. Antimicrobial resistance and pathogenicity determination of community-acquired hypervirulent Klebsiella pneumoniae. Microb. Drug Resist. 26(10), 1195–1200. Hansen, S.K., Kaya, H., Roer, L., Hansen, F., Skovgaard, S., Justesen, U.S. and Hammerum, A.M. 2020. Molecular characterization of Danish ESBL/AmpC-producing Klebsiella pneumoniae from bloodstream infections, 2018. J. Glob. Antimicrob. Resist. 22, 562–567. Hu, T., Dai, Q., Chen, H., Zhang, Z., Dai, Q., Gu, X. and Zhu, L. 2021. Geographic pattern of antibiotic resistance genes in the metagenomes of the giant panda. Microb. Biotechnol. 14, 186–197. Hu, Y., Anes, J., Devineau, S. and Fanning, S. 2021. Klebsiella pneumoniae: prevalence, reservoirs, antimicrobial resistance, pathogenicity, and infection: a hitherto unrecognized zoonotic bacterium. Foodborne Path. Dis. 18(2), 63–84. Khalid, T.W.B.M. and Ghaima, K.K. 2022. Effect the natural efflux pump inhibitor (berberine) in multidrug resistant Kleibsiella pneumoniae isolated from urinary tract infections in several Baghdad Hospitals. Egyptian J. Hospital Med. 89(2), 6882–6888. Liza, N.A., Hossain, H., Rahman Chowdhury, M.S., Al Naser, J., Lasker, R.M., Rahman, A. and Rahman, M.M. 2024. Molecular epidemiology and antimicrobial resistance of extended-spectrum β-lactamase (ESBL)-producing Klebsiella pneumoniae in retail Cattle Meat. Vet. Med. Int. 2024(1), 3952504. Mansour, A.M., Zaki, H.M., Hassan, N.A. and Al-Humiany, A.A. 2014. Molecular characterization and immunoprotective activity of capsular polysaccharide of Klebsiella Pneumoniae isolated from farm animals at Taif Governorate. American J. Infect. Dis. 10(1), 11–10. Marques, C., Belas, A., Aboim, C., Cavaco-Silva, P., Trigueiro, G., Gama, L.T. and Pomba, C. 2019. Evidence of sharing of Klebsiella pneumoniae strains between healthy companion animals and cohabiting humans. J. Clin. Microbiol. 57(6), 10–1128. Mekibib, B., Mikir, T., Fekadu, A. and Abebe, R. 2019. Prevalence of pneumonia in sheep and goats slaughtered at Elfora Bishoftu export abattoir, Ethiopia: a pathological investigation. J. Vet. Med. 2019, 1–10. Mohammed, A.K. 2023. Investigation of bacterial contamination with Klebsilla and E. coli in the prepucal cavity of pubertal and adult age in caprine. Iran. J. Vet. Sci. Technol. 15(3), 27–32. Mukherjee, S., Naha, S., Bhadury, P., Saha, B., Dutta, M., Dutta, S. and Basu, S. 2020. Emergence of OXA-232-producing hypervirulent Klebsiella pneumoniae ST23 causing neonatal sepsis. J. Antimicrob. Chemother. 75(7), 2004–2006. Mukuna, W., Aniume, T., Pokharel, B., Khwatenge, C., Basnet, A. and Kilonzo-Nthenge, A. 2023. Antimicrobial susceptibility profile of pathogenic and commensal bacteria recovered from Cattle and Goat farms. Antibiotics 12(2), 1–19. Munaff, J.A. and Chmagh, A.A. 2014. Molecular genetic study confirming the transmission of nasopharyngeal bacteria to middle ear in patients with chronic suppurative otitis media, including new global strains in GenBank: MunAala1, MunAala2, IRQBAS5 and IRQBAS6. Int. J. Pharm. Res. Bio. Sci. 3(5), 379–397. Nejiban, Z.S. and Al-Amery, M.A. 2018. Clinical and diagnostic study of sheep pneumonic pasteurellosis in Basrah, Iraq. Al-Qadisiyah J. Vet. Med. Sci. 17(1), 1–5. Padmini, N., Ajilda, A.A.K., Sivakumar, N. and Selvakumar, G. 2017. Extended spectrum β-lactamase producing Escherichia coli and Klebsiella pneumoniae: critical tools for antibiotic resistance pattern. J. Basic Microbiol. 57(6), 460–470. Potter, R.F., Lainhart, W, Twentyman, J., Wallace, M.A., Wang, B., Burnham, C.A.D. and Dantas, G. 2018. Population structure, antibiotic resistance, and uropathogenicity of Klebsiella variicola. mBio 9(6), 10–1128. Priyanka, A., Akshatha, K., Deekshit, V.K., Prarthana, J. and Akhila, D.S. 2020. Klebsiella pneumoniae infections and antimicrobial drug resistance. Model organisms for microbial pathogenesis, biofilm formation and antimicrobial drug discovery. Singapore: Springer Nature Singapore Pte Ltd., pp: 195–225. Quinn, P.J., Markey, B.K., Leonard, F.C., Hartigan, P., Fanning, S. and Fitzpatrick, E. 2011. Veterinary microbiology and microbial disease. Newyork, NY: John Wiley & Sons, p: 74. Rawy, D.K., El-Mokhtar, M.A., Hemida, S.K., Askora, A. and Yousef, N. 2020. Isolation, characterization and identification of Klebsiella pneumoniae from assiut university hospital and sewage water in Assiut governorate, Egypt. Assiut Univer. J. Botany Microbiol. 49(2), 60–76. Russo, T.A. and Marr, C.M. 2019. Hypervirulent Klebsiella pneumoniae. Clin. Microbiol. Rev. 32(3), 10–1128. Saleh, N.S. and Allam, T.S. 2014. Pneumonia in sheep: bacteriological and clinicopathological studies. Am. J. Res. Commun., 2(11), 70–88. Sathyavathy, K. and Madhusudhan, B.K. 2020. Review on clinical diseases caused by Klebsiella. J. Pharmaceutic. Res. Int. 32(21), 12–19. Shankar, C., Veeraraghavan, B., Nabarro, L.E.B., Ravi, R., Ragupathi, N.K.D. and Rupali, P. 2018. Whole genome analysis of hypervirulent Klebsiella pneumoniae isolates from community and hospital acquired bloodstream infection. BMC Microbiol. 18, 1–9. Su, X., Yan, X., Li, Y., Zhang, D,. Li, L., Geng, Y. and Liu, S. 2022. Identification of extended-spectrum beta-lactamase (CTX-M)-producing Klebsiella pneumoniae belonging to ST37, ST290, and ST2640 in captive giant pandas. BMC Vet. Res. 18(1), 186–196. Wahab, B.A.A., Merah, M.H., Latif, A.D. and Gharban, H.A. 2024. Alternative therapeutic approach of ovine subclinical mastitis using the ethanolic roots extract of Capparis spinosa. Open Vet. J. 14(3), 814–821. Wang, G., Zhao, G., Chao, X., Xie, L. and Wang, H. 2020. The characteristic of virulence, biofilm and antibiotic resistance of Klebsiella pneumoniae. Int. J. Environ. Res. Public Health 17(17), 1–17. Williams, P., Lambert, P.A., Brown, M.R. and Jones, R.J. 1983. The role of the O and K antigens in determining the resistance of Klebsiella aerogenes to serum killing and phagocytosis. Microbiology 129(7), 2181–2191. Wu, D., Ding, J., Jia, Y., Liu, H., Xiao, J. and Peng, J. 2021. Predictors of mortality in acute pancreatitis complicated with multidrug-resistant Klebsiella pneumoniae infection. BMC Infect. Dis. 21, 1–8. Yaseen, S.A.S., Al-Maaly, N.M. and Al-Rubaie, E.M.M. 2019. Isolation and identification of pathogenic bacteria from vaginal cavity of sheep and goats in Iraq. J. Pure Appl. Microbiol. 13(4), 2295–2299. Younis, G., Awad, A., El-Gamal, A. and Hosni, R. 2016. Virulence properties and antimicrobial susceptibility profiles of Klebsiella species recovered from clinically diseased broiler chicken. Adv. Anim. Vet. Sci. 4(10), 536–542. Zhang, Z., Wang, H., Guo, Y., Liu, Z. and Chang, Z. 2022. Metagenome analysis of the bacterial characteristics in invasive Klebsiella pneumoniae liver abscesses. Front. Cell. Infect. Microbiol. 12, 1–11. Zhou, Z., Zhang, M., Li, H., Yang, H., Li, X., Song, X. and Wang, Z. 2017. Prevalence and molecular characterization of Staphylococcus aureus isolated from goats in Chongqing, China. BMC Vet. Res. 13, 1–8. Zhu, J., Wang, T., Chen, L. and Du, H. 2021. Virulence factors in hypervirulent Klebsiella pneumoniae. Front. Microbiol. 12, 1–14. | ||
| How to Cite this Article |
| Pubmed Style Ibraheim HK, Madhi KS, Jasim AS, Gharban HA. Molecular identification of Klebsiella species from pneumonic goats, Iraq. Open Vet. J.. 2024; 14(11): 2980-2988. doi:10.5455/OVJ.2024.v14.i11.26 Web Style Ibraheim HK, Madhi KS, Jasim AS, Gharban HA. Molecular identification of Klebsiella species from pneumonic goats, Iraq. https://www.openveterinaryjournal.com/?mno=216304 [Access: January 25, 2026]. doi:10.5455/OVJ.2024.v14.i11.26 AMA (American Medical Association) Style Ibraheim HK, Madhi KS, Jasim AS, Gharban HA. Molecular identification of Klebsiella species from pneumonic goats, Iraq. Open Vet. J.. 2024; 14(11): 2980-2988. doi:10.5455/OVJ.2024.v14.i11.26 Vancouver/ICMJE Style Ibraheim HK, Madhi KS, Jasim AS, Gharban HA. Molecular identification of Klebsiella species from pneumonic goats, Iraq. Open Vet. J.. (2024), [cited January 25, 2026]; 14(11): 2980-2988. doi:10.5455/OVJ.2024.v14.i11.26 Harvard Style Ibraheim, H. K., Madhi, . K. S., Jasim, . A. S. & Gharban, . H. A. (2024) Molecular identification of Klebsiella species from pneumonic goats, Iraq. Open Vet. J., 14 (11), 2980-2988. doi:10.5455/OVJ.2024.v14.i11.26 Turabian Style Ibraheim, Hanaa Khaleel, Khadeeja S. Madhi, Alyaa Sabti Jasim, and Hasanain A.j. Gharban. 2024. Molecular identification of Klebsiella species from pneumonic goats, Iraq. Open Veterinary Journal, 14 (11), 2980-2988. doi:10.5455/OVJ.2024.v14.i11.26 Chicago Style Ibraheim, Hanaa Khaleel, Khadeeja S. Madhi, Alyaa Sabti Jasim, and Hasanain A.j. Gharban. "Molecular identification of Klebsiella species from pneumonic goats, Iraq." Open Veterinary Journal 14 (2024), 2980-2988. doi:10.5455/OVJ.2024.v14.i11.26 MLA (The Modern Language Association) Style Ibraheim, Hanaa Khaleel, Khadeeja S. Madhi, Alyaa Sabti Jasim, and Hasanain A.j. Gharban. "Molecular identification of Klebsiella species from pneumonic goats, Iraq." Open Veterinary Journal 14.11 (2024), 2980-2988. Print. doi:10.5455/OVJ.2024.v14.i11.26 APA (American Psychological Association) Style Ibraheim, H. K., Madhi, . K. S., Jasim, . A. S. & Gharban, . H. A. (2024) Molecular identification of Klebsiella species from pneumonic goats, Iraq. Open Veterinary Journal, 14 (11), 2980-2988. doi:10.5455/OVJ.2024.v14.i11.26 |