| Research Article | ||
Open Vet. J.. 2024; 14(11): 3026-3036 Open Veterinary Journal, (2024), Vol. 14(11): 3026-3036 Research Article Detection of the methicillin-resistant Staphylococcus aureus gene encoding mecA in nasal swabs of cats in Surabaya, IndonesiaSafira Ramadhani1, Aswin Rafif Khairullah2, Mustofa Helmi Effendi3, Mohammad Sukmanadi4, Wiwiek Tyasningsih5*, Saumi Kirey Millannia1, Daniah Ashri Afnani6, Ikechukwu Benjamin Moses7, M. Thoriq Ihza Farizqi1 and Sergius Erikson Kaben81Profession Program of Veterinary Medicine, Faculty of Veterinary Medicine, Universitas Airlangga, Surabaya, Indonesia 2Research Center for Veterinary Science, National Research and Innovation Agency (BRIN), Bogor, Indonesia 3Division of Veterinary Public Health, Faculty of Veterinary Medicine, Universitas Airlangga, Surabaya, Indonesia 4Division of Basic Veterinary Medicine, Faculty of Veterinary Medicine, Universitas Airlangga, Surabaya, Indonesia 5Division of Veterinary Microbiology, Faculty of Veterinary Medicine, Universitas Airlangga, Surabaya, Indonesia 6Department of Microbiology and Parasitology, Faculty of Veterinary Medicine, Universitas Pendidikan Mandalika, Mataram, Indonesia 7Department of Applied Microbiology, Faculty of Science, Ebonyi State University, Abakaliki, Nigeria 8Master Program of Veterinary Medicine and Public Health Science, Faculty of Veterinary Medicine, Universitas Airlangga, Surabaya, Indonesia *Corresponding Author: Wiwiek Tyasningsih. Division of Veterinary Microbiology, Faculty of Veterinary Medicine, Universitas Airlangga, Surabaya, Indonesia. Email: wiwiek-t [at] fkh.unair.ac.id Submitted: 29/08/2024 Accepted: 16/10/2024 Published: 30/11/2024 © 2024 Open Veterinary Journal
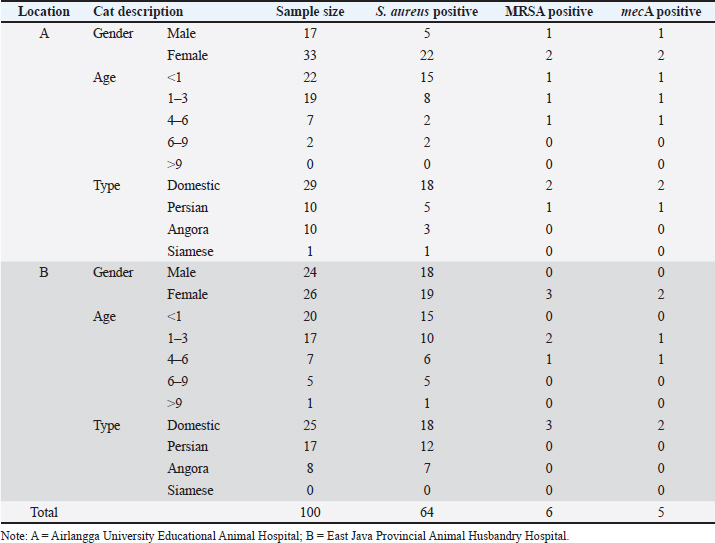
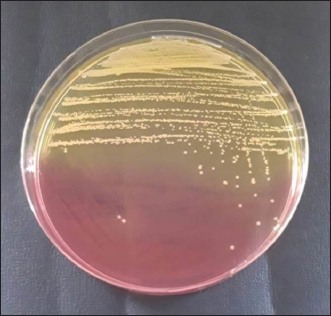
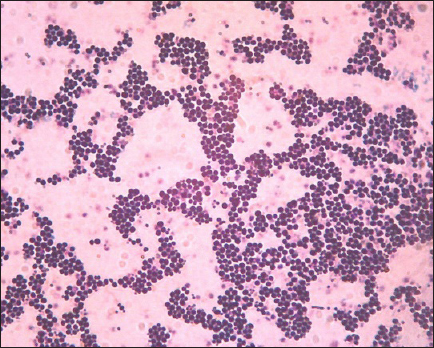
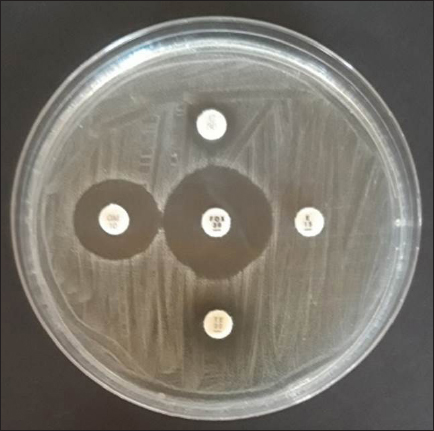
AbstractBackground: The most susceptible group of people to spread methicillin-resistant Staphylococcus aureus (MRSA) among domestic cats is their owners’ relatives. Aim: Considering the aforementioned, research at the Surabaya City Animal Hospital is necessary to determine whether the mecA gene may be detected in cat nasal swabs. Methods: Samples were taken using a sterile cotton swab, and the transport medium was buffered peptone water. Standard operating procedures in microbiology were then employed to identify the samples. Using five distinct antibiotic discs, the Kirby-Bauer diffusion method was used to determine the antibiotic resistance profile of S. aureus. All cefoxitin-resistant S. aureus isolates were tested for MRSA and then the mecA gene was detected. Results: Based on morphological culture features, Gram staining, and biochemical assays, 64 samples (64%) of the 100 isolated cat nose swab samples were positive for S. aureus, according to the results of the biochemical tests. The profile of antibiotic resistance developed from the results of the S. aureus resistance test to antibiotics revealed that five (7.81%) of the 64 S. aureus isolates were confirmed to be multidrug resistant. Out of the 6 MRSA isolates analyzed, the mecA gene was detected in 5 isolates. Conclusion: Based on the findings of this study, a thorough assessment of the impact of using antibiotics to prevent the spread of MRSA in veterinary hospitals on the general public’s health is required in light of the detection of MRSA isolates bearing the mecA gene in the Surabaya veterinary hospital. Keywords: Antibiotic, Cat, MDR, MRSA, Public health. IntroductionOne of the animals that is frequently kept as a pet is the cat. The fact that cats can benefit their owners by lowering the effects of stress, creating a pleasant environment, and regaining mental stability is a common reason why people desire to keep cats (Afnani et al., 2022). Cats are a popular pet due to their non-noisy nature, ease of maintenance, and exceptional ability to adapt to their surroundings. To keep cats healthy and less prone to illness, it is important to take good care of them (Hesselmar et al., 2018; Kaben et al., 2024). A bacterial infection in cats is one potential illness. In cats and other animals, Staphylococcus aureus is a typical microorganism of the natural bacterial flora that can be detected on the skin and nasal mucosa (Bierowiec et al., 2019). Despite the fact that the bacteria are present in clinically healthy people, they can nevertheless cause a number of infections when the immune system is compromised (Khairullah et al., 2019; Khairullah et al., 2022a). Staphylococcus aureus is a Gram-positive bacterium, non-motile, spherical, 0.8–1 µm in diameter, grouped in clusters resembling grapes, and does not produce spores (Ramandinianto et al., 2020; Khairullah et al., 2020a). This bacterium is commonly found in infections in cats. Companion animals, especially cats, can act as a reservoir for the propagation of bacterial illnesses brought on by S. aureus, endangering public health (Gwenzi et al., 2021; Llor and Bjerrum, 2014). It is currently known that S. aureus bacteria originating from animals can infect humans or other animals, given that cats are frequently treated as family members to enable physical interaction between people and animals, including skin-to-skin contact, aerosols from coughing, sneezing, and saliva (Elmoslemany et al., 2021; Rahmaniar et al., 2020). Methicillin-resistant Staphylococcus aureus (MRSA) is associated with resistance of S. aureus isolates to β-lactam antibiotics and with recurrent nosocomial infections (Green et al., 2014; Nimer, 2022). It is believed that all β-lactam antibiotics, including carbapenems and cephalosporins, are useless against cefoxitin-resistant MRSA bacteria (Gajdács, 2019). Compared to human MRSA isolates, animal MRSA isolates were shown to have significantly greater levels of ampicillin, ciprofloxacin, cotrimoxazole, erythromycin, clindamycin, azithromycin, and tetracycline resistance (Fitranda et al., 2023). In MRSA strains, the mecA gene is located on a cellular genetic component known as the staphylococcal cassette chromosome mec (SCCmec) (Rajabiani et al., 2014; Saber et al., 2017). This gene produces Penicillin-binding protein 2a (PBP2a), which has a lesser affinity than regular PBP for β-lactam antibiotics (Fergestad et al., 2020). Strains of S. aureus with the mecA gene will have resistance to β-lactam antibiotics (Ramandinianto et al., 2020). The city of Surabaya is one of the big cities in Indonesia which has animal hospitals including the Airlangga University Educational Animal Hospital and Animal Husbandry Service Animal Hospital of East Java Province. Veterinary Hospital in Surabaya still conducts very little research on the detection mecA gene in cats (Decline et al., 2020). Since MRSA is one of the primary sources of infection in cats, this will negatively impact the selection of treatments to be used in the management of the illness (Costa et al., 2022). The purpose of this study was to detect the mecA gene from cat nasal swabs at the Animal Hospital in Surabaya. The results of this study are important as a reference to understand how resistance develops and how to apply detection methods in animal hospitals. Materials and MethodsStudy area and sample collectionThe study has been carried out in March–April 2024. Cat nose swab sampling was done in Surabaya at the Airlangga University Teaching Animal Hospital and the East Java Provincial Animal Husbandry Hospital, while bacterial isolation and sensitivity tests were done at the Microbiology Laboratory, Airlangga University. The cat’s nasal mucosa was sampled with a sterile cotton swab (OneMed) and Amies transport medium (Charuzu). The cotton swab is placed inside the Amies transport medium tube, which is then labeled and preserved in a coolbox. Isolation and identification of S. aureusIsolates that have grown on Amies media are taken using an inoculum needle then streaked on MSA media and then incubated at 37°C for 24 hours. Bacterial colonies that grow yellow on MSA media are suspected as S. aureus (Yembise et al., 2020). The Gram stain, coagulase, and catalase tests were performed for confirmation. The following procedures are the coagulase and catalase assays, which detect the S. aureus bacteria (Effendi et al., 2019). Gram staining is done by mixing one loop of bacterial culture from MSA with physiological NaCl that has been dropped on a glass object, dried, and fixed on alcohol. The preparation is dripped with crystal violet dye for 2 minutes, then washed with running water, dripped with Lugol after that left for 1 minute, and washed with 95% alcohol for 10 seconds. Furthermore, the alcohol is washed with water and given safranin dye for 2 minutes, the preparation is washed with running water and then dried. The staining results are observed using a microscope with a magnification of 1000x. The morphology of S. aureus will appear round or coccus-like grapes and purple because it absorbs crystal violet dye (Gebremedhin et al., 2022). The catalase test was performed on a single loop of bacterial culture on MSA media that was considered to be S. aureus. After that, a layer of the bacterial culture is distributed over a glass object and mixed after being drip-applied with 3% hydrogen peroxide. The catalase test was successful if there were gas bubbles produced by the genus Staphylococcus (Teo et al., 2015). In the coagulase test, rabbit blood is centrifuged for 15 minutes at 10,000 rpm after plasma has been collected using the anticoagulant EDTA. The bacterium culture on the MSA media that was suspected to be S. aureus was taken out using a loop and put in 1 ml of nutrient broth for additional incubation for 24 hours at 37°C. The next step is to add 1 ml of rabbit plasma, which is already infused with microorganisms, to the nutritional broth using a syringe. Then let it grow for 4–24 hours (Saputra et al., 2017). Antibiotic sensitivity testAn antibiotic sensitivity test was performed using the Kirby Bauer disc diffusion method. After being isolated, recognized, and made into a suspension, the S. aureus bacteria were prepared by transferring the bacterial culture using a loop into a test tube filled with 9 ml of physiological NaCl and homogenizing with a vortex. Suspension turbidity is compared to the Mc Farland threshold of 0.5. The bacterial suspension was evenly smeared on the surface of the MHA media. Paper discs containing the antibiotics erythromycin, tetracycline, gentamicin, ampicillin, and cefoxitin were placed on the surface of the media using tweezers. The bacteria were cultivated for 24 hours at 37°C (Tyasningsih et al., 2022). The zone of inhibition is the area around the antibiotic where no bacterial growth was observed. The diameter of the area that prevents bacterial growth and the antibiotic disc’s circumference in millimeters (Effendi et al., 2018). The antibiotic susceptibility of the isolates was determined using the disc diffusion method according to Clinical and Laboratory Standards Institute guidelines (Widodo et al., 2022). Confirmatory MRSA testAll cefoxitin-resistant S. aureus isolates were tested for MRSA. Oxacillin Resistance Screening Agar Base (ORSAB) media combined with Oxacillin Resistance Selective Supplement was used to inoculate bacterial colonies from MHA media to confirm the presence of MRSA. The ORSAB medium was then incubated for 24 hours at 37°C. mecA detectionTo determine if the isolates have presented the mecA gene with molecular detection, 5 µl was used as a template. The primers used were specific primers for the mecA gene (Table 1). Mixture for the PCR reaction with a volume of 25 µl consisting of 12.5 µl PCR Mastermix (0.2 mM dNTP, 0.5 U Taq Polymerase, Buffer, and 1.5 mM MgCl2), 1.25 µl for each primer and 5 µl DNA template. The PCR was conducted under the following conditions: initial denaturation at 94°C for 7 minutes, 35 cycles of 96°C for 50 seconds of denaturation, 50°C for 40 seconds of annealing, 72°C for 1 minute of extension, and finally 10 minutes of extension at 72°C. The PCR product was visualized in 1.5% agarose. A total of 5 µl of the PCR product were combined with 1 µl of the marker, placed on parafilm, and submerged in an ethium bromide solution for 10 minutes in a dark place. Observe the PCR product formed with a UV transilluminator with a wavelength of 360 nm. ResultsBacterial isolatesBased on morphological culture features, Gram staining, and biochemical assays, 64 samples (64%) of the 100 isolated cat nose swab samples were found to be positive for S. aureus (Table 2). The yellow bacterial colonies on MSA media were indicative of S. aureus (Fig. 1). Regarding Gram stain, the presence of bacteria is indicated by purple colonies and clustered cocci (Fig. 2). In the catalase test, the growth of bubbles on the object glass indicates a positive biochemical result for S. aureus, as well as the coagulase test showing a wallop at the bottom of the microtube. Antibiotic resistance of S. aureusFor the antibiotic susceptibility, 64 S. aureus isolates were analyzed; based on the profile of antibiotic resistance created from the results of the S. aureus antibiotic resistance test, 8 isolates (12.5%) were found to be resistant to one class of antibiotics tested, 6 isolates (9.37%) to two classes of antibiotics, and 5 isolates (7.81%) to be multidrug resistant (MDR). In the observation of resistance patterns, two isolates had a GM–E–TE (Gentamicin, Erythromycin, Tetracycline) antibiotic resistance pattern and two isolates had an E–TE–FOX–C (Erythromycin, Tetracycline, Cefoxitin, Chloramphenicol) resistance pattern, and one isolate had a GM–E–TE–FOX–C (Gentamicin, Erythromycin, Tetracycline, Cefoxitin, Chloramphenicol) antibiotic resistance pattern (Fig. 3) and was resistant to three to four groups of antibiotics (Table 3). Three MDR S. aureus isolates from the Animal Husbandry Service Animal Hospital and two MDR S. aureus isolates were discovered in cat nose swab samples from the Airlangga University Animal Teaching Hospital, respectively (Table 4). Thus, a total of 5 MDR S. aureus isolates were obtained from 100 samples of cat nasal discharge analyzed in Surabaya. Confirmatory MRSAAccording to Table 2, six of the 64 S. aureus isolates tested positive for the ORSAB test. The six isolates that the ORSAB test were identified as MRSA and then genotyped to ascertain the presence of the mecA gene. Based on the electrophoresis results of the six isolates examined, it was discovered that five of the isolates carried the mecA gene (Table 2). The appearance of a single band on the electrophoresis findings indicates a positive mecA gene (Fig. 4). DiscussionStaphylococcus aureus, a common animal flora generally found in animals, is linked to the discovery of S. aureus in the cat’s nose (Mala et al., 2021). These bacteria are a typical commensal bacterium that lives in both human and many animal species noses, throats, and skin (Heaton et al., 2020; Aslam et al., 2021). The results of this study obtained 64 isolates of S. aureus in nasal swabs of cats in Surabaya. S. aureus bacteria can infect cats and be transmitted to humans, especially those with weakened or compromised immune systems (Bierowiec et al., 2019). The majority of infected cats do not exhibit health issues, according to clinical detection of S. aureus colonization in cats, albeit opportunistic infections may start to manifest (Waruwu et al., 2023). Physical contact, particularly by touching infected blood or bodily fluids, can spread Staphylococcus infections (Afnani et al., 2022). Table 1. Details of primers used in this study.
Table 2. Isolation of S. aureus, MRSA, and mecA gene from cat nose swabs.
Fig. 1. Colonies of S. aureus in MSA. It is not realized that transmission of S. aureus from cats can occur due to frequent direct contact between humans and cats, this is because cases of S. aureus infection in humans are more widely known in the community (Khairullah et al., 2023). Because cats are frequently maintained as pets and are therefore a reservoir for the spread of S. aureus, there is a chance of zoonotic transmission (Khairullah et al., 2020). Cats are susceptible to infection because S. aureus is present on their skin and fur (Bierowiec et al., 2016). Recent research has indicated that cats may have a role in the transmission of S. aureus within the home (Afnani et al., 2022; Millannia et al., 2023; Waruwu et al., 2023).
Fig. 2. Colonies of S. aureus stained with Gram under a microscope.
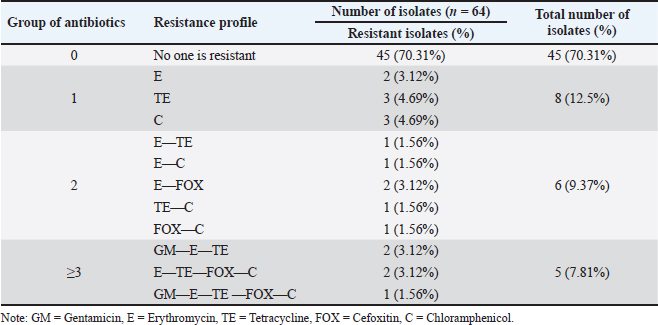
Fig. 3. Analyze the antibiotic sensitivity of a S. aureus isolate grown on MHA. The S. aureus isolate sensitivity results showed some resistance to the antibiotics erythromycin, tetracycline, cefoxitin, gentamicin, and chloramphenicol, each tested with variable degrees of resistance. The findings revealed that 29.69% (19/64) of the S. aureus isolates from cat nose swab samples at the Surabaya City Animal Hospital were resistant to antibiotics. These isolates included 11 from the Airlangga University Animal Hospital and 8 from the East Java Animal Husbandry Service Animal Hospital. These findings support the claims made by Qekwana et al. (2017) who isolated 63% (24/38) of S. aureus from cats at the Teaching Animal Hospital in South Africa has antibiotic resistance. Table 3. Profile of antibiotic resistance in isolated S. aureus.
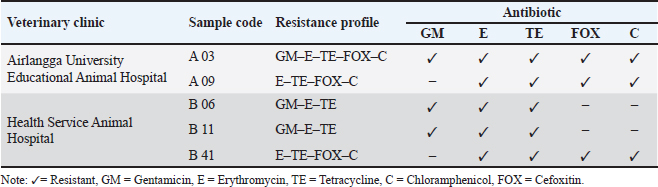
Table 4. Isolates of S. aureus with an MDR profile.
There are several ways that can trigger the resistance mechanisms, including the alteration of antibiotic targets, upregulation of efflux pumps, and enzymatic inactivation of antibiotics (Serwecińska, 2020; Lade and Kim, 2021). One factor that is related to antibiotic resistance is the of antibiotics in an earlier stage of the S. aureus infection in cats. Long-term use of antibiotics can induce mutations and genetic changes in bacterial cells, and also lead to the ineffectiveness of antibiotic treatment and the development of resistance to that antibiotic (Baym et al., 2016). The two antibiotics that are most frequently used to treat cats are tetracycline and chloramphenicol, resistance develops as a result of this condition. In Indonesia, tetracycline and chloramphenicol antibiotics are most commonly used to treat infections, which reflects the resistance associated with these antibiotics (Waruwu et al., 2023). Additionally, the results of this study agree with those of Morris et al. (2006), who found that 13 S. aureus isolates from cats had significant levels of resistance to tetracycline (94%), erythromycin (82%), and chloramphenicol (100%). S. aureus isolates from nasal swabs from animals, including cats, were shown to be resistant to the antibiotics erythromycin, chloramphenicol, and tetracycline in Taiwan, according to Wan et al. (2012). According to the study’s findings, S. aureus has a lower level of gentamicin resistance than other antibiotics, namely 4.69% (3/64). The findings of this investigation are consistent with those of Rubin et al. (2011), who found that S. aureus isolated from cats in veterinary clinics has a low level of gentamicin resistance. The use of gentamicin still needs to be closely monitored, even though the pattern of gentamicin resistance in this study was only 4.69% because the results of this study showed that there were already 3 isolates of S. aureus that were resistant to gentamicin. This antibiotic is frequently used in clinical settings in combination with other antibiotics to treat Gram-positive and Gram-negative bacteria (Serio et al., 2018).
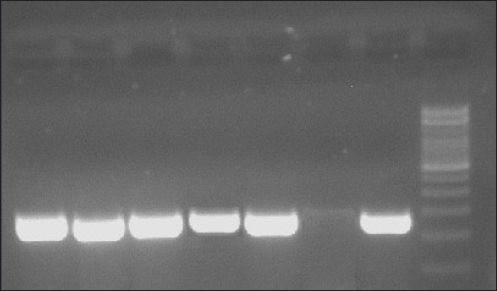
Fig. 4. mecA gene detection results. A03, A09, B06, B11, B41=Positive mecA; B49=Negative mecA; K+=Positive control; M=Marker. In addition, 9.37% (6/64) of S. aureus isolates in this study were found to be resistant to cefoxitin. The results of this investigation are also consistent with the study by Millannia et al. (2023), which found that cefoxitin resistance from cats in Animal Hospital in Indonesia was less common than resistance to methicillin and oxacillin. Cefoxitin is a second-generation antibiotic that is particularly efficient against Staphylococcus species and several Gram-negative bacteria like Eschericia coli, Klebsiella, and Proteus (Bazaid et al., 2021). Staphylococcus aureus isolates that are resistant to cefoxitin can be suspected to be MRSA isolates because cefoxitin is a β-lactam antibiotic. In this study, 7.81% (5/64) of S. aureus bacteria were found to carry the mecA gene, namely in isolates A03 and A09 from the Teaching Animal Hospital of Airlangga University and B06, B11, and B41 from the Animal Husbandry Service Animal Hospital East Java. These results agree with those of Qekwana et al. (2017), which found that MDR S. aureus was more common in cats than S. intermedius, with a prevalence of 28.57% (2/7) compared to 11.76% (2/17). Additionally, Ferradas et al. (2022) found a 50% (12/24) MDR of S. aureus from cats in the mid-Atlantic region. A multidrug-resistant bacterium is one that is resistant to three or more drugs. MDR events can occur for a number of reasons, including the administration of antibiotics at the wrong dose, an incorrect diagnosis, and the wrong bacteria (Yunita et al., 2020). One of two molecular pathways can lead to MDR in bacteria. First, these bacteria can have multiple drug-resistance genes in a single cell; this accumulation typically occurs on resistance plasmids (Nikaido, 2009). MDR can also result from an increase in the expression of genes encoding efflux multidrug pumps, which expel a variety of antibiotics (Spengler et al., 2017; Khairullah et al., 2022b). With only 6 MRSA isolates found out of 100 samples examined, this indicates that the MRSA infection rate at the Surabaya Veterinary Hospital is still low. A significant DNA insertion between 20 and 100 kb, known as SCCmec, caused S. aureus to evolve into a methicillin-resistant form of the bacterium (Rolo et al., 2017). Due to modifications in the native penicillin-binding protein (PBP), specifically from PBP2 to PBP2a, MRSA isolates are no longer susceptible to any β-lactam antibiotic (Otto, 2013). Although PBP2a is cultivated in environments with elevated levels of β-lactams, the MRSA strain can persist, which can be attributed to PBP2a’s relatively poor affinity for β-lactam antibiotics (Fergestad et al., 2020). Several resistance genes, notably the mecA gene, are encoded by the pathogenic bacteria S. aureus that exhibits MRSA characteristics (Hiramatsu et al., 2013). The mecA gene on the SCCmec chromosome of MRSA encodes a specific transpeptidase that confers antibiotic resistance to bacteria against methicillin and β-lactams (Fishovitz et al., 2014). The PBP2a protein is produced and the β-lactam class of antibiotics has a moderate affinity for this protein (Hiramatsu et al., 2013; Ramandinianto et al., 2020). All types of β-lactam antibiotics will not affect the bacteria that generate this protein (Fisher and Mobashery, 2016). In the MRSA isolates, mecA gene is directly transmissible via touch, aerosols, and inanimate objects (Ramandinianto et al., 2020). Molecular identification of the mecA gene in the same MRSA strains in cats is linked to the people living in the same household and the transfer of bacterial strains between pets and their owners (Afnani et al., 2022). Humans and pets are more likely to be colonized than infected because they act as reservoirs for the mecA gene (Khairullah et al., 2022). To control this cross-contamination, hospitals, veterinary clinics, and animal care facilities must strictly follow established health regulations (Elnageh et al., 2021). Good hygiene practices, including hand washing and environmental cleaning, can help prevent transmission (Avberšek et al., 2021). The use of barrier measures, such as masks and gloves, and the separation of infected animals is important when animals with MRSA infections are present (Algammal et al., 2020). Early molecular identification is crucial to identify MRSA-infected animals carrying the mecA gene (Dey et al., 2023). The overall conclusion of this study is that tetracycline, chloramphenicol, erythromycin, gentamicin, and cefoxitin drugs can still be used as a treatment option for S. aureus infection in cats at the Surabaya City Veterinary Hospital because there are still S. aureus isolates that are susceptible to these antibiotics. However, it is important to be aware of bacteria that have developed resistance when using tetracycline, cefoxitin, chloramphenicol, erythromycin, and gentamicin. In addition, a thorough assessment of the public health implications of the use of antibiotics to control the spread of MRSA in veterinary hospitals is required in light of the detection of MRSA isolates expressing the mecA gene in the Surabaya Veterinary Hospital. ConclusionThis study concludes that from 100 cat nasal swab samples in Surabaya, 6 isolates were confirmed to be MRSA and 5 of them contained the mecA gene. Early detection of MRSA in pets is essential to identify the spread of MRSA that threatens public health. AcknowledgmentsThe authors are thankful to the Universitas Airlangga for providing the necessary facilities for this study. Conflict of interestThe authors declare that there is no conflict of interest. FundingThis study was partly supported by the International Research Consortium, Lembaga Penelitian dan Pengabdian Masyarakat, Universitas Airlangga, Surabaya, Indonesia Year 2024 with grant number: 171/UN3.LPPM/PT.01.03/2024. Author’s contributionsSR and SKM: Conceived, designed, and coordinated the study. MHE and ARK: Designed data collections tools, supervised the field sample and data collection, and laboratory work as well as data entry. WT and IBM: Validation, supervision, and formal analysis. MS and DAA: Contributed reagents, materials, and analysis tools. SEK and MTIF: Carried out the statistical analysis and interpretation and participated in the preparation of the manuscript. All authors have read, reviewed, and approved the final manuscript. Data availabilityAll data are available in the manuscript. ReferencesAfnani, D.A., Fatih, N., Effendi, M.H., Tyasningsih, W., Kairullah, A.R., Kurniawan, S.C., Silaen, O.S.M., Ramandianto, S.C., Widodo, A., Hendriana, K. and Riwu, K.H.P. 2022. Profile of multidrug resistance and methicillin-resistant Staphylococcus aureus (MRSA) isolated from cats in Surabaya, Indonesia. Biodiversitas 23(11), 5703–5709. Algammal, A.M., Hetta, H.F., Elkelish, A., Alkhalifah, D.H.H., Hozzein, W.N., Batiha, G.E., El Nahhas, N. and Mabrok, M.A. 2020. Methicillin-resistant Staphylococcus aureus (MRSA): one health perspective approach to the bacterium epidemiology, virulence factors, antibiotic-resistance, and zoonotic impact. Infect. Drug Resist. 13(1), 3255–3265. Aslam, B., Khurshid, M., Arshad, M.I., Muzammil, S., Rasool, M., Yasmeen, N., Shah, T., Chaudhry, T.H., Rasool, M.H., Shahid, A., Xueshan, X. and Baloch, Z. 2021. Antibiotic resistance: one health one world outlook. Front. Cell. Infect. Microbiol. 11(1), 771510. Avberšek, J., Papić, B., Kušar, D., Erjavec, V., Seme, K., Golob, M. and Zdovc, I. 2021. Feline otitis externa caused by methicillin-resistant Staphylococcus aureus with mixed hemolytic phenotype and overview of possible genetic backgrounds. Antibiotics (Basel) 10(5), 599. Baym, M., Stone, L.K. and Kishony, R. 2016. Multidrug evolutionary strategies to reverse antibiotic resistance. Science 351(6268), aad3292. Bazaid, A.S., Saeed, A., Alrashidi, A., Alrashidi, A., Alshaghdali, K., Hammam, S.A., Alreshidi, T., Alshammary, M., Alarfaj, A., Thallab, R. and Aldarhami, A. 2021. Antimicrobial surveillance for bacterial uropathogens in Ha’il, Saudi Arabia: a five-year multicenter retrospective study. Infect. Drug Resist. 14(1), 1455–1465. Bierowiec, K., Korzeniowska-Kowal, A., Wzorek, A., Rypuła, K. and Gamian, A. 2019. Prevalence of staphylococcus species colonization in healthy and sick cats. BioMed Res. Int. 2019(1), 4360525. Bierowiec, K., Płoneczka-Janeczko, K. and Rypuła, K. 2016. Is the Colonisation of Staphylococcus aureus in pets associated with their close contact with owners? PLoS One 11(5), e0156052. Costa, S.S., Ribeiro, R., Serrano, M., Oliveira, K., Ferreira, C., Leal, M., Pomba, C. and Couto, I. 2022. Staphylococcus aureus causing skin and soft tissue infections in companion animals: antimicrobial resistance profiles and clonal lineages. Antibiotics 11(5), 599. Decline, V., Effendi, M.H., Rahmaniar, R.P., Yanestria, S.M. and Harijani, N. 2020. Profile of antibiotic-resistant and presence of methicillin-resistant Staphylococcus aureus from nasal swab of dogs from several animal clinics in Surabaya, Indonesia. Int. J. One Health 6(1), 90–94. Dey, T.K., Shome, B.R., Bandyopadhyay, S., Goyal, N.K., Lundkvist, Å., Deka, R.P., Shome, R., Venugopal, N., Grace, D., Sharma, G., Rahman, H. and Lindahl, J.F. 2023. Molecular characterization of methicillin-resistant staphylococci from the dairy value chain in two Indian States. Pathogens 12(2), 344. Effendi, M.H., Hisyam, M.A.M., Hastutiek, P. and Tyasningsih, W. 2019. Detection of coagulase gene in Staphylococcus aureus from several dairy farms in East Java, Indonesia, by polymerase chain reaction. Vet. World 12(1), 68–71. Effendi, M.H., Oktavianto, A. and Hastutiek, P. 2018. Tetracycline resistance gene in Streptococcus agalactiae isolated from bovine subclinical mastitis in Surabaya, Indonesia. Philip. J. Vet. Med. 55(Special Issue), 115–120. Elmoslemany, A., Elsohaby, I., Alorabi, M., Alkafafy, M., Al-Marri, T., Aldoweriej, A., Alaql, F. A., Almubarak, A. and Fayez, M. 2021. Diversity and risk factors associated with multidrug and methicillin-resistant staphylococci isolated from cats admitted to a veterinary clinic in Eastern Province, Saudi Arabia. Antibiotics 10(4), 367. Elnageh, H.R., Hiblu, M.A., Abbassi, M.S., Abouzeed, Y.M. and Ahmed, M.O. 2021. Prevalence and antimicrobial resistance of Staphylococcus species isolated from cats and dogs. Open Vet. J. 10(4), 452–456. Fergestad, M.E., Stamsås, G.A., Angeles, D.M., Salehian, Z., Wasteson, Y. and Kjos, M. 2020. Penicillin-binding protein PBP2a provides variable levels of protection toward different β-lactams in Staphylococcus aureus RN4220. Microbiologyopen 9(8), e1057. Ferradas, C., Cotter, C., Shahbazian, J.H., Iverson, S.A., Baron, P., Misic, A.M., Brazil, A.M., Rankind, S.C., Nachamkin, I., Ferguson, J.M., Peng, R.D., Bilker, W.B., Lautenbach, E., Morris, D.O., Lescano, A.G. and Davis, M.F. 2022. Risk factors for antimicrobial resistance among Staphylococcus isolated from pets living with a patient diagnosed with methicillin-resistant Staphylococcus aureus infection. Zoonoses Public Health 69(5), 550–559. Fisher, J.F. and Mobashery, S. 2016. β-lactam resistance mechanisms: gram-positive bacteria and mycobacterium tuberculosis. Cold Spring Harb. Perspect. Med. 6(5), a025221. Fishovitz, J., Hermoso, J.A., Chang, M. and Mobashery, S. 2014. Penicillin-binding protein 2a of methicillin-resistant Staphylococcus aureus. IUBMB Life 66(8), 572–577. Fitranda, M., Salasia, S.I.O., Sianipar, O., Dewananda, D.A., Arjana, A.Z., Aziz, F., Wasissa, M., Lestari, F.B. and Santosa, C.M. 2023. Methicillin-resistant Staphylococcus aureus isolates derived from humans and animals in Yogyakarta, Indonesia. Vet. World 16(1), 239–245. Gajdács, M. 2019. The continuing threat of methicillin-resistant Staphylococcus aureus. Antibiotics (Basel) 8(2), 52. Gebremedhin, E.Z., Ararso, A.B., Borana, B.M., Kelbesa, K.A., Tadese, N.D., Marami, L.M. and Sarba, E.J. 2022. Isolation and identification of Staphylococcus aureus from milk and milk products, associated factors for contamination, and their antibiogram in Holeta, Central Ethiopia. Vet. Med. Int. 2022(1), 6544705. Green, B.N., Johnson, C.D., Egan, J.T., Rosenthal, M., Griffith, E.A. and Evans, M.W. 2012. Methicillin-resistant Staphylococcus aureus: an overview for manual therapists. J. Chiropr. Med. 11(1), 64–76. Gwenzi, W., Chaukura, N., Muisa-Zikali, N., Teta, C., Musvuugwa, T., Rzymski, P. and Abia, A.L.K. 2021. Insects, rodents, and pets as reservoirs, vectors, and sentinels of antimicrobial resistance. Antibiotics 10(1), 68. Heaton, C.J., Gerbig, G.R., Sensius, L.D., Patel, V. and Smith, T.C. 2020. Staphylococcus aureus epidemiology in Wildlife: a systematic review. Antibiotics (Basel) 9(2), 89. Hesselmar, B., Hicke-Roberts, A., Lundell, A.C., Adlerberth, I., Rudin, A., Saalman, R., Wennergren, G. and Wold, A.E. 2018. Pet-keeping in early life reduces the risk of allergy in a dose-dependent fashion. PLoS One 13(1), e0208472. Hiramatsu, K., Ito, T., Tsubakishita, S., Sasaki, T., Takeuchi, F., Morimoto, Y., Katayama, Y., Matsuo, M., Kuwahara-Arai, K., Hishinuma, T. and Baba, T. 2013. Genomic basis for methicillin resistance in Staphylococcus aureus. Infect. Chemother. 45(2), 117–136. Kaben, S.E., Al-Arif, M.A., Ma’ruf, A., Effendi, M.H., Kurniawan, S.C., Riwu, K.H.P., Afnani, D.A., Silaen, O.S.M., Millania, S.K., Ramadhani, S., Widodo, A., Farizqi, M.T.I. and Khairullah, A.R. 2024. Detection mecA gene and Staphylococcus aureus resistance to several antibiotics isolated from cat ear swabs at a veterinary hospital located at Surabaya—Indonesia. Braz. J. Vet. Res. Anim. Sci. 61(1), e209027. Khairullah, A.R., Rahardjo, D., Rahmahani, J., Suwarno, Tyasningsih, W. and Harijani, N. 2019. Antibiotics resistant at Staphylococcus aureus and Steptococcus sp isolated from bovine mastitis in Karangploso, East Java, Indonesia. Indian J. Forensic Med. Toxicol. 13(4), 439–444. Khairullah, A.R., Ramandinianto, S.C. and Effendi, M.H. 2020a. A review of livestock-associated methicillin-resistant Staphylococcus aureus (LA-MRSA) on bovine mastitis. Syst. Rev. Pharm. 11(7), 172–183. Khairullah, A.R., Rehman, S., Sudjarwo, S.A., Effendi, M.H., Ramandinianto, S.C., Gelolodo, M.A., Widodo, A., Riwu, K.H.P. and Kurniawati, D.A. 2022a. Detection of mecA gene and methicillin-resistant Staphylococcus aureus (MRSA) isolated from milk and risk factors from farms in Probolinggo, Indonesia. F1000Res. 11(1), 722. Khairullah, A.R., Sudjarwo, S.A., Effendi, M.H., Ramandinianto, S.C., Gelolodo, M.A., Widodo, A., Riwu, K.H.P., Kurniawati, D.A. and Rehman, S. 2022b. Profile of multidrug resistance and methicillin-resistant Staphylococcus aureus (MRSA) on dairy cows and risk factors from farmer. Biodiversitas 23(6), 2853–2858. Khairullah, A.R., Sudjarwo, S.A., Effendi, M.H., Harijani, N., Tyasningsih, W., Rahmahani, J., Permatasari, D.A., Ramandinianto, S.C., Widodo, A. and Riwu, K.H.P. 2020b. A review of methicillin-resistant Staphylococcus aureus (MRSA) on milk and milk products: public health importance. Syst. Rev. Pharm. 11(8), 59–69. Khairullah, A.R., Sudjarwo, S.A., Effendi, M.H., Ramandinianto, S.C., Gelolodo, M.A., Widodo, A., Riwu, K.H.P. and Kurniawati, D.A. 2023. Pet animals as reservoirs for spreading methicillin-resistant Staphylococcus aureus to human health. J. Adv. Vet. Anim. Res. 10(1), 1–13. Lade, H. and Kim, J.S. 2021. Bacterial targets of antibiotics in methicillin-resistant Staphylococcus aureus. Antibiotics (Basel) 10(4), 398. Llor, C. and Bjerrum, L. 2014. Antimicrobial resistance: risk associated with antibiotic overuse and initiatives to reduce the problem. Ther. Adv. Drug Saf. 5(6), 229–241. Mala, L., Lalouckova, K. and Skrivanova, E. 2021. Bacterial skin infections in livestock and plant-based alternatives to their antibiotic treatment. Animals (Basel) 11(8), 2473. Millannia, S.K., Khairullah, A.R., Effendi, M.H., Utama, S., Kurniawan, S.C., Afnani, D.A., Silaen, O.S.M., Ramadhani, S., Ramandinianto, S.C., Waruwu, Y.K.K., Widodo, A., Putra, G.D.S., Farizqi, M.T.I. and Riwu, K.H.P. 2023. Phenotypic detection strategies of multidrug-resistant Staphylococcus aureus isolated from cat nasal swab in Madiun city, Indonesia. Biodiversitas 24(2), 940–946. Morris, D.O., Mauldin, E.A., O’Shea, K., Shofer, F.S. and Rankin, S.C. 2006. Clinical, microbiological, and molecular characterization of methicillin-resistant Staphylococcus aureus infections of cats. Am. J. Vet. Res. 67(8), 1421–1425. Nikaido, H. 2009. Multidrug resistance in bacteria. Ann. Rev. Biochem. 78(1), 119–146. Nimer, N.A. 2022. Nosocomial infection and antibiotic-resistant threat in the middle east. Infect. Drug Resist. 15(1), 631–639. Noto, M.J., Kreiswirth, B.N., Monk, A.B. and Archer, G.L. 2008. Gene acquisition at the insertion site for SCCmec, the genomic island conferring methicillin resistance in Staphylococcus aureus. J. Bacteriol. 190(4), 1276–1283. Otto, M. 2013. Coagulase-negative staphylococci as reservoirs of genes facilitating MRSA infection: staphylococcal commensal species such as Staphylococcus epidermidis are being recognized as important sources of genes promoting MRSA colonization and virulence. Bioessays 35(1), 4–11. Qekwana, D.N., Sebola, D., Oguttu, J.W. and Odoi, A. 2017. Antimicrobial resistance patterns of Staphylococcus species isolated from cats presented at a veterinary academic hospital in South Africa. BMC Vet. Res. 13(1), 286. Rahman, M.T., Sobur, M.A., Islam, M.S., Ievy, S., Hossain, M.J., El Zowalaty, M.E., Rahman, A.T. and Ashour, H.M. 2020. Zoonotic diseases: etiology, impact, and control. Microorganisms 8(9), 1405. Rahmaniar, R.P., Yunita, M.N., Effendi, M.H. and Yanestria, S.M. 2020. Encoding gene for methicillin resistant Staphylococcus aureus (MRSA) isolated from nasal swab of dogs. Indian Vet. J. 97(2), 37–40. Rajabiani, A., Kamrani, F., Boroumand, M.A. and Saffar, H. 2014. mec-A-mediated resistance in Staphylococcus aureus in a referral Hospital, Tehran, Iran. Jundishapur J. Microbiol. 7(4), e9181. Ramandinianto, S.C., Khairullah, A.R. and Effendi, M.H. 2020. MecA gene and methicillin-resistant Staphylococcus aureus (MRSA) isolated from dairy farms in East Java, Indonesia. Biodiversitas 21(8), 3562–3568. Rolo, J., Worning, P., Nielsen, J.B., Bowden, R., Bouchami, O., Damborg, P., Guardabassi, L., Perreten, V., Tomasz, A., Westh, H., de Lencastre, H. and Miragaia, M. 2017. Evolutionary origin of the staphylococcal cassette chromosome mec (SCCmec). antimicrob. Agents Chemother. 61(6), e02302–16. Rubin, J.E., Ball, K.R. and Chirino-Trejo, M. 2011. Antimicrobial susceptibility of Staphylococcus aureus and Staphylococcus pseudintermedius isolated from various animals. Can. Vet. J. 52(2), 153–157. Saber, H., Jasni, A.S., Jamaluddin, T.Z.M.T. and Ibrahim, R. 2017. A review of staphylococcal cassette chromosome mec (SCCmec) types in coagulase-negative staphylococci (CoNS) Species. Malays. J. Med. Sci. 24(5), 7–18. Saputra, S., Jordan, D., Worthing, K.A., Norris, J.M., Wong, H.S., Abraham, R., Trott, D.J. and Abraham, S. 2017. Antimicrobial resistance in coagulase-positive staphylococci isolated from companion animals in Australia: a one year study. PLoS One 12(1), e0176379. Serio, A .W., Keepers, T., Andrews, L. and Krause, K.M. 2018. Aminoglycoside revival: review of a historically important class of antimicrobials undergoing rejuvenation. EcoSal Plus. 8(1), 10.1128. Serwecińska, L. 2020. Antimicrobials and antibiotic-resistant bacteria: a risk to the environment and to public health. Water 12(12), 3313. Spengler, G., Kincses, A., Gajdács, M. and Amaral, L. 2017. New roads leading to old destinations: efflux pumps as targets to reverse multidrug resistance in bacteria. Molecules 22(3), 468. Teo, J.W., Kum, S., Jureen, R. and Lin, R.T. 2015. Molecular characterization of a catalase-negative Staphylococcus aureus blood culture isolate. J. Clin. Microbiol. 53(11), 3699–3701. Tyasningsih, W., Ramandinianto, S.C., Ansharieta, R., Witaningrum, A.M., Permatasari, D.A., Wardhana, D.K., Effendi, M.H. and Ugbo, E.N. 2022. Prevalence and antibiotic resistance of Staphylococcus aureus and Escherichia coli isolated from raw milk in East Java, Indonesia. Vet. World 15(8), 2021–2028. Wan, M.T., Fu, S.Y., Lo, Y.P., Huang, T.M., Cheng, M.M. and Chou, C.C. 2012. Heterogeneity and phylogenetic relationships of community-associated methicillin-sensitive/resistant Staphylococcus aureus isolates in healthy dogs, cats and their owners. J. Appl. Microbiol. 112(1), 205–213. Waruwu, Y.K.K., Khairullah, A.R., Effendi, M.H., Lukiswanto, B.S., Afnani, D.A., Kurniawan, S.C., Silaen, O.S.M., Riwu, K.H.P., Widodo, A. and Ramandinianto, S.C. 2023. Detection of methicillin-resistant Staphylococcus aureus and multidrug resistance isolated from cats in animal clinic at Sidoarjo District, East Java, Indonesia. Biodiversitas 24(1), 106–111. Widodo, A., Lamid, M., Effendi, M.H., Khairullah, A.R., Riwu, K.H.P., Yustinasari, L.R., Kurniawan, S.C., Ansori, A.N.M., Silaen, O.S.M. and Dameanti, F.N.A.E.P. 2022. Antibiotic sensitivity profile of multidrug-resistant (MDR) Escherichia coli isolated from dairy cow’s milk in Probolinggo, Indonesia. Biodiversitas 23(10), 4971–4976. Yembise, D., Ariestanti, C.A. and Budiarso, T.Y. 2020. Isolation and identification of biofilm-forming Staphylococcus aureus in commercial cow milk products. Sciscitatio 1(2), 87–93. Yunita, M.N., Effendi, M.H., Rahmaniar, R.P., Arifah, S. and Yanestria, S.M. 2020. Identification of spa gene for strain typing of methicillin resistant Staphylococcus aureus (MRSA) isolated from nasal swab of dogs. Biochem. Cell. Arch. 20(1), 2999–3004. | ||
| How to Cite this Article |
| Pubmed Style Ramadhani S, Khairullah AR, Effendi MH, Sukmanadi M, Tyasningsih W, Millannia SK, Afnani DA, Moses IB, Farizqi MTI, Kaben SE. Detection of the methicillin-resistant Staphylococcus aureus (MRSA) gene encoding mecA in nasal swabs of cats in Surabaya, Indonesia. Open Vet. J.. 2024; 14(11): 3026-3036. doi:10.5455/OVJ.2024.v14.i11.31 Web Style Ramadhani S, Khairullah AR, Effendi MH, Sukmanadi M, Tyasningsih W, Millannia SK, Afnani DA, Moses IB, Farizqi MTI, Kaben SE. Detection of the methicillin-resistant Staphylococcus aureus (MRSA) gene encoding mecA in nasal swabs of cats in Surabaya, Indonesia. https://www.openveterinaryjournal.com/?mno=217834 [Access: December 02, 2025]. doi:10.5455/OVJ.2024.v14.i11.31 AMA (American Medical Association) Style Ramadhani S, Khairullah AR, Effendi MH, Sukmanadi M, Tyasningsih W, Millannia SK, Afnani DA, Moses IB, Farizqi MTI, Kaben SE. Detection of the methicillin-resistant Staphylococcus aureus (MRSA) gene encoding mecA in nasal swabs of cats in Surabaya, Indonesia. Open Vet. J.. 2024; 14(11): 3026-3036. doi:10.5455/OVJ.2024.v14.i11.31 Vancouver/ICMJE Style Ramadhani S, Khairullah AR, Effendi MH, Sukmanadi M, Tyasningsih W, Millannia SK, Afnani DA, Moses IB, Farizqi MTI, Kaben SE. Detection of the methicillin-resistant Staphylococcus aureus (MRSA) gene encoding mecA in nasal swabs of cats in Surabaya, Indonesia. Open Vet. J.. (2024), [cited December 02, 2025]; 14(11): 3026-3036. doi:10.5455/OVJ.2024.v14.i11.31 Harvard Style Ramadhani, S., Khairullah, . A. R., Effendi, . M. H., Sukmanadi, . M., Tyasningsih, . W., Millannia, . S. K., Afnani, . D. A., Moses, . I. B., Farizqi, . M. T. I. & Kaben, . S. E. (2024) Detection of the methicillin-resistant Staphylococcus aureus (MRSA) gene encoding mecA in nasal swabs of cats in Surabaya, Indonesia. Open Vet. J., 14 (11), 3026-3036. doi:10.5455/OVJ.2024.v14.i11.31 Turabian Style Ramadhani, Safira, Aswin Rafif Khairullah, Mustofa Helmi Effendi, Mohammad Sukmanadi, Wiwiek Tyasningsih, Saumi Kirey Millannia, Daniah Ashri Afnani, Ikechukwu Benjamin Moses, M. Thoriq Ihza Farizqi, and Sergius Erikson Kaben. 2024. Detection of the methicillin-resistant Staphylococcus aureus (MRSA) gene encoding mecA in nasal swabs of cats in Surabaya, Indonesia. Open Veterinary Journal, 14 (11), 3026-3036. doi:10.5455/OVJ.2024.v14.i11.31 Chicago Style Ramadhani, Safira, Aswin Rafif Khairullah, Mustofa Helmi Effendi, Mohammad Sukmanadi, Wiwiek Tyasningsih, Saumi Kirey Millannia, Daniah Ashri Afnani, Ikechukwu Benjamin Moses, M. Thoriq Ihza Farizqi, and Sergius Erikson Kaben. "Detection of the methicillin-resistant Staphylococcus aureus (MRSA) gene encoding mecA in nasal swabs of cats in Surabaya, Indonesia." Open Veterinary Journal 14 (2024), 3026-3036. doi:10.5455/OVJ.2024.v14.i11.31 MLA (The Modern Language Association) Style Ramadhani, Safira, Aswin Rafif Khairullah, Mustofa Helmi Effendi, Mohammad Sukmanadi, Wiwiek Tyasningsih, Saumi Kirey Millannia, Daniah Ashri Afnani, Ikechukwu Benjamin Moses, M. Thoriq Ihza Farizqi, and Sergius Erikson Kaben. "Detection of the methicillin-resistant Staphylococcus aureus (MRSA) gene encoding mecA in nasal swabs of cats in Surabaya, Indonesia." Open Veterinary Journal 14.11 (2024), 3026-3036. Print. doi:10.5455/OVJ.2024.v14.i11.31 APA (American Psychological Association) Style Ramadhani, S., Khairullah, . A. R., Effendi, . M. H., Sukmanadi, . M., Tyasningsih, . W., Millannia, . S. K., Afnani, . D. A., Moses, . I. B., Farizqi, . M. T. I. & Kaben, . S. E. (2024) Detection of the methicillin-resistant Staphylococcus aureus (MRSA) gene encoding mecA in nasal swabs of cats in Surabaya, Indonesia. Open Veterinary Journal, 14 (11), 3026-3036. doi:10.5455/OVJ.2024.v14.i11.31 |