| Research Article | ||
Open Vet. J.. 2025; 15(3): 1440-1445 Open Veterinary Journal, (2025), Vol. 15(3): 1440-1445 Research Article Evaluation of a commercial real-time polymerase chain reaction kit for the detection of avian orthoavulavirus type IHasnae Zekhnini1,2*, Fatiha El Mellouli2, Mohamed Rida Salam1, Faiza Bennis1, and Fatima Chegdani11Laboratory of Immunology and Biodiversity, Faculty of Science Ain Chock, Hassan II University of Casablanca, Casablanca, Morocco 2Regional Laboratory of Analysis and Research Casablanca, National Office for Food Safety “ONSSA”, Casablanca, Morocco *Corresponding Author: Hasnae Zekhnini. Regional Laboratory of Analysis and Research Casablanca, National Office for Food Safety “ONSSA”, Casablanca, Morocco. Email: ha.zekhnini [at] gmail.com Submitted: 25/09/2024 Accepted: 15/02/2025 Published: 31/03/2025 © 2025 Open Veterinary Journal
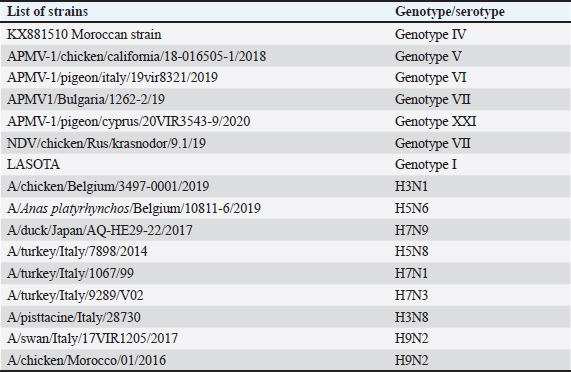
AbstractBackground: Newcastle disease (ND) is a highly contagious and often fatal viral disease that affects a wide range of avian species, regardless of age or sex. It continues to represent a significant challenge to the productivity and survival of both commercial and traditional poultry, particularly in developing countries. Despite advancements in vaccination strategies, ND outbreaks remain a recurring issue in many areas, including Morocco. Early detection of Newcastle Disease Virus (NDV) is critical for effective disease management, but conventional virus isolation methods are labor-intensive and time-consuming. Aim: This study aimed to evaluate and validate the performance of the ID Gene™ Newcastle Disease Duplex real-time RT-PCR kit for the rapid and sensitive detection of NDV in poultry samples. The validation was performed in accordance with international OIE standards to ensure reliability and applicability in veterinary diagnostics. Methods: The study utilized spiked biological samples and NDV strains, including Moroccan field strains and reference strains from the Isntituto Zooprofilattico Sperimentale delle Venezie proficiency panel. RNA extraction was performed using the NucleoSpin® RNA Virus kit. Negative tracheal and cloacal swab supernatants, as well as homogenized tissue samples from the liver and lungs, were spiked with JEL/Morocco Newcastle virus, genotype VI at different concentrations. Real-time RT-PCR targeting the NDV M gene was performed with an exogenous internal control for duplex amplification. The analytical specificity was assessed for inclusivity with various NDV genotypes and exclusivity against other orthomyxoviruses. Sensitivity was determined using 10-fold serial dilutions of NDV, with detection limits evaluated for individual and pooled samples. Repeatability was assessed by calculating intra-assay and inter-assay coefficients of variation based on Ct values. Results: The ID Gene™ kit demonstrated excellent inclusivity and reliably detected all tested NDV strains irrespective of genotype while showing no cross-reactivity with non-NDV orthomyxoviruses. The analytical sensitivity was validated at 102 DIO50/ml for individual samples and 103 DIO50/ml for pools of five samples, respectively, across all tested matrices. The intraassay and interassay repeatability coefficients of variation (CV) were consistently below 10%, confirming the robustness of the method. Conclusion: The ID Gene™ Newcastle Disease Duplex Kit is a rapid, accurate, and sensitive diagnostic alternative to traditional virus isolation methods. The performance of this method, validated against international and national standards, highlights its potential as a reliable tool for the early detection and effective management of NDV infections in Moroccan poultry populations. Keywords: Avian orthoavulavirus type I, Polymerase chain reaction (PCR), Validation. IntroductionNewcastle disease (ND) is a fatal viral disease that often affects a wide range of avian hosts, regardless of age or sex. A major obstacle to the development, survival, and productivity of both commercial and traditional poultry has been reported to be this disease. The virus belongs to the Paramyxoviridae family with a single serotype: the avian paramyxovirus serotype 1 (APMV-1) (Bujarski et al., 2019). It is classified into two classes, based on the variability of the F fusion protein or the entire genome. Class I comprises virulent strains found in poultry, whereas Class II comprises virulent strains exhibiting significant genetic diversity (Diel et al., 2012). Until the recent upsurge in avian influenza, ND was the most important viral disease in poultry, especially in developing countries (Nwanta et al., 2008), with potentially devastating effects on the productivity and survival of both the commercial poultry industry and traditional breeding (Awan et al., 1994). In recent years, the Moroccan poultry sector has experienced a great boom, mainly due to investments made within the framework of the Green Morocco Plan. Despite significant progress in poultry vaccination programs and their effectiveness, outbreaks of infectious diseases, especially ND, are still recorded in many areas where both vaccinated and unvaccinated herds are bred (Rauw et al., 2009). Although the exact economic losses due to infectious diseases remain uncertain, estimates suggest that over 250 million chicks, broilers, and ducklings perish annually in Africa due to various infections (Sonaiya et al., 1999). Therefore, it was imperative to develop diagnostic methods that would allow for the early detection of infected birds. Although virus isolation remains the gold standard for diagnosis, this method requires a high level of laboratory security and takes at least 6–12 days to produce results. During the last decade, different RT-PCR methods have been described for NDV, generally focusing on the M, L, and F genes, sometimes followed by genetic typing. In addition to tracheal and cloacal swabs taken from live and dead animals, organ samples may be used for the detection of viral RNA. Our objective in conducting this study was to test and validate the ID Gene NDV real-time RT-PCR kit (IDvet, Grabels, France) according to the procedures specified by the World Organisation of Animal Health (WOAH) standard for testing diagnostic assays in veterinary medicine and by Moroccan standards equivalent to the French standard for testing PCR tools in animal health. Materials and MethodsSamplesVirus strains The NDV strains selected for testing were included in the proficiency test panel 2021 provided by the Isntituto Zooprofilattico Sperimentale delle Venezie (IZSVe) reference laboratory for IA and NDV or field strains representing different NDV genotypes (Table 1). Other orthomyxoviruses were used to evaluate the exclusivity of the RT-PCR Kit. Standard procedures were followed to propagate the virus strains in chicken embryonated eggs (WOAH, 2021; Kleij, 2023). Sterile water was used as a no-template control. Spiked biological samples Negative tracheal and cloacal swab supernatants and homogenized tissue samples were spiked with JEL/Morocco Newcastle virus, genotype VI (Genbank accession number KX881510) at different concentrations. The virus was obtained from the Virology Department of the National Expertise and Control Service in Rabat, Morocco. The NDV strain was isolated from a Moroccan field and titrated in ovoculture at 108 DIO50/ml according to the OIE standards. The spiked biological samples were prepared and used to determine the analytical sensitivity of the test. RT-PCR assaysRNA extraction According to the manufacturer’s instructions, RNA was extracted using a Nucleospin RNA Virus kit (Machery-Nagel, Duren, Germany). We extracted viral RNA from 150 µl of water, eluted it in 50 µl of nuclease-free water supplied in the kit, and stored it at –80°C for a maximum of 1 month before analysis. One-step duplex real-time PCRThe kit consists of a real-time PCR kit that is ready for use and provides qualitative results indicating whether or not a sample is detected. This kit targets the M gene of NDV and was developed by IDvet. This method consists of converting NDV RNA to complementary DNA, followed by amplification of cDNA by PCR using primers specific to NDV and Taqman probes labeled with Fluorescein Amidite (FAM). As a result of the one-step RT-PCR assay, both reactions took place in the same tube. The reaction mix also contains primers and a fluorochrome-labeled probe equivalent to VIC (A proprietary fluorescent dye developed by Applied Biosystems (Thermo Fisher Scientific), commonly used in real-time PCR (qPCR) as a reporter fluorophore) or HEX, specific for an exogenous internal control (EIC) provided with the kit, which is added to each sample before RNA extraction. Both target and EIC gene amplifications occurred in a single tube (duplex PCR). On the AriaMX real-time PCR thermocycler (Agilent Technologies, Santa Clara, CA), RT-PCR assays were performed using 96-well plates. The Aria Mx software packages were used to automatically analyze the data, which were then manually verified. Evaluation of RT-PCR performanceThe study was conducted according to the Moroccan and French standards, NM 0.08.538 and UF 4600 part 2, respectively. Analytical specificityTo assess analytical specificity, two steps were taken. As a first step, inclusivity was evaluated using the NDV strains provided by IZSVe, Italy, and the Moroccan field strains. A qualitative assessment of exclusivity was then performed using results obtained with other orthomyxoviruses that cause avian infectious diseases (Table 1). Analytical sensitivityThe analytical sensitivity or detection limit was determined for each matrix separately (tracheal, cloacal swabs, homogenized tissues). The first assessment involved the analysis of 10-fold serial dilutions of the titrated strain to determine the detection limit. The cloacal swab supernatant matrix, considered the richest in inhibitors, was chosen to perform this step. Each dilution was analyzed in triplicate. The last dilution with a positive result in triplicate and 10-fold dilution was checked as the detection limit for the three matrices studied. Each dilution for each matrix was analyzed in quadruplicate in two independent sessions with two different operators. Table 1. List of virus strains.
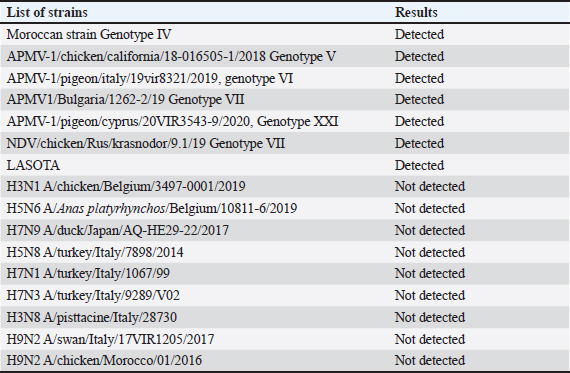
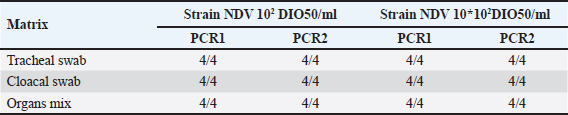
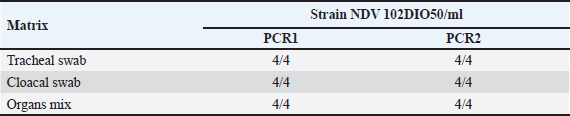
The detection limit was determined for individual samples and up to five pooled samples for each matrix. Preparations of a pool of five at the detection limit For each type of analytical matrix, we diluted the NDV 108 DIO50/ml strain 10-fold in Phosphate-Buffered Saline (PBS) supernatant of negative samples to obtain a suspension at the previously determined detection limit. Then, we further diluted this suspension1/5 in PBS supernatant of negative samples and processed it as described above. Intra-y and inter-assay repeatabilityThe CV was calculated by dividing the standard deviations of the samples by the mean of the cycle threshold values and multiplying the result by 100 to estimate the inter-assay and intra-assay repeatability. CVs below 10% were considered satisfactory (Vandemeulebroucke et al., 2010). Results Analytical specificityThe experimental analytical specificity of the ID Gene™ Newcastle Disease Duplex kit was verified in terms of inclusivity and exclusivity. All selected NDV strains were scored positive regardless of their genotype, confirming that the ID Gene™ Newcastle Disease Duplex kit allows the detection of Moroccan NDV strains, as well as other NDV genotypes. Samples of other orthomyxoviruses were negative at PCR, excluding cross-reactivity with highly pathogenic avian orthomyxoviruses, such as influenza A virus subtypes H5 and H7 (Table 2). Analytical sensitivityLimit detection to be checked The analytical sensitivity was determined for each type of matrix (tracheal swab, cloacal swab, and tissues) and for the extraction method (NucleoSpin® RNA Virus, Macherey-Nagel). For this purpose, pools of PBS supernatants of each type of matrix were formed. We performed extractions using the extraction kit with 150 µl of each dilution of Strain NDV 10 DIO50/ml to Strain NDV 107 DIO50/ml, adding exogenous Non-target positive control (NTPC) from the ID Gene™ Newcastle Disease Duplex kit. After assessing several NDV dilutions, the analytical sensitivity was checked for each matrix and for each extraction method, as the last dilution point at which a positive Reverse Transcription Polymerase Chain Reaction (RT-PCR) result was obtained, was 102 and 103 IOD50/ml for individual and pooled samples of five, respectively. Limit of detection for individual samples With the NucleoSpin® RNA Virus kit (Macherey-Nagel), the validated limit detection method for individual samples was “NDV 102 DIO50/ml” for tracheal and cloacal swabs, as well as for organs (Table 3). Limit detection in pools of five samples With the NucleoSpin® RNA Virus kit, MachereyNagel, the validated “LDMethod for pools is “NDV 102 DIO50/ml” for tracheal and cloacal swabs, as well as for organs (Table 4). Intra-assay and inter-assay repeatabilityThe intra-assay repeatability was between 2% and 4% and 1% for 102 and 103 DIO50/ml, respectively. The inter-assay repeatability was 3% and 1% for 102 and 103 DIO50/ml, respectively. The results are considered satisfactory. Table 2. Strains used to test inclusivity and exclusivity.
Table 3. Determination of analytical sensitivity in individual samples.
Table 4. Determination of analytical sensitivity in pools of five samples.
DiscussionAvian orthoavulavirus type 1 is the most significant concern for commercial and rural poultry in developing countries. Unfortunately, no data were available for the neighboring regions of Morocco. More than 20 genotypes have been identified worldwide (Absalón et al., 2019), and classification has been determined according to the variability of the F protein or the whole genome (Liu, 2017). This variability requires surveillance and monitoring of AOAV-1 viruses in avian species to better understand their ecology. The situation in Africa is alarming, as different AOAV-1 viruses have been detected and notified in different areas of the continent. In Egypt, outbreaks are caused by genotypes II, VI, and VII (El Khantour et al., 2017; Megahed et al., 2018). In the Eastern African countries (Tanzania), genotypes V, VII, and XIII were found. In the central and western African countries, in addition to other genotypes such as II, VII, and V, newly circulating genotypes (XIV, XVII, and XVIII) have been isolated, but for the moment, they are restricted to this area. Genotypes II, VII, VIII, XI, and XIII have been detected in Southern African countries (Belák, 2005; Byarugaba et al., 2015; Damena et al., 2016; Elmardi et al., 2016; Ewies et al., 2017). In Morocco, only genotype V was reported to be present in 1995 (no published data) and in 2015 (El Khantour et al., 2017). Strengthening biosecurity measures is crucial to limit the introduction of novel NDV genotypes. Specifically, implementing controlled farm access, stringent disinfection protocols, and vaccination programs targeting diverse genotypes can enhance disease prevention. Furthermore, viral circulation must be monitored, and an efficient, rapid, and reliable analytical method is needed to detect the viral genome in every infected avian species. High sensitivity is required for early detection of the circulating virus and the quick implementation of countermeasures to limit potential damage to individual farms and the bird industry. The aim of this study was to validate an AOAV-1 real-time RT-PCR kit for diagnostic purposes. The validation process is necessary to ascertain whether the performance of the proposed method satisfies the requirements for routine tests (Belák, 2005). Different NDV strains, such as a Moroccan strain, and other genotypes of the virus also have high and low pathogenic avian orthomyxovirus and were tested to verify that the ID Gene™ Newcastle Disease Duplex kit allows the detection of any strains and does not cross with orthomyxovirus. All selected NDV strains regardless of their genotype were scored positive, whereas the samples of other orthomyxoviruses were negative. The experimental analytical specificity of the ID Gene™ Newcastle Disease Duplex kit was confirmed in terms of inclusivity and exclusivity. Due to the great diversity of AOAV-1 genotypes, analytical sensitivity was assessed by detecting the Moroccan NDV strain in different matrices. The results presented are comparable to those reported in some studies on the detection limit of the AOAV-1 M gene (Wise et al., 2004). The analysis of pooled samples on different matrices showed that AOAV-1 detection was similar to that of individual samples, making this real-time PCR kit suitable for mass diagnostics. In conclusion, based on the satisfactory analytical specificity and sensitivity of the measured method, the IDgene NDV real-time PCR kit has been validated for routine diagnostic or surveillance purposes and recommended to veterinary laboratories in the local area. AcknowledgmentsWe sincerely thank the Innovative Diagnostics Society for their invaluable support and collaboration throughout this research. Our gratitude also goes to the Istituto Zooprofilattico Sperimentale (IZS) Influenza Virus Reference Laboratory (Italy) for generously providing positive controls. Conflict of interestThe authors declare no conflicts of interest. FundingThis research received no external funding. Authors’ contributionsHasnae Zekhnini: Conceptualization, Methodology, Writing - original draft. Fatiha El Mellouli: Conceptualization, Methodology. Mohammed Rida Salam: Conceptualization, Methodology. Faiza Bennis: Conceptualization, Supervision. Fatima Chegdani: Conceptualization, Supervision. Data availabilityThe primary data used to support the findings of this study are available from the corresponding author upon request. ReferencesAbsalón, A.E., Cortés-Espinosa, D.V., Lucio, E., Miller, P.J. and Afonso, C.L. 2019. Epidemiology, control, and prevention of Newcastle disease in endemic regions: Latin America. Trop. Anim. Health Prod. 51(5), 1033–1048; doi:10.1007/s11250-019-01843-z. Awan, M.A., Otte, M.J. and James, A.D. 1994. The epidemiology of Newcastle disease in rural poultry: a review. Avian Pathol. 23(3), 405–423; doi:10.1080/03079459408419012. Belák, S. 2005. The molecular diagnosis of porcine viral diseases: a review. Acta Vet. Hung. 53(1), 113–124. Bujarski, J., Gallitelli, D., García-Arenal, F., Pallás, V., Palukaitis, P., Reddy, M.K., Wang, A. and ICTV Report Consortium. 2019. ICTV virus taxonomy profile: bromoviridae. J. Gen. Virol. 100(8), 1206–1207; doi:10.1099/jgv.0.001282. Byarugaba, D.K., Mugimba, K.K., Omony, J.B, Okitwi, M., Wanyana, A., Otim, M.O., Kirunda, H., Nakavuma, J.L., Teillaud, A. and Paul, M. 2015. Virulence et faible évolution génétique de virus de la maladie de Newcastle vélogènes isolés en Ouganda. 11èmes Journées de la Recherches Avicole, Tours, FRA, 2015-03-25-2015-03-26 [Internet]. Available via https://www.academia.edu/download/93467792/document.pdf (Accessed 18 December 2024). Damena, D., Fusaro, A., Sombo, M., Belaineh, R., Heidari, A., Kebede, A., Kidane, M. and Chaka H. 2016. Characterization of Newcastle disease virus isolates obtained from outbreak cases in commercial chickens and wild pigeons in Ethiopia. SpringerPlus 5(1), 476; doi:10.1186/s40064-016-2114-8. Diel, D.G., da Silva, L.H., Liu, H., Wang, Z., Miller, P.J. and Afonso, C.L. 2012. Genetic diversity of avian paramyxovirus type 1: proposal for a unified nomenclature and classification system of Newcastle disease virus genotypes. Infect. Genet. Evol. 12(8), 1770–1779. El Khantour, A., Darkaoui, S., Tatár-Kis, T., Mató, T., Essalah-Bennani, A., Cazaban, C. and Palya V. 2017. Immunity elicited by a turkey herpesvirus-vectored Newcastle disease vaccine in Turkey against challenge with a recent genotype IV Newcastle disease virus field strain. Avian Dis. 61(3), 378–386. Elmardi, N.A., Bakheit, M.A. and Khalafalla, A.I. 2016. Phylogenetic analysis of some Newcastle disease virus isolates from the Sudan. Open Vet. J. 6(2), 89–97. Ewies, S.S., Ali, A., Tamam, S.M. and Madbouly, H.M. 2017. Molecular characterization of Newcastle disease virus (genotype VII) from broiler chickens in Egypt. Beni-Suef Univ. J. Basic Appl. Sci. 6(3), 232–237. Kleij, L. 2023. Identification and validation of the virulence determinants of circulating equine influenza viruses [Internet]. PhD Thesis, Université Paris-Saclay. Available via https://theses.hal.science/tel-04618015/ (Accessed 6 January 2025). Liu, H. 2017. Generating a new vaccine for protecting poultry from Newcastle disease and controlling viral shedding [Internet]. PhD Thesis, Université Montpellier. Available via https://theses.hal.science/tel-01684253/ (Accessed 18 December 2024). Megahed, M.M., Eid, A.A., Mohamed, W. and Hassanin, O. 2018. Genetic characterization of Egyptian Newcastle disease virus strains isolated from flocks vaccinated against Newcastle disease virus, 2014-2015 [Internet]. Available via https://www.academia.edu/download/94738517/127.pdf (Accessed 18 December 2024). Nwanta, J.A., Egege, S.C., Alli-Balogun, J.K. and Ezema, W.S. 2008. Evaluation of prevalence and seasonality of Newcastle disease in chicken in Kaduna, Nigeria. World Poult. Sci. J. 64(3), 416–423. Rauw, F., Gardin, Y., van den Berg, T. and Lambrecht, B. 2009. La vaccination contre la maladie de Newcastle chez le poulet (Gallus gallus). BASE [Internet]. Available via https://popups.uliege.be/1780-4507/index.php?id=4758 (Accessed 6 January 2025). Sonaiya, E.B., Branckaert, R.D.S. and Gueye, E.F. 1999. The scope and effect of family poultry research and development. In: Research and Development Options for Family Poultry: First INTERNATIONAL NETWORK FOR FAMILY POULTRY DEVELOPMENT (INFPD/FAO) Electronic Conference on Family Poultry. FAO, Rome, Italy. http://www.fao.org/ag/againfo/subjects/en/infpd/econf_scope.html Vandemeulebroucke, E., De Clercq, K., Van der Stede, Y. and Vandenbussche, F. 2010. A proposed validation method for automated nucleic acid extraction and RT-qPCR analysis: an example using Bluetongue virus. J. Virol. Methods 165(1), 76–82. Wise, M.G., Suarez, D.L., Seal, B.S., Pedersen, J.C., Senne, D.A., King, D.J., Kapczynski, D.R. and Spackman, E. 2004. Development of a real-time reverse-transcription PCR for detection of Newcastle disease virus RNA in clinical samples. J. Clin. Microbiol. 42(1), 329–338; doi:10.1128/JCM.42.1.329-338.2004. World Organisation for Animal Health (WOAH). 2021. Newcastle disease virus. In: Manual of Diagnostic Tests and Vaccines for Terrestrial Animals. Paris: WOAH. Available from: https://www.woah.org | ||
| How to Cite this Article |
| Pubmed Style Zekhnini H, Mellouli FE, Salam MR, Bennis F, Chegdani F. Evaluation of a commercial real-time polymerase chain reaction kit for the detection of avian orthoavulavirus type I. Open Vet. J.. 2025; 15(3): 1440-1445. doi:10.5455/OVJ.2025.v15.i3.34 Web Style Zekhnini H, Mellouli FE, Salam MR, Bennis F, Chegdani F. Evaluation of a commercial real-time polymerase chain reaction kit for the detection of avian orthoavulavirus type I. https://www.openveterinaryjournal.com/?mno=222020 [Access: January 25, 2026]. doi:10.5455/OVJ.2025.v15.i3.34 AMA (American Medical Association) Style Zekhnini H, Mellouli FE, Salam MR, Bennis F, Chegdani F. Evaluation of a commercial real-time polymerase chain reaction kit for the detection of avian orthoavulavirus type I. Open Vet. J.. 2025; 15(3): 1440-1445. doi:10.5455/OVJ.2025.v15.i3.34 Vancouver/ICMJE Style Zekhnini H, Mellouli FE, Salam MR, Bennis F, Chegdani F. Evaluation of a commercial real-time polymerase chain reaction kit for the detection of avian orthoavulavirus type I. Open Vet. J.. (2025), [cited January 25, 2026]; 15(3): 1440-1445. doi:10.5455/OVJ.2025.v15.i3.34 Harvard Style Zekhnini, H., Mellouli, . F. E., Salam, . M. R., Bennis, . F. & Chegdani, . F. (2025) Evaluation of a commercial real-time polymerase chain reaction kit for the detection of avian orthoavulavirus type I. Open Vet. J., 15 (3), 1440-1445. doi:10.5455/OVJ.2025.v15.i3.34 Turabian Style Zekhnini, Hasnae, Fatiha El Mellouli, Mohamed Rida Salam, Faiza Bennis, and Fatima Chegdani. 2025. Evaluation of a commercial real-time polymerase chain reaction kit for the detection of avian orthoavulavirus type I. Open Veterinary Journal, 15 (3), 1440-1445. doi:10.5455/OVJ.2025.v15.i3.34 Chicago Style Zekhnini, Hasnae, Fatiha El Mellouli, Mohamed Rida Salam, Faiza Bennis, and Fatima Chegdani. "Evaluation of a commercial real-time polymerase chain reaction kit for the detection of avian orthoavulavirus type I." Open Veterinary Journal 15 (2025), 1440-1445. doi:10.5455/OVJ.2025.v15.i3.34 MLA (The Modern Language Association) Style Zekhnini, Hasnae, Fatiha El Mellouli, Mohamed Rida Salam, Faiza Bennis, and Fatima Chegdani. "Evaluation of a commercial real-time polymerase chain reaction kit for the detection of avian orthoavulavirus type I." Open Veterinary Journal 15.3 (2025), 1440-1445. Print. doi:10.5455/OVJ.2025.v15.i3.34 APA (American Psychological Association) Style Zekhnini, H., Mellouli, . F. E., Salam, . M. R., Bennis, . F. & Chegdani, . F. (2025) Evaluation of a commercial real-time polymerase chain reaction kit for the detection of avian orthoavulavirus type I. Open Veterinary Journal, 15 (3), 1440-1445. doi:10.5455/OVJ.2025.v15.i3.34 |