| Research Article | ||
Open Vet. J.. 2025; 15(3): 1340-1348 Open Veterinary Journal, (2025), Vol. 15(3): 1340-1348 Research Article Dissemination and phenotypic characterization of ESBL-producing Escherichia coli in IndonesiaTati Ariyanti*, Suhaemi Suhaemi, Sri Mulyati, Sukatma Sukatma, Sumirah Sumirah, Susan Maphilindawati Noor, Faidah Rachmawati, Prima Mei Widiyanti, Eddy Sukmawinata, Andriani Andriani, Eni Kusumaningtyas and Aswin Rafif KhairullahResearch Center for Veterinary Science, National Research and Innovation Agency (BRIN), Bogor, Indonesia *Corresponding Author: Tati Ariyanti. Research Center for Veterinary Science, National Research and Innovation Agency (BRIN), Bogor, Indonesia. Email: tatiariyanti23 [at] gmail.com Submitted: 20/12/2024 Accepted: 17/02/2025 Published: 31/03/2025 © 2025 Open Veterinary Journal
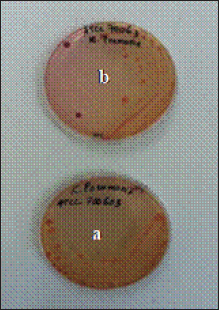
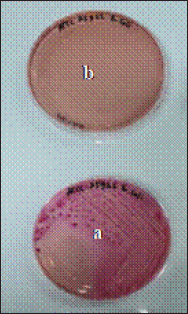
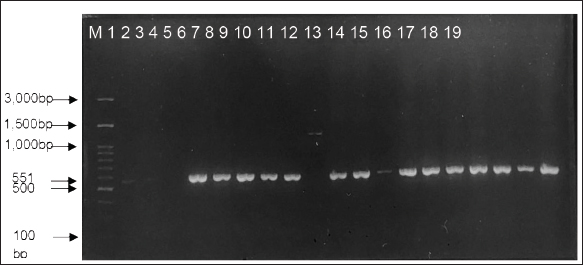
AbstractBackground: The alarming rise in infections caused by extended-spectrum beta-lactamase (ESBL)-producing Escherichia coli in animals and humans poses a serious threat due to its escalating antibiotic resistance. Unveiling this problematic bacteria’s prevalence and resistance patterns in animals is crucial for formulating effective control strategies and safeguarding public health. Aim: The purpose of this study was to analyze the expression of three main genes: blaCTX-M, blaSHV, and blaTEM, in ESBL-producing E. coli isolates from The Research Center for Veterinary Science and the National Research and Innovation Agency. Moreover, their resistance profiles against various antibiotics should be systematically evaluated. Methods: Ninety-seven E. coli isolates from the bacteriology laboratory of The Research Center for Veterinary Science were identified on MacConkey medium supplemented with cefotaxime. The isolates were verified for the existence of the blaCTX-M, blaSHV, and blaTEM genes using PCR. Antimicrobial susceptibility testing was conducted using antibiotic discs following the CLSI standards. Results: The prevalence of ESBL-producing E. coli in chicken ceca, eggs, and fish intestines was 16.5% (16/97). The specific genes detected were blaCTX-M gene at 93.75% (15/16), followed by the blaTEM gene, at 81.25% (13/16), and blaSHV at 25% (4/16). The antimicrobial sensitivity test results revealed that all ESBL-producing E. coli isolates had multidrug resistance 81.25% to 1–5 antibiotics and 18.75% to 6–7 antibiotics. The isolate exhibited 100% resistance to ampicillin and sulfamethoxazole, with exclusive sensitivity to chloramphenicol. Conclusion: The dominant gene in the ESBL-producing isolates was blaCTX-M. This bacterium is completely resistant to ampicillin and sulfamethoxazole, whereas it displays multidrug resistance to 1–7 different types of antibiotics. Keywords: Escherichia coli, ESBL, blaCTX-M, blaTEM, blaSHV. IntroductionAntibiotics, once heralded as magic bullets against bacterial foes, are now facing their nemesis: superbugs (Baral and Mozafari, 2020; Ramandinianto et al., 2020). These evolved microorganisms, like extended-spectrum beta-lactamase (ESBL)-producing Escherichia coli (ESBL-E. coli), have developed armor against our most potent weapons, making infections more difficult, costly, and sometimes even fatal (Ramadan et al., 2019; Castanheira et al., 2021). This growing threat looms large over human and animal health worldwide, and in Indonesia, the situation is particularly concerning (Siahaan et al., 2022). The thriving agriculture industry and high population density in Indonesia provide an ideal environment for the proliferation of antibiotic-resistant bacteria such as ESBL-E. coli (Yanestria et al., 2022; Widodo et al., 2024). The close proximity between humans and animals, combined with the potential overuse of antibiotics in both sectors, fosters the transmission and evolution of resistant bacteria (Okeke et al., 2005; McEwen and Collignon, 2017). This creates a ticking time bomb: a reservoir of superbugs capable of jumping from animals to humans, potentially compromising even critical medical procedures (Chishimba et al., 2016). Understanding the dissemination and characteristics of ESBL-E. coli in Indonesia, it is no longer a luxury but a critical necessity (Severin et al., 2010). It is the key to unlocking effective control strategies and protecting public health before it is too late. Our study embarks on this vital mission, venturing into the diverse world of Indonesian animal samples to shed light on the lurking superbug. Through meticulous genetic analysis and rigorous antibiotic susceptibility testing, we aim to paint a detailed picture of the enemy we face. By identifying the prevalence and resistance patterns of ESBL-producing E. coli in different animal sources, we can anticipate its behavior and design targeted interventions. Acquiring this understanding will provide us with the necessary resources to effectively counteract the transmission of this highly resistant bacterium, ensuring the well-being of both animals and humans in Indonesia and other regions (Khairullah et al., 2019; Endale et al., 2023). This study is not just an academic exercise; it is a pivotal step toward reclaiming the magic of antibiotics and securing a future where even the most formidable superbugs can be eradicated. The objective of this research was to determine the frequency of E. coli bacteria that produce extended-spectrum beta-lactamases in animals and to analyze resistance to antimicrobial drugs. The insights gained will inform effective antibiotic strategies and infection control plans for animals, ultimately preventing the dissemination of these strains to both animals and humans. Materials and MethodsMaterialsThe materials used in this study comprised 97 animal isolates of E. coli sourced from the Research Center for Veterinary Science of the National Research and Innovation Agency of Indonesia (BRIN). Confirmation of ESBL and antibiotic sensitivity of E. coli isolatesThe presence of ESBL was confirmed using culture methods on agar media and PCR. E. coli isolates were cultured in Tryptic Soy Broth supplemented with 2 mg/l cefotaxime and incubated at 35°C for 24 hours. The culture was transferred to a new medium on MacConkey agar supplemented with Cefotaxime powder at a concentration of 2 mg/l. The culture was then placed in an incubator at 35°C for 24 hours. The negative control for ESBL synthesis was Escherichia coli ATCC 25922, whereas the positive control was either E. coli ATCC 35218 or Klebsiella pneumoniae ATCC 700603. After identifying the ESBL-producing E. coli isolates, they were cultivated on nutrient agar for additional testing. PCR was used to confirm the presence of ESBL-producing E. coli by detecting the genes that encode ESBL. We used specific primers for blaCTX-M, blaSHV, and blaTEM for this purpose (Mondal et al., 2018). The disc diffusion method was used to perform sensitivity testing for ESBL-producing E. coli isolates, following the guidelines provided by the Clinical and Laboratory Standards Institute (CLSI, 2022). Pure cultures were prepared as suspensions with a turbidity of 0.5 McFarland (1–2 × 108 cfu/ml). The culture was transferred onto Mueller Hinton agar (MHA) using a sterilized cotton swab. Afterward, paper disks containing antibiotics were positioned on the MHA agar plate with a separation of 25–30 mm and then kept in an incubator at 35°C for 24 hours. The antibiotics used in the trial comprised Ampicillin, Sulfamethoxazole, Ceftriaxone, Gentamicin, Tetracycline, Amoxicillin, Cefoxitin, Ceftazidime, Ciprofloxacin, and Chloramphenicol. The E. coli strain ATCC 25922 was used as the negative control, whereas either E. coli ATCC 35218 or Klebsiella pneumoniae strain ATCC 700603 was used as the positive control. These controls were employed to assess the effectiveness of ESBL-producing organism detection and serve as controls in each antibiotic sensitivity test. Data analysisThe research data were analyzed descriptively, and the results are presented in tables and figures regarding the resistance of ESBL-producing E. coli to antibiotics. Ethical approvalEthical permission is not required for this project, according to the institute’s ethics committee. ResultsReidentification of E. coli producing ESBLThe re-identification of 97 isolates of ESBL-producing E. coli from the RCVS collection, which came from various animals and their products, revealed 16 isolates of ESBL-producing E. coli that were found in the area. There were 13 isolates from chickens and 3 isolates from fish (Table 1). The native strains of ESBL-producing E. coli were able to grow on MacConkey agar that contained cefotaxime. Figure 1 shows observable colonies. The colony growth of E. coli indicates resistance to cefotaxime, a characteristic feature of ESBL-producing isolates. Colony growth of the 16 local E. coli isolates on MacConkey medium with the addition of cefotaxime aligns with the ESBL-producing reference isolate, Klebsiella pneumoniae ATCC 700603 (Fig. 2). Escherichia coli isolates that do not produce ESBL did not exhibit growth on MacConkey medium with the addition of cefotaxime, consistent with the negative control, E. coli ATCC 25922 (Fig. 3). Detection of ESBL-producing E. coli resistance genesPCR was used to confirm the presence of certain ESBL-producing genes, namely blaCTX-M, blaSHV, and blaTEM, in 16 E. coli isolates. Positive and negative ESBL-producing controls were also included in the analysis. Figure 4 presents the research findings of the 16 ESBL-producing E. coli isolates analyzed. Among these, 15 isolates (93.75%) were found to be positive for the presence of the blaCTX-M gene, with an amplicon size of 551 bp. One sample, specifically sample number 9, did not contain the blaCTX-M gene. Controls 1 and 2 exhibited visible albeit thin DNA bands, whereas control 3 (negative) did not contain the blaCTX-M gene. In Figure 5, the research results of the examined 16 ESBL-producing E. coli isolates are presented. Among these, 14 isolates (87.5%) were found to be positive for the blaTEM gene, with an amplicon size of 860 bp. Two samples, specifically samples 7 and 18, did not contain the blaTEM gene. Controls 1 and 3 do not exhibit visible DNA bands, indicating the absence of blaCTX-M. Table 1. ESBL-producing E. coli isolates collected from RCVS in 2022.
Fig. 1. Growth of local E. coli isolates on (a) MacConkey without cefotaxime (a) and on MacConkey media with cefotaxime (b).
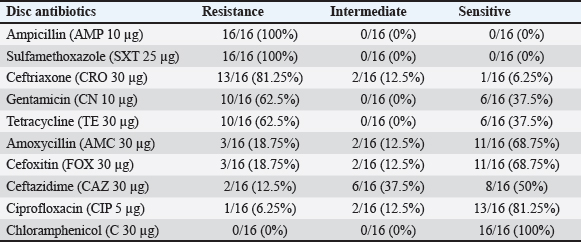
Fig. 2. Growth of Klebsiella pneumoniae ATCC 700603 on (a) MacConkey without cefotaxime (a) and on MacConkey media with cefotaxime (b). The research findings suggest that of the 16 ESBL-producing E. coli isolates that were analyzed, 4 isolates (25%) were found to have the SHV gene (Fig. 6). The results of the Antimicrobial Sensitivity Test indicated that all 16 ESBL-producing E. coli isolates exhibited resistance to nine types of antibiotics: Ampicillin (100%), Sulfamethoxazole (100%), Ceftriaxone (81.25%), Gentamicin (62.5%), Tetracycline (62.5%), Ciprofloxacin (62.5%), Amoxicillin (18.75%), Cefoxitin (18.75%), and Ceftazidime (12.5%). Furthermore, these isolates were sensitive only to Chloramphenicol (Table 2). The inhibitory zones against antibiotics on Mueller–Hinton Agar, where ESBL-producing E. coli was cultured, are shown in Figure 7.
Fig. 3. Growth of E. coli ATCC 25922 on MacConkey media without cefotaxime (a) and no growth on MacConkey media with cefotaxime (b).
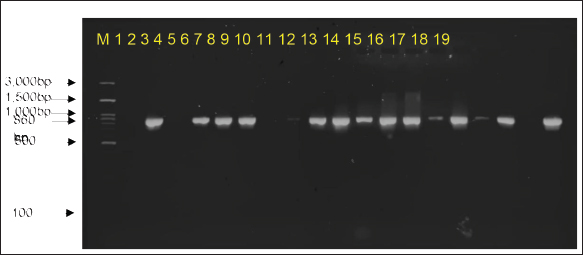
Fig. 4. PCR amplification of blaCTX-M with an amplicon size of 551 bp. M marker (100–3,000 bp), controls (1–3), Samples (4–19), and details of controls and samples are provided in Table 2.
Fig. 5. PCR amplification of blaTEM with an amplicon size of 860 bp. M marker (100–1,000 bp), controls (1–3), Samples (4–19), and details of controls and samples are provided in Table 2.
Fig. 6. PCR amplification of blaSHV with an amplicon size of 787 bp. M marker (100–1,000 bp), controls (1–3), Samples (4–19), and details of controls and samples are provided in Table 2. Table 2. Results of antimicrobial sensitivity test on 16 ESBL-producing E. coli isolates against 10 antibiotics.
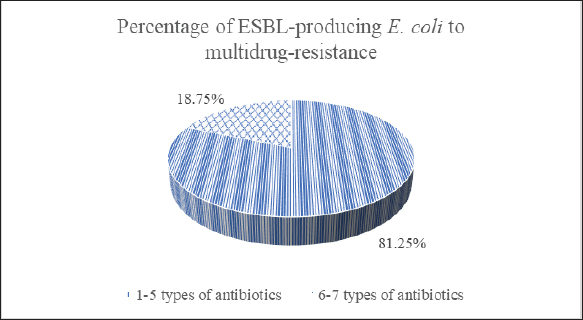
Isolates that show resistance to three or more types of antibiotics are classified as multidrug-resistant (MDR) (Al-Ansari et al., 2021). Among the 16 E. coli isolates that produce ESBL, 81.25% showed MDR, meaning they were resistant to 1–5 different antibiotics. The remaining 18.75% were resistant to 6–7 types of antibiotics (Fig. 8). DiscussionA recent study conducted in Indonesia highlighted significant concerns regarding the existence of animals infected with ESBL-producing E. coli. The study, which examined samples collected until 2021, found that a substantial number of animal samples contained germs that were resistant to a broad spectrum of antibiotics. This has important consequences for the health of both animals and humans. It was surprising to find that 16.5% (16 of 97) of the isolates tested positive for ESBL-producing E. coli. These isolates were obtained from different sources, including the cecum, eggs, and intestines of chickens and fish. This finding underscores the widespread distribution of ESBL-producing E. coli across various animal species and geographical regions, with positive samples identified in diverse areas of Indonesia, including West Java, Yogyakarta, and East Java. Inadequate sanitation measures at animal farms and slaughterhouses facilitate the transmission of antibiotic-resistant microorganisms from animals to people. Additionally, inadequate sanitation and wastewater management in communities can lead to environmental contamination, escalating the risk of exposure to resistant bacteria (Wellington et al., 2013; Manyi-Loh et al., 2018). This study contributes to the growing body of evidence demonstrating the global prevalence of ESBL-producing E. coli in animals. Similar troubling trends have been observed in Africa, where studies in Zambia reported a 20.1% prevalence of ESBL-producing E. coli in poultry samples (Chishimba et al., 2016). Likewise, studies in Ghana documented a 29% prevalence of broilers and a staggering range of 0%–85% on farms (Falgenhauer et al., 2019). These regional comparisons highlight the urgency of addressing this widespread threat. PCR analysis yielded valuable information about the genetic composition of ESBL-producing E. coli in animals, revealing three genes that are responsible for robust antibiotic resistance. The CTX-M gene, found in 93.75% of E. coli isolates, aligns with previous research in Indonesia and other countries, emphasizing its dominance among resistant strains (Sudarwanto et al., 2017; Falgenhauer et al., 2019; Wibisono et al., 2020). The TEM gene is present in 81.25% of bacteria, and it is another well-known resistance marker, although it is not as prevalent as CTX-M. The less frequent defender, SHV, was identified in only 25% of resistant E. coli. These findings depict a concerning and adaptable enemy, with the dominance of the CTX-M gene suggesting an efficient mechanism for resistance dissemination. The potential spread of these bacteria through contaminated environments, particularly in areas with poor hygiene and sanitation, highlights the urgency of addressing this growing threat (Kumar et al., 2014).
Fig. 7. Inhibition zones against antibiotics in ESBL-producing E. coli cultures.
Fig. 8. Percentage of multidrug resistance in ESBL-producing E. coli. Antimicrobial sensitivity test results further highlight the alarming resistance of ESBL-producing E. coli (Onduru et al., 2021). Ampicillin and Sulfamethoxazole faced complete resistance, with 100% of the isolates demonstrating resistance. Similar concerns have been raised in other regions, such as Zambia and South Africa, where poultry isolates exhibit a 100% resistance rate to ampicillin and cefotaxime (Chishimba et al., 2016). Although ceftriaxone managed to resist the attack in only 18.75% of isolates, concerns about resistance against third-generation cephalosporins, like ceftriaxone, were observed in Ghana (Falgenhauer et al., 2019). The results highlight the immediate need to tackle the use of antibiotics in poultry and their possible function as a source for ESBL-producing E. coli, which poses concerns for the health of both animals and humans (Chishimba et al., 2016). The 16.5% prevalence of ESBL-producing E. coli in Indonesia, isolated from diverse animal sources and geographical regions, can be attributed to several factors, such as antibiotic overuse in animal agriculture, transmission through the food chain and environment, and a lack of proper hygiene and sanitation. Frequent antibiotic administration in poultry and fish farming creates selective pressure that favors the survival and proliferation of resistant bacteria, thereby contributing to resistance development. Improper antibiotic storage and usage, environmental contamination through manure and wastewater, and transmission through direct contact or contamination with food further amplify the risks (Polianciuc et al., 2020). The observation that 81.25% of ESBL-producing E. coli isolates in this study were MDR, resisting 1–5 types of antibiotics, with an additional 18.75% exhibiting resistance to a staggering 6–7 types, paints a concerning picture with far-reaching implications. This finding aligns with a global trend of increased MDR in ESBL-producing E. coli. Studies in Africa, for instance, report similar alarming rates: 94,6% in Nigeria (Aworh et al., 2020), 70.7% in Zambia (Chishimba et al., 2016), 51,6% in Tanzania (Kimera et al., 2021), and resistance was observed against various antibiotics commonly used in poultry farming, including ampicillin, tetracycline, trimethoprim-sulfamethoxazole, and cephalosporins. This finding is particularly worrisome because resistance to multiple antibiotics, including powerful ones, suggests the emergence of “superbugs” with limited vulnerability. This further emphasized the global threat of MDR in these bacteria and the need for urgent action. The existence of ESBL-producing E. coli in animals poses a significant health risk to both animals and humans (Farizqi et al., 2023). This renders conventional antibiotic treatments ineffective and increases the risk of transmission to humans (Khairullah et al., 2024). Further investigation is essential to clarify the frequency and methods of transmission of ESBL-producing E. coli in animals and to devise efficient measures to prevent their dissemination within and between populations. Implement focused antibiotic stewardship initiatives. ConclusionA concerning finding has emerged from the world of animal bacteria: a significant portion of E. coli, over 16%, obtained from chickens and fish, possess a “super shield” against antibiotics. This shield, formed by special genes, renders them resistant not only to cefotaxime but also to common antibiotics like Ampicillin and Sulfamethoxazole. This development makes treating infections caused by these resistant bacteria extremely challenging, with nearly 73% able to shrug off attacks from 6 to 7 different antibiotics, leaving few treatment options in our arsenal. The consequences are clear: properly controlling these illnesses has become considerably more challenging, presenting a potential danger to both animal and human health. This discovery underscores the urgent need for careful antibiotic use in animals to prevent the spread of super-shielded bacterial warriors. AcknowledgmentsThe authors are grateful to the following organizations: Organisasi Riset Kesehatan, Badan Riset dan Inovasi Nasional (BRIN). Conflict of interestThe authors declare no conflict of interest. Author’s contributionsTA and SS: Conceptualization and design. SM and SS: Acquisition of data. SMN and FR: Formal analysis and interpretation of data. PMW, ARK, and ES: Writing and preparation of the original draft. AA, SS, and EK: Writing, reviewing, and editing. All authors have read and agreed to the publication of the final version of the manuscript. FundingThe authors are thankful to Organisasi Riset Kesehatan, Badan Riset dan Inovasi Nasional (BRIN) for funding the research. Data availabilityAll data are available in the manuscript. ReferencesAl-Ansari, M., Aljubali, M., Somily, A.M., Albarrag, A.M. and Masood, A. 2021. Isolation and molecular characterization of multidrug-resistant Salmonella enterica serovars. J. Infect. Public Health 14(12), 1767–1776. Aworh, M.K., Kwaga, J., Okolocha, E., Harden, L., Hull, D., Hendriksen, R.S. and Thakur, S. 2020. Extended-spectrum β-lactamase production of Escherichia coli in humans, chickens, and poultry environments in Abuja, Nigeria. One Health Outlook 2(1), 8. Baral, B. and Mozafari, M.R. 2020. Strategic Moves of “Superbugs” against available chemical scaffolds: signaling, regulation, and challenges. ACS Pharmacol. Transl. Sci. 3(3), 373–400. Castanheira, M., Simner, P.J. and Bradford, P.A. 2021. Extended-spectrum β-lactamases: an update on their characteristics, epidemiology, and detection. JAC Antimicrob. Resist. 3(3), dlab092. Chishimba, K., Hang’ombe, B.M., Muzandu, K., Mshana, S.E., Matee, M.I., Nakajima, C. and Suzuki, Y. 2016. Detection of extended-spectrum Beta-lactamase-producing Escherichia coli in market-ready chickens in Zambia. Int. J. Microbiol. 2016(1), 5275724. CLSI (Clinical and Laboratory Standards Institute). 2022. Performance Standards for Antimicrobial Susceptibility Testing. 32nd ed. CLSI supplement M100. Wayne, Pennsylvania. CLSI. Available via https://clsi.org/standards/products/microbiology/documents/m100/. (Aceessed 20 December 2023). Endale, H., Mathewos, M. and Abdeta, D. 2023. Potential causes of the spread of antimicrobial resistance and preventive measures in one health perspective-a review. Infect. Drug Resist. 16(1), 7515–7545. Falgenhauer, L., Imirzalioglu, C., Oppong, K., Akenten, C.W., Hogan, B., Krumkamp, R., Poppert, S., Levermann, V., Schwengers, O., Sarpong, N., Owusu-Dabo, E., May, J. and Eibach, D. 2019, Detection and characterization of ESBL-producing Escherichia coli from humans and poultry in Ghana. Front. Microbiol. 9(1), 3358. Farizqi, M.T.I., Effendi, M.H., Adikara, R.T.S., Yudaniayanti, I.S., Putra, G.D.S., Khairullah, A.R., Kurniawan, S.C., Silaen, O.S.M., Ramadhani, S., Millannia, S.K., Kaben, S.E. and Waruwu, Y.K.K. 2023. Detection of extended-spectrum β-lactamase-producing Escherichia coli genes isolated from cat rectal swabs at Surabaya Veterinary Hospital, Indonesia. Vet. World 16(9), 1917–1925. Khairullah, A.R., Kurniawan, S.C., Sudjarwo, S.A., Effendi, M.H., Widodo, A., Moses, I.B., Hasib, A., Zahra, R.L.A., Gelolodo, M.A., Kurniawati, D.A., Riwu, K.H.P., Silaen, O.S.M., Afnani, D.A. and Ramandinianto, S.C. 2024. Kinship analysis of mecA of methicillin-resistant Staphylococcus aureus isolated from milk and risk factors from farmers in Blitar, Indonesia. Vet. World 17(1), 216–225. Khairullah, A.R., Raharjo, D., Rahmahani, J., Suwarno, Tyasningsih, W. and Harijani, N. 2019. Antibiotics resistant at Staphylococcus aureus and Streptococcus sp isolated from bovine mastitis in Karangploso, East Java, Indonesia. Indian J. Forensic Med. Toxicol. 13(4), 439–444. Kimera, Z.I., Mgaya, F.X., Misinzo, G., Mshana, S.E., Moremi, N. and Matee, M.I.N. 2021. Multidrug-resistant, including extended-spectrum beta lactamase-producing and quinolone-resistant, Escherichia coli isolates from domestic and poultry pigs in Dar es Salaam, Tanzania. Antibiotics 10(4), 406. Kumar, D., Singh, A.K., Ali, M.R. and Chander, Y. 2014. Antimicrobial susceptibility profile of extended spectrum β-lactamase (ESBL) producing Escherichia coli from various clinical samples. Infect. Dis. Res. Treat. 7(1), 1–8. Manyi-Loh, C., Mamphweli, S., Meyer, E. and Okoh, A. 2018. Antibiotic use in agriculture and its consequences in environmental sources: potential public health implications. Molecules 23(4), 795. McEwen, S.A. and Collignon, P.J. 2017. Antimicrobial resistance: a one health perspective. Trans. R. Soc. Trop. Med. Hyg. 111(6), 255–260. Mondal, A.H., Siddiqui, M.T., Sultan, I. and Haq, Q.M.R. 2018. Prevalence and diversity of blaTEM, blaSHV, and blaCTX-M variants among multidrug-resistant Klebsiella spp. from an urban riverine environment in India. Int. J. Environ. Health Res. 29(2), 117–129. Okeke, I.N., Laxminarayan, R., Bhutta, Z.A., Duse, A.G., Jenkins, P., O’Brien, T.F., Pablos-Mendez, A. and Klugman, K.P. 2005. Antimicrobial resistance in developing countries. Part I: recent trends and current status. Lancet Infect. Dis. 5(8), 481–493. Onduru, O.G., Aboud, S., Nyirenda, T.S., Rumisha, S.F. and Mkakosya, R.S. 2021. Antimicrobial susceptibility testing profiles of ESBL-producing Enterobacterales isolated from hospital and community adult patients in Malawi. IJID Reg. 1(1), 47–52. Polianciuc, S.I., Gurzău, A.E., Kiss, B., Ștefan, M.G. and Loghin, F. 2020. Antibiotics in the environment: causes and consequences. Med. Pharm. Rep. 93(3), 231–240. Ramadan, A.A., Abdelaziz, N.A., Amin, M.A. and Aziz, R.K. 2019. Novel blaCTX-M variants and genotype-phenotype correlations among clinical isolates of extended-spectrum beta-lactamase-producing Escherichia coli. Sci Rep 9(1), 4224. Ramandinianto, S.C., Khairullah, A.R., Effendi, M.H., Tyasningsih, W. and Rahmahani, J. 2020. Detection of enterotoxin type B gene on methicillin resistant Staphylococcus aureus (MRSA) isolated from raw milk in East Java, Indonesia. Syst. Rev. Pharm. 11(7), 290–298. Severin, J.A., Mertaniasih, N.M., Kuntaman, K., Lestari, E.S., Purwanta, M., Toom, N.L.D., Duerink, D.O., Hadi, U., van Belkum, A., Verbrugh, H.A., Goessens, W.H. and Study Group ‘Antimicrobial resistance in Indonesia: prevalence and prevention’ (AMRIN). 2010. Molecular characterization of extended-spectrum beta-lactamases in clinical Escherichia coli and Klebsiella pneumoniae isolates from Surabaya, Indonesia. J. Antimicrob. Chemother. 65(3), 465–469. Siahaan, S., Herman, M.J. and Fitri, N. 2022. Antimicrobial resistance situation in Indonesia: a challenge of multisector and global coordination. J. Trop. Med. 2022(1), 2783300. Sudarwanto, M.B., Akineden, Ö., Sukmawinata, E., Pisestyani, H., Lukman, D.W. and Latif, H. 2017. CTX-M-15 and CTX-M-55 produce Escherichia coli in milk from dairy farms in West Java, Indonesia. Int. J. Sci. Basic Appl. Res. 31(1), 409–416. Wellington, E.M.H., Boxall, A.B.A., Cross, P., Feil, E.J., Gaze, W.H., Hawkey, P.M., Johnson-Rollings, A.S., Jones, D.L., Lee, N.M., Otten, W., Thomas, C.M. and Williams, A.P. 2013. The role of the natural environment in the emergence of antibiotic resistance in Gram-negative bacteria. Lancet Infect. Dis. 13(2), 155–165. Wibisono, F.J., Sumiarto, B., Untari, T., Effendi, M.H., Permatasari, D.A. and Witaningrum, A.M. 2020. Short communication: antibiotic resistance in extended-spectrum beta-lactamases genes producing Escherichia coli in laying hens in Blitar, Indonesia. Biodiversitas 21(10), 4631–4635. Widodo, A., Khairullah, A.R., Effendi, M.H., Moses, I.B. and Agustin, A.L.D. 2024. Extended-spectrum β-lactamase-producing Escherichia coli from poultry: a review. Vet. World 17(9), 2017–2027. Yanestria, S.M., Dameanti, F.N.A.E.P., Musayannah, B.G., Pratama, J.W.A., Witaningrum, A.M., Effendi, M.H. and Ugbo, E.N. 2022. Antibiotic resistance pattern of extended-spectrum β-lactamase (ESBL) producing Escherichia coli isolated from a broiler farm environment in Pasuruan district, Indonesia. Biodiversitas 23(9), 4460–4465. | ||
| How to Cite this Article |
| Pubmed Style Ariyanti T, Suhaemi S, Mulyati S, Sukatma S, Sumirah S, Noor SM, Rachmawati F, Widiyanti PM, Sukmawinata E, Andriani A, Kusumaningtyas E, Khairullah AR. Dissemination and phenotypic characterization of ESBL-producing Escherichia coli in Indonesia. Open Vet. J.. 2025; 15(3): 1340-1348. doi:10.5455/OVJ.2025.v15.i3.25 Web Style Ariyanti T, Suhaemi S, Mulyati S, Sukatma S, Sumirah S, Noor SM, Rachmawati F, Widiyanti PM, Sukmawinata E, Andriani A, Kusumaningtyas E, Khairullah AR. Dissemination and phenotypic characterization of ESBL-producing Escherichia coli in Indonesia. https://www.openveterinaryjournal.com/?mno=233841 [Access: January 25, 2026]. doi:10.5455/OVJ.2025.v15.i3.25 AMA (American Medical Association) Style Ariyanti T, Suhaemi S, Mulyati S, Sukatma S, Sumirah S, Noor SM, Rachmawati F, Widiyanti PM, Sukmawinata E, Andriani A, Kusumaningtyas E, Khairullah AR. Dissemination and phenotypic characterization of ESBL-producing Escherichia coli in Indonesia. Open Vet. J.. 2025; 15(3): 1340-1348. doi:10.5455/OVJ.2025.v15.i3.25 Vancouver/ICMJE Style Ariyanti T, Suhaemi S, Mulyati S, Sukatma S, Sumirah S, Noor SM, Rachmawati F, Widiyanti PM, Sukmawinata E, Andriani A, Kusumaningtyas E, Khairullah AR. Dissemination and phenotypic characterization of ESBL-producing Escherichia coli in Indonesia. Open Vet. J.. (2025), [cited January 25, 2026]; 15(3): 1340-1348. doi:10.5455/OVJ.2025.v15.i3.25 Harvard Style Ariyanti, T., Suhaemi, . S., Mulyati, . S., Sukatma, . S., Sumirah, . S., Noor, . S. M., Rachmawati, . F., Widiyanti, . P. M., Sukmawinata, . E., Andriani, . A., Kusumaningtyas, . E. & Khairullah, . A. R. (2025) Dissemination and phenotypic characterization of ESBL-producing Escherichia coli in Indonesia. Open Vet. J., 15 (3), 1340-1348. doi:10.5455/OVJ.2025.v15.i3.25 Turabian Style Ariyanti, Tati, Suhaemi Suhaemi, Sri Mulyati, Sukatma Sukatma, Sumirah Sumirah, Susan Maphilindawati Noor, Faidah Rachmawati, Prima Mei Widiyanti, Eddy Sukmawinata, Andriani Andriani, Eni Kusumaningtyas, and Aswin Rafif Khairullah. 2025. Dissemination and phenotypic characterization of ESBL-producing Escherichia coli in Indonesia. Open Veterinary Journal, 15 (3), 1340-1348. doi:10.5455/OVJ.2025.v15.i3.25 Chicago Style Ariyanti, Tati, Suhaemi Suhaemi, Sri Mulyati, Sukatma Sukatma, Sumirah Sumirah, Susan Maphilindawati Noor, Faidah Rachmawati, Prima Mei Widiyanti, Eddy Sukmawinata, Andriani Andriani, Eni Kusumaningtyas, and Aswin Rafif Khairullah. "Dissemination and phenotypic characterization of ESBL-producing Escherichia coli in Indonesia." Open Veterinary Journal 15 (2025), 1340-1348. doi:10.5455/OVJ.2025.v15.i3.25 MLA (The Modern Language Association) Style Ariyanti, Tati, Suhaemi Suhaemi, Sri Mulyati, Sukatma Sukatma, Sumirah Sumirah, Susan Maphilindawati Noor, Faidah Rachmawati, Prima Mei Widiyanti, Eddy Sukmawinata, Andriani Andriani, Eni Kusumaningtyas, and Aswin Rafif Khairullah. "Dissemination and phenotypic characterization of ESBL-producing Escherichia coli in Indonesia." Open Veterinary Journal 15.3 (2025), 1340-1348. Print. doi:10.5455/OVJ.2025.v15.i3.25 APA (American Psychological Association) Style Ariyanti, T., Suhaemi, . S., Mulyati, . S., Sukatma, . S., Sumirah, . S., Noor, . S. M., Rachmawati, . F., Widiyanti, . P. M., Sukmawinata, . E., Andriani, . A., Kusumaningtyas, . E. & Khairullah, . A. R. (2025) Dissemination and phenotypic characterization of ESBL-producing Escherichia coli in Indonesia. Open Veterinary Journal, 15 (3), 1340-1348. doi:10.5455/OVJ.2025.v15.i3.25 |