| Research Article | ||
Open Vet. J.. 2025; 15(3): 1397-1406 Open Veterinary Journal, (2025), Vol. 15(3): 1397-1406 Research Article Molecular and serological incidences of Bordetella bronchiseptica in pet dogs with urinary infectionsHadaf Mahdi Kadhim1, Ahlam Ali Soghi Al-Galebi1, Mithal K. A. Al-Hassani1 and Hasanain A. J. Gharban2*1Department of Biology, College of Education, University of Al-Qadisiyah, Al-Diwaniyah, Iraq 2Department of Internal and Preventive Veterinary Medicine, College of Veterinary Medicine, University of Wasit, Wasit, Iraq *Corresponding Author: Hasanain A. J. Gharban. Department of Internal and Preventive Veterinary Medicine, College of Veterinary Medicine, University of Wasit, Wasit, Iraq. Email: hghirban [at] uowasit.edu.iq Submitted: 27/12/2024 Accepted: 17/02/2025 Published: 31/03/2025 © 2025 Open Veterinary Journal
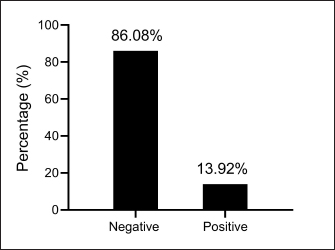
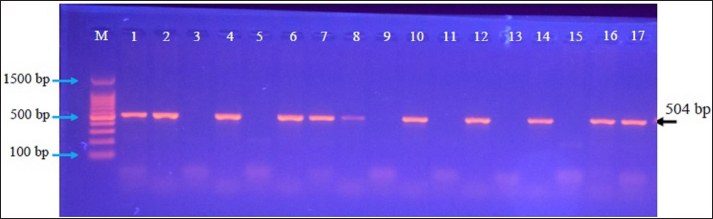
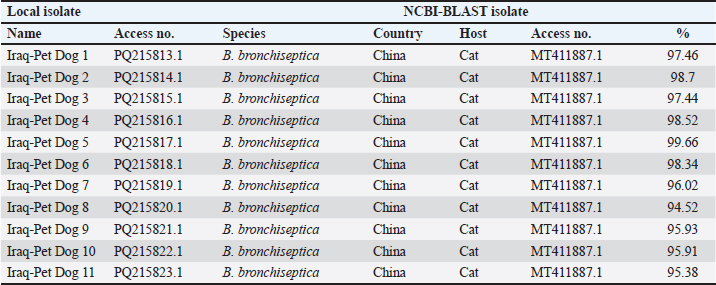
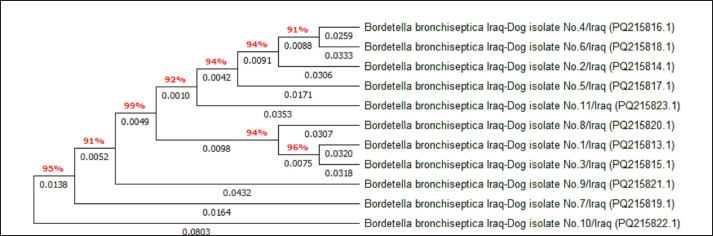
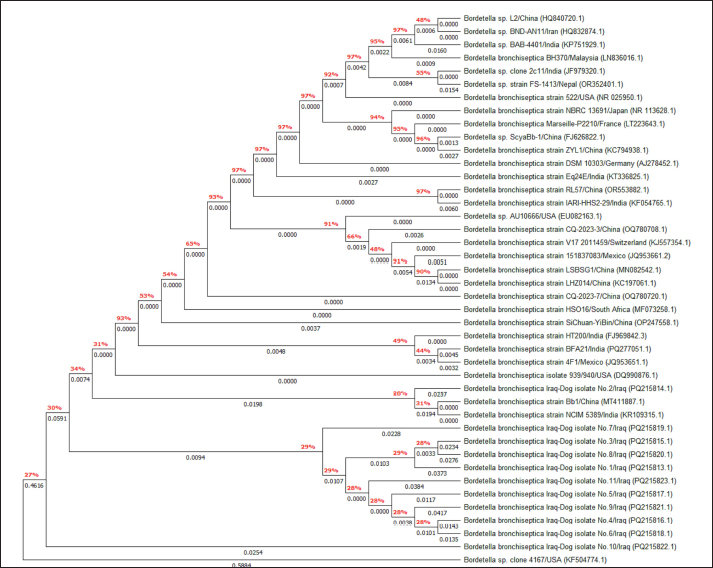
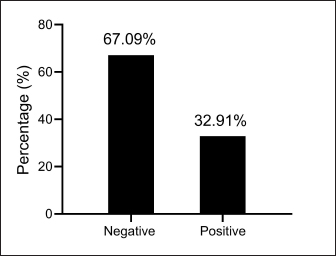
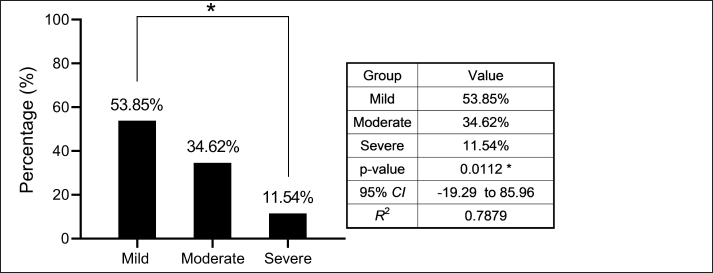
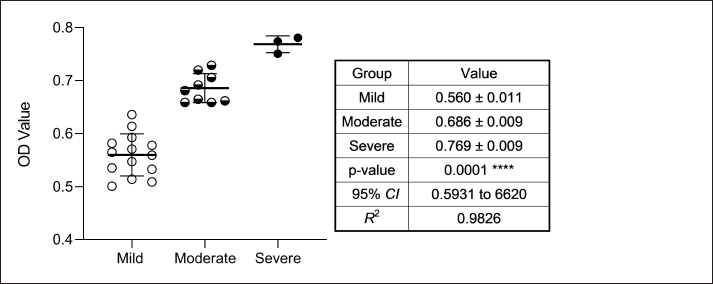
AbstractBackground: Bordetella bronchiseptica is an important bacterial pathogen that is linked primarily to canine infectious respiratory disease complex and rarely to other health issues, including urinary tract infections. Aims: This study aimed to molecularly identify B. bronchiseptica in the urine of diseased dogs with urinary infections and to document local isolates in NCBI. The serological prevalence of anti-B. bronchiseptica antibodies in the sera of the study dogs were also assessed. Methods: A total of 129 pet dogs with urinary tract infections were attended to private clinics in Baghdad province (Iraq) and subjected aseptically to collection of fresh urine and venous blood. Molecular testing of urine samples was performed using the PCR assay, and positive DNA was sequenced, submitted to the NCBI database, and analyzed phylogenetically. Serological testing of sera was performed using ELISA. Results: Targeting the 16S RNA gene, PCR assay revealed that 13.92% of study dogs were positive. Phylogenetic analysis of study B. bronchiseptica isolates showed the presence of identity with global NCBI-BLAST Chinese B. bronchiseptica strain (NCBI-GenBank ID: MT411887.1) at 94.52%–99.66% as the range of similarity and 0.01%–0.0001% as the range of mutation/changes. Serologically, the prevalence rate of anti-B. bronchiseptica antibodies by ELISA was 32.91%; in which, the mild, moderate, and severe infections occurring in 53.85%, 34.62%, and 11.54% of cases, respectively. Subsequently, the ODs of mild, moderate, and severe seropositive ODs were 0.560 ± 0.011, 0.686 ± 0.009, and 0.769 ± 0.009, respectively. Conclusion: This study revealed the prevalence of B. bronchiseptica in dogs by molecular testing of urine and serological examination of blood. Keywords: Canine infectious respiratory disease complex (CIRDIC), Enzyme-linked immunosorbent assay (ELISA), Polymerase chain reaction (PCR), National Center for Biotechnology Information (NCBI), Iraq. IntroductionBordetella bronchiseptica is a small, Gram-negative, rod-shaped, coccobacillus bacterium that belongs to the Alcaligenaceae Family in the Burkholderiales Order under the Class of Betaproteibacteria (Bhardwaj, 2013; Lin et al., 2017). Commonly, B. bronchiseptica exists within the respiratory tracts of different mammals, such as dogs, with well-known contributions to CIRDIC (Reagan and Sykes, 2020; Reagan, 2021). Humans are an uncommon host to B. bronchiseptica; however, exposure to diseased or live-vaccinated animals may increase the chance of infection (Miguelena Chamorro et al., 2023). The bacterium spreads between dogs either directly via licking, nuzzling, coughing, and sneezing, or indirectly by contaminated fomites (Desforges, 2024; Palillo et al., 2024). Worldwide, several reports suggested that the lifecycle of different species of Bordetella is significant when it is found inside and/or outside the respiratory tract (Dewan and Harvill, 2019). Briefly, Bordetella has several virulence-activated genes (vags) that encode the factors of adhesion (e.g., filamentous hemagglutinin, fimbriae, and pertactin) and enable the adhering of bacteria to ciliated epithelial cells of the respiratory tract. Additionally, the regulatory locus of vags is crucial for expressing virulence factors and controlling their production in response to environmental signals (Mekalanos, 1992). Conversion of genes may potentially initiate the environmental source of frequent unexplained outbreaks caused by different Bordetella species (Dewan and Harvill, 2019; Fenwick, 2022; Miguelena Chamorro et al., 2023). In contrast to other bacteria that act as a food source for ameba, B. bronchiseptica can evade the predation of ameba, survive within it for a long time, and incorporate and disseminate along with ameba (Ma et al., 2022). Taylor-Mulneix et al. (2017) mentioned the capability of B. bronchiseptica for growing and disseminating by a complex lifecycle and transferring continuously within a type of ameba known as Dictyostelium discoideum. This suggests that the presence of a stable association might permit the bacterium to expand to different geographical environments (Locht et al., 2020; Nugraha et al., 2023). Other researchers have indicated that ameba may act as environmental vectors by which different Bordetella species could be expanded and disseminated to infect new mammalian hosts (Rivera, 2020; Samba-Louaka, 2021). For diagnosis, the sensitivity and specificity of any diagnostic technique are of great importance to enabling the rapid control of infection (Gharban et al., 2023). Although isolation using appropriate media remains the most reliable method, it is usually challenging and requires confirmation using other advanced laboratory techniques (Kadlec and Schwarz, 2018; Ivaska et al., 2022). In recent years, molecular and sequencing tools have considerably enabled the diagnosis of different pathogens (Schmitz et al., 2022; Matys et al., 2024). Therefore, we aimed to identify B. bronchiseptica in the urine of diseased dogs with various urinary infections molecularly by PCR assay, phylogenetic analysis of study isolates, and estimating the serological prevalence of anti-B. bronchiseptica antibodies in the sera of study dogs by ELISA. Materials and MethodsSamplesTotally, 79 pet dogs with urinary tract symptoms were attended to private veterinary clinicians in Baghdad province (Iraq) during January (2023) and May (2024) and were subjected to the present study. Under aseptic conditions, fresh urine was collected from each study dog into labeled plastic containers and kept frozen (−20°C) until molecular testing. Additionally, approximately 1.5 ml of venous blood was drained into a free-anticoagulant glass-gel tube and centrifuged at 5,000 rpm for 5 minutes. The obtained sera were pipetted into labeled Eppendorf tubes and kept frozen (−4°C) until serological examination. Molecular assayingAccording to Protocol (A) of the G-SpinTM Total DNA Extraction Kit (Intron, Korea), urine samples were processed, and the DNAs of all study samples were tested using the Nanodrop System (Thermo Scientific, UK) to detect their concentration and purity. Targeting the 16S rRNA gene, one set of primers was designed [F: (5´- AGC GGT GGA TGA TGT GGT TT-3´) and R: (5´- AGG CTA CCT ACT TCT GGC GA-3´)] based on the NCBI-GenBank B. bronchiseptica isolate (JX129161.2), and served to prepare the MasterMix tubes at a final volume of 20 µl. For the PCR reaction, a Thermal Cycler System (Bio Rad, USA) was applied at the following conditions: 1 cycle for initial denaturation (95°C/5 minute), 35 cycles for denaturation (95°C/40 second), annealing (56°C/40 second) and extension (72°C/40 second), and 1 cycle for final extension (72°C/7 minute). For PCR analysis, electrophoresis of agarose gel (1.5%) stained with Ethidium Bromide was conducted at 100 voltage and 80 Am for 90 minutes. The band sizes of the PCR products were visualized under a UV transilluminator (Clinx Science, China), and the positive samples were detected at approximately 504 bp. PhylogenyThe DNA of all positive B. bronchiseptica isolates was sequenced by the Macrogen Company (Korea). The sequence data of the study isolates were submitted to NCBI GenBank to obtain specific access numbers. Then, multiple sequence alignment analysis, phylogenetic tree analysis, and homology sequence identity were performed using MEGA-11 Software and NCBI-Viewer to identify significant identity between the study local B. bronchiseptica isolates and the NCBI-BLAST B. bronchiseptica isolates/strains. Serological testingAccording to the manufacturer’s instructions for the B. bronchiseptica antibody ELISA kit (EVL, Spain), the sera in addition to the contents of kits were prepared and processed, and the absorbency values [optical density (OD)] were measured using the Microplate Reader system (BioTek, USA) at 450 nm. The results were then validated and interpreted qualitatively. Statistical analysisA one-way ANOVA and t-test (GraphPad Prism Software, version 8.0.2) were applied to evaluate significant differences between the study values at p < 0.05 (Gharban et al., 2023). Ethical approvalApproval No. 121/CVM-UQ/18-10-2023 was obtained from the Scientific Committee of the Department of Biology in the College of Education (University of Al-Qadisiyah). ResultsAmong the 79 pet dogs, molecular testing of urine samples using the conventional PCR assay revealed that 11 (13.92%) dogs were positive (Figs. 1 and 2). The sequencing data of positive study isolates were submitted to NCBI GenBank at specific access numbers (PQ215813.1, PQ215814.1, PQ215815.1, PQ215816.1, PQ215817.1, PQ215818.1, PQ215819.1, PQ215820.1, PQ215821.1, PQ215822.1, and PQ215823.1). Phylogenetic analysis of the study B. bronchiseptica isolates showed the presence of identity with the global NCBI-BLAST Chinese B. bronchiseptica strain (NCBI-GenBank ID: MT411887.1) at 94.52%–99.66% as the range of similarity and 0.01%–0.0001% as the range of mutations/changes (Table 1, Figs. 3–7). Serological examination of 79 pet dogs by ELISA revealed 26 (32.91%) positive samples; in which, the mild, moderate, and severe infections were observed in 14 (53.85%), 9 (34.62%), and 3 (11.54%), respectively. Subsequently, the ODs of the mild, moderate, and severe seropositive groups were 0.560 ± 0.011, 0.686 ± 0.009, and 0.769 ± 0.009, respectively (Figs. 8–10).
Fig. 1. Total results for testing urine samples by molecular PCR.
Fig. 2. Agarose-gel electrophoresis of PCR products at 100 Volt and 80 Am for 90 minutes. M: Ladder marker (100–1,500 bp); Lanes (3, 5, 9, 11, 13, and 15): Negative samples; Lanes (1, 2, 4, 6, 7, 8, 10, 12, 14, 16, and 17): Positive samples at ≈ 504bp. Table 1. Homology sequence identity (%) of local and NCBI-BLAST B. bronchiseptica isolates.
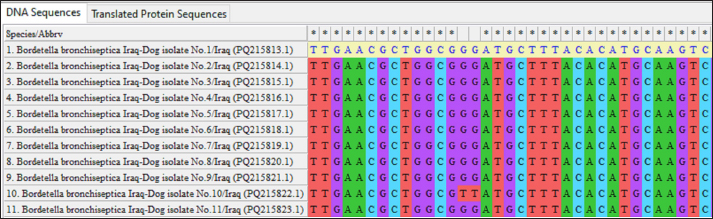
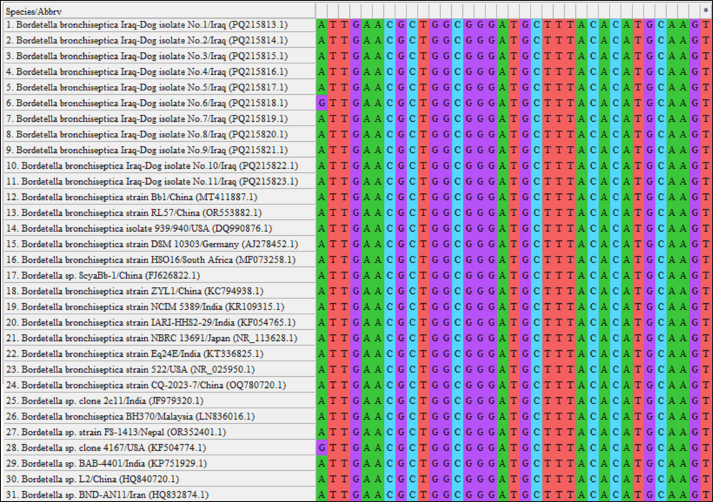
Fig. 3. Multiple sequence alignment of local B. bronchiseptica isolates using MEGA-11 software.
Fig. 4. Multiple sequence alignment of local B. bronchiseptica isolates and NCBI-BLAST B. bronchiseptica isolates/strains using MEGA-11 software. DiscussionBacterial infections represent a major obstacle to public health, linked to several morbidities and mortality in animals and humans. Although great advancements have been made in preventive and therapeutic measures, bacteria remain a cause of great economic losses and social burdens (Gharban and Yousif, 2020; Janik et al., 2020; Mestrovic et al., 2022). In the present study, the molecular and serological data indicated that the prevalence rates of B. bronchiseptica were 13.92% and 32.91%, respectively. In comparison to other worldwide studies, the percentages were 7.4% in Japan (Mochizuki et al., 2008), 45.6% in Germany (Schulz et al., 2014), 1.94% in India (Rose et al., 2024), and 15.47%–36.23% in China (Shang et al., 2024). This variation in the prevalence of B. bronchiseptica infection might be attributed to several factors, including demographic characteristics, environmental conditions, and health care practices. Although traditional diagnostic techniques based on culture and isolation remains the gold standard, they are difficult to perform, interpret, and are time-consuming. Therefore, many serological techniques have been used for the rapid assessment of B. bronchiseptica in various animals and humans (Di Bonaventura et al., 2021; Zhang et al., 2021; Calderaro et al., 2022). During outbreaks, although these techniques are widely used as fast and simple tools for detecting different infections, they have a reduced degree of specificity by yielding false-positive results due to cross-reaction and past exposure (Al-Graibawi et al., 2021; Smolejová et al., 2023). For this reason, PCR exploits to sustaining the accurate and rapid identification of B. bronchiseptica in different clinical specimens by targeting genus- and species-specific genes (Fastrès et al., 2020; Hashish et al., 2021; Cole et al., 2024). When both detection methods, ELISA and PCR, were compared regarding their ability to detect B. bronchiseptica in dogs, PCR was found to be more sensitive than ELISA in the detection of B. bronchiseptica (Batrinou et al., 2022; Bhardwaj et al., 2024). The high sensitivity and specificity of PCR may be attributable to its ability to amplify small amounts of DNA, with detection even at low copy numbers for target sequences (Palka-Santini et al., 2009; Mirabile et al., 2024). Additionally, PCR targeting specific DNA sequences can lead to high specificity by differentiating between closely related bacteria or genetic variants (Leonardo et al., 2021; Smolejová et al., 2023).
Fig. 5. Phylogenetic tree analysis of local B. bronchiseptica isolates using MEGA-11 software.
Fig. 6. Phylogenetic tree analysis of local B. bronchiseptica and NCBI-BLAST B. bronchiseptica isolates/strains using MEGA-11 software.
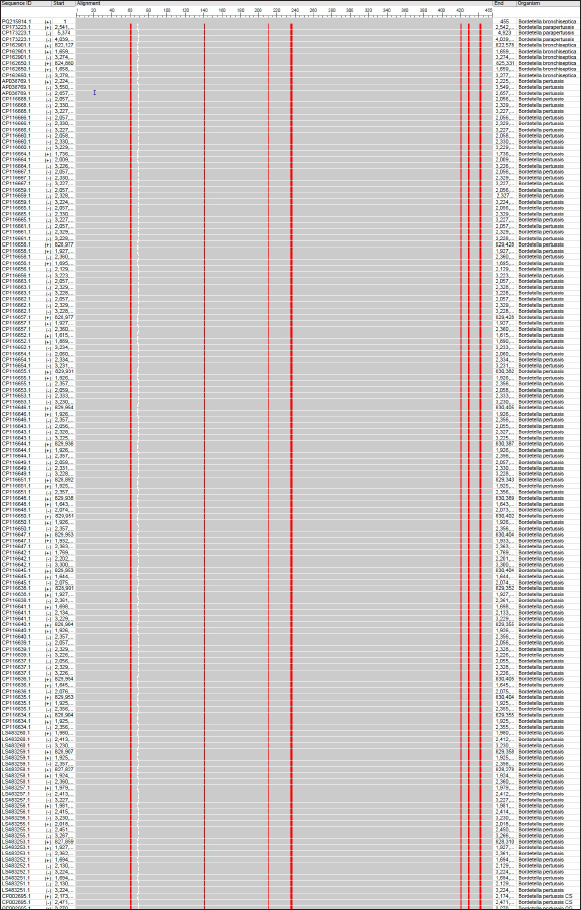
Fig. 7. Multiple sequence alignment of local and NCBI-BLAST B. bronchiseptica isolates/strains using the NCBI MSA Viewer
Fig. 8. Total results for testing the serum samples by serological ELISA.
Fig. 9. Prevalence rate (%) of mild, moderate, and severe seropositive infections according to the severity of OD.
Fig. 10. Values (Mean ± Standard error) of seropositive ODs according to severity. Phylogenetic analyses have already been combined with the data of various researchers to evaluate genetic relatedness among canine B. bronchiseptica isolates (Nicholson and Shore, 2024). Targeting the 16S rRNA gene, phylogenetic analysis of B. bronchiseptica isolates demonstrated their identity to the Chinese NCBI-BLAST B. bronchiseptica strain. In bacteria, the 16S rRNA gene encodes the small subunit ribosomal RNA molecules of ribosomes, which are responsible for the essential process of converting genetic messages to functional cell components via the translation of mRNA to proteins (Chiaruttini and Guillier, 2020; Gong and Koudelka, 2023). Since the advent of high-throughput sequencing, PCR-amplified 16S rRNA genes have typically been clustered based on their similarity to generate operational taxonomic units (OTUs), and then, representing the OTU sequences compared with the reference databases to infer likely taxonomy (Yeh et al., 2021; Egenriether et al., 2022). ConclusionThis may represent the first Iraqi study to elucidate the prevalence rate of B. bronchiseptica in dogs. Molecular PCR testing of urine and serological identification of anti-B. bronchiseptica antibodies by ELISA confirm the circulation of infection in study dogs and their areas. Phylogenetic analysis of isolates targeting the 16S rRNA gene provides important information about the possible source of infection. However, further studies are necessary to estimate the prevalence rate of infection in other local areas and to demonstrate the existence of other Bordetella species in various livestock and wildlife animals. AcknowledgmentsThe authors thank the veterinarians of the private clinic for facilitating sample collection. Conflict of interestNo. FundingNo funds were received to complete this work. Authors’ contributionHMK: Molecular testing of urine samples. AASA: Serological testing of serum samples. MKAA: Collection of urine and blood samples. HAJG: Phylogenetic and statistical analyses of obtained data. All authors contributed to study design and manuscript writing. Data availabilityThe obtained data were included in this manuscript. ReferencesAl-Graibawi, M.A., Yousif, A.A., Gharban, H.A. and Zinsstag, J. 2021. First serodetection and molecular phylogenetic documentation of Coxiella burnetii isolates from female camels in Wasit governorate, Iraq. Iraq J. Vet. Sci. 35 (Supplement III), 47–52. Batrinou, A., Strati, I.F., Tsantes, A.G., Papaparaskevas, J., Dimou, I., Vourvidis, D. and Houhoula, D. 2022. The importance of complementary PCR analysis in addition to serological testing for the detection of transmission sources of Brucella spp. in Greek Ruminants. Vet. Sci. 9(4), 1–8. Bhardwaj, M. 2013. Evaluation of molecular and serological techniques for diagnosis of bordetellosis in dogs. MSc Thesis, Indian Veterinary Research Institute, Izatnagar, India. Bhardwaj, M., Singh, B.R., Sinha, D.K., Kumar, S. and Karthik, K. 2024. Comparative evaluation of micro-agglutination test (MAT), whole cell antigen ELISA (wcELISA) and precipitated protein antigen ELISA (ppELISA) for detection of Bordetella bronchiseptica antibodies in dog population of India. Int. J. Vet. Sci. Anim. Husb. 9(2), 420–424. Calderaro, A., Buttrini, M., Farina, B., Montecchini, S., De Conto, F. and Chezzi, C. 2022. Respiratory tract infections and laboratory diagnostic methods: a review with a focus on syndromic panel-based assays. Microorganisms 10(9), 1856. Chiaruttini, C. and Guillier, M. 2020. On the role of mRNA secondary structure in bacterial translation. Wiley Interdiscip. Rev. RNA 11(3), 1–35. Cole, M., Simon, A.K., Faulkner, A., Skoff, T., Tondella, M.L., Montero, C. and Williams, M. 2024. Comparison of Bordetella species identification among differing rt-PCR assays in the United States. Microbiol. Spect. 12(8), 1–12. Desforges, E. 2024. The domestic cat. The UFAW Handbook on the care and management of laboratory and other research animals. Hoboken, NJ: John Wiley & Sons Ltd. Pp: 546–569. Dewan, K.K. and Harvill, E.T. 2019. Did new transmission cycles in anthropogenic, dense, host populations encourage the emergence and speciation of pathogenic Bordetella?. PLoS Pathog. 15(3), 1–6. Di Bonaventura, G., Angeletti, S., Ianni, A., Petitti, T. and Gherardi, G. 2021. Microbiological laboratory diagnosis of human brucellosis: an overview. Pathog. 10(12), 1–14. Egenriether, S., Sanford, R., Yang, W.H. and Kent, A.D. 2022. Nitrogen cycling microbial diversity and operational taxonomic unit clustering: when to prioritize accuracy over speed. Front. Microbiol. 13(5), 1–14. Fastrès, A., Canonne, M.A., Taminiau, B., Billen, F., Garigliany, M.M., Daube, G. and Clercx, C. 2020. Analysis of the lung microbiota in dogs with Bordetella bronchiseptica infection and correlation with culture and quantitative polymerase chain reaction. Vet. Res. 51, 1–12. Fenwick, B.W. 2022. Bordetella. Chapter 13. Vet. Microbiol., John Wiley & Sons, Inc. New Jersey, USA. pp: 136–150. Gharban, H.A. and Yousif, A.A. 2020. Serological, clinical and hematological prevalence of Coxiella burnetii in adult cows, Iraq. Biochem. Cell. Ach. 20(1), 67–74. Gharban, H.A., Al-Shaeli, S.J. and Hussen, T.J. 2023. Molecular genotyping, histopathological and immunohistochemical studies of bovine papillomatosis. Open Vet. J. 13(1), 26–41. Gong, C. and Koudelka, G.B. 2023. The Shiga toxin (Stx)-phage encoded ribosomal RNA methyltransferase regulates Stx-producing Escherichia coli (STEC) virulence by blocking Stx-mediated inactivation of bacterial ribosomes. bioRxiv 9, 1–38. Hashish, A., Sinha, A., Mekky, A., Sato, Y., Macedo, N.R. and El-Gazzar, M. 2021. Development and validation of two diagnostic real-time PCR (TaqMan) assays for the detection of Bordetella avium from clinical samples and comparison to the currently available real-time TaqMan PCR assay. Microorganisms 9(11), 1–18. Ivaska, L., Barkoff, A.M., Mertsola, J. and He, Q. 2022. Macrolide resistance in Bordetella pertussis: current situation and future challenges. Antibiotics 11(11), 1–11. Janik, E., Ceremuga, M., Niemcewicz, M. and Bijak, M. 2020. Dangerous pathogens as a potential problem for public health. Medicina 56(11), 1–23. Kadlec, K. and Schwarz, S. 2018. Antimicrobial resistance in Bordetella bronchiseptica. Microbiol. Spect. 6(4), 1–11. Leonardo, S., Toldrà, A. and Campàs, M. 2021. Biosensors based on isothermal DNA amplification for bacterial detection in food safety and environmental monitoring. Sensors 21(2), 1–24. Lin, J.Y., Hobson, W.J. and Wertz, J.T. 2017. Saccharedens versatilis gen. nov., sp. nov., a sugar-degrading member of the Burkholderiales isolated from Cephalotes rohweri ant guts. Int. J. Syst. Evol. Microbiol. 67(2), 447–453. Locht, C., Carbonetti, N.H., Cherry, J.D., Damron, F.H., Edwards, K.M., Fernandez, R. and Mascart, F. 2020. Highlights of the 12th international Bordetella symposium. Clin. Infect. Dis. 71(9), 2521–2526. Ma, L., Linz, B., Caulfield, A.D., Dewan, K.K., Rivera, I. and Harvill, E.T. 2022. Naturalhistory and ecology of interactions between Bordetella species and amoeba. Front. Cell. Infect. Microbiol. 12(2), 1–12. Matys, J., Kensy, J., Gedrange, T., Zawiślak, I., Grzech-Leśniak, K. and Dobrzyński, M. 2024. A Molecular approach for detecting bacteria and fungi in healthcare environment aerosols: a systematic review. Int. J. Mol. Sci. 25(4), 1–23. Mekalanos, J.J. 1992. Environmental signals controlling expression of virulence determinants in bacteria. J. Bacteriol. 174(1), 1–7. Mestrovic, T., Aguilar, G.R., Swetschinski, L.R., Ikuta, K.S., Gray, A.P., Weaver, N.D. and Naghavi, M. 2022. The burden of bacterial antimicrobial resistance in the WHO European region in 2019: a cross-country systematic analysis. Lancet Public Health 7(11), e897–e913. Miguelena Chamorro, B., De Luca, K., Swaminathan, G., Longet, S., Mundt, E. and Paul, S. 2023. Bordetella bronchiseptica and Bordetella pertussis: similarities and differences in infection, immuno-modulation, and vaccine considerations. Clin. Microbiol. Rev. 36(3), 1–38. Mirabile, A., Sangiorgio, G., Bonacci, P.G., Bivona, D., Nicitra, E., Bonomo, C. and Musso, N. 2024. Advancing pathogen identification: the role of digital PCR in enhancing diagnostic power in different settings. Diagn. 14(15), 1–18. Mochizuki, M., Yachi, A., Ohshima, T., Ohuchi, A. and Ishida, T. 2008. Etiologic study of upper respiratory infections of household dogs. J. Vet. Med. Sci. 70(6), 563–569. Nicholson, T.L. and Shore, S.M. 2024. Comparative analysis of antimicrobial resistance and genetic diversity of Bordetella bronchiseptica isolates obtained from swine within the United States. Front. Microbiol. 15, 1–11. Nugraha, D.K., Nishida, T., Tamaki, Y., Hiramatsu, Y., Yamaguchi, H. and Horiguchi, Y. 2023. Survival of Bordetella bronchiseptica in Acanthamoeba castellanii. Microbiol. Spect. 11(2), 1–14. Palillo, M.B., Mishkin, N., Atmane, M., Palillo, J.A., Carrasco, S., Woods, C. and Arbona, R.R. 2024. Assessing the biosecurity risk of footwear as a fomite for transmission of adventitious infectious agents to mice. bioRxiv 11, 1–28. Palka-Santini, M., Cleven, B.E., Eichinger, L., Krönke, M. and Krut, O. 2009. Large scale multiplex PCR improves pathogen detection by DNA microarrays. BMC Microbiol. 9, 1–14. Reagan, K.L. 2021. Bordetellosis. In Greene’s infectious diseases of the dog and cat. WB Saunders., St. Louis MO, USA. pp: 669–678. Reagan, K.L. and Sykes, J.E. 2020. Canine infectious respiratory disease. Vet. Clin. Small Anim. Pract. 50(2), 405–418. Rivera, I. 2020. Characterization and evolution of intracellular survival in Bordetella. PhD dissertation, University of Georgia, Georgia, USA. Rose, T.K., Rajesh, J.B., Marwein, S.C., Kar, P., Behera, S.K., Chethan, G E. and Prasad, H. 2024. Hospital based incidence of Bordetella bronchiseptica in Canines in Mizoram. J. Anim. Res. 14(1), 95–97. Samba-Louaka, A. 2021. Amoebae as targets for toxins or effectors secreted by mammalian pathogens. Toxins 13(7), 1–13. Schmitz, J.E., Stratton, C.W., Persing, D.H. and Tang, Y.W. 2022. Forty years of molecular diagnostics for infectious diseases. J. Clin. Microbiol. 60(10), 1–17. Schulz, B.S., Kurz, S., Weber, K., Balzer, H.J. and Hartmann, K. 2014. Detection of respiratory viruses and Bordetella bronchiseptica in dogs with acute respiratory tract infections. Vet. J. 201(3), 365–369. Shang, Q., Gao, W., Zhang, X., Zhao, J., Wu, Y., Li, H. and Zhao, L. 2024. Investigation of the epidemiology, pathogenicity and immunogenicity of Bordetella bronchiseptica isolated from cats and dogs in China from 2021 to 2023. Anim. Dis. 4(1), 1–17. Smolejová, M., Krčmáriková, J., Cihová, I. and Sulo, P. 2023. Are ELISA and PCR discrepancies in the identification of Chlamydia pneumoniae caused by the presence of “Chlamydia-related bacteria”?. Microorganisms 11(1), 1–13. Taylor-Mulneix, D.L., Bendor, L., Linz, B., Rivera, I., Ryman, V.E., Dewan, K.K. and Harvill, E.T. 2017. Bordetella bronchiseptica exploits the complex life cycle of Dictyostelium discoideum as an amplifying transmission vector. PLoS Biol. 15(4), 1–28. Yeh, Y.C., McNichol, J., Needham, D.M., Fichot, E.B., Berdjeb, L. and Fuhrman, J.A. 2021. Comprehensive single-PCR 16S and 18S rRNA community analysis validated with mock communities, and estimation of sequencing bias against 18S. Environ. Microbiol. 23(6), 3240–3250. Zhang, Y., Yang, H., Guo, L., Zhao, M., Wang, F., Song, W. and Wu, B. 2021. Isolation, antimicrobial resistance phenotypes, and virulence genes of Bordetella bronchiseptica from pigs in China, 2018–2020. Front. Vet. Sci. 8, 1–10. | ||
| How to Cite this Article |
| Pubmed Style Kadhim HM, Al-galebi AAS, Al-hassani MKA, Gharban HAJ. Molecular and serological incidences of Bordetella bronchiseptica in pet dogs with urinary infections. Open Vet. J.. 2025; 15(3): 1397-1406. doi:10.5455/OVJ.2025.v15.i3.31 Web Style Kadhim HM, Al-galebi AAS, Al-hassani MKA, Gharban HAJ. Molecular and serological incidences of Bordetella bronchiseptica in pet dogs with urinary infections. https://www.openveterinaryjournal.com/?mno=234989 [Access: January 25, 2026]. doi:10.5455/OVJ.2025.v15.i3.31 AMA (American Medical Association) Style Kadhim HM, Al-galebi AAS, Al-hassani MKA, Gharban HAJ. Molecular and serological incidences of Bordetella bronchiseptica in pet dogs with urinary infections. Open Vet. J.. 2025; 15(3): 1397-1406. doi:10.5455/OVJ.2025.v15.i3.31 Vancouver/ICMJE Style Kadhim HM, Al-galebi AAS, Al-hassani MKA, Gharban HAJ. Molecular and serological incidences of Bordetella bronchiseptica in pet dogs with urinary infections. Open Vet. J.. (2025), [cited January 25, 2026]; 15(3): 1397-1406. doi:10.5455/OVJ.2025.v15.i3.31 Harvard Style Kadhim, H. M., Al-galebi, . A. A. S., Al-hassani, . M. K. A. & Gharban, . H. A. J. (2025) Molecular and serological incidences of Bordetella bronchiseptica in pet dogs with urinary infections. Open Vet. J., 15 (3), 1397-1406. doi:10.5455/OVJ.2025.v15.i3.31 Turabian Style Kadhim, Hadaf Mahdi, Ahlam Ali Soghi Al-galebi, Mithal K. A. Al-hassani, and Hasanain A. J. Gharban. 2025. Molecular and serological incidences of Bordetella bronchiseptica in pet dogs with urinary infections. Open Veterinary Journal, 15 (3), 1397-1406. doi:10.5455/OVJ.2025.v15.i3.31 Chicago Style Kadhim, Hadaf Mahdi, Ahlam Ali Soghi Al-galebi, Mithal K. A. Al-hassani, and Hasanain A. J. Gharban. "Molecular and serological incidences of Bordetella bronchiseptica in pet dogs with urinary infections." Open Veterinary Journal 15 (2025), 1397-1406. doi:10.5455/OVJ.2025.v15.i3.31 MLA (The Modern Language Association) Style Kadhim, Hadaf Mahdi, Ahlam Ali Soghi Al-galebi, Mithal K. A. Al-hassani, and Hasanain A. J. Gharban. "Molecular and serological incidences of Bordetella bronchiseptica in pet dogs with urinary infections." Open Veterinary Journal 15.3 (2025), 1397-1406. Print. doi:10.5455/OVJ.2025.v15.i3.31 APA (American Psychological Association) Style Kadhim, H. M., Al-galebi, . A. A. S., Al-hassani, . M. K. A. & Gharban, . H. A. J. (2025) Molecular and serological incidences of Bordetella bronchiseptica in pet dogs with urinary infections. Open Veterinary Journal, 15 (3), 1397-1406. doi:10.5455/OVJ.2025.v15.i3.31 |