| Original Article | ||
Open Vet. J.. 2020; 10(2): 189-197 doi: 10.4314/ovj.v10i2.8 Open Veterinary Journal, (2020), Vol. 10(2): 189–197 Original Research DOI: http://dx.doi.org/10.4314/ovj.v10i2.8 Unexpected cases in field diagnosis of African swine fever virus in Vietnam: The needs consideration when performing molecular diagnostic testsAnh Duc Truong, Duc Viet Ly, Thi Hao Vu, Van Tuan Hoang, Thi Chinh Nguyen, Thi Nhu Chu, Huyen Thi Nguyen, The Vinh Nguyen, Ngoc Thi Pham, Ha Thi Thanh Tran and Hoang Vu Dang*Department of Biochemistry and Immunology, National Institute of Veterinary Research (NIVR), Hanoi, Vietnam *Corresponding Author: Hoang Vu Dang. Department of Biochemistry and Immunology, National Institute of Veterinary Research (NIVR), Hanoi, Vietnam. Email: dangnivr [at] yahoo.com Submitted: 06/12/2019 Accepted: 29/05/2020 Published: 21/06/2020 © 2020 Open Veterinary Journal
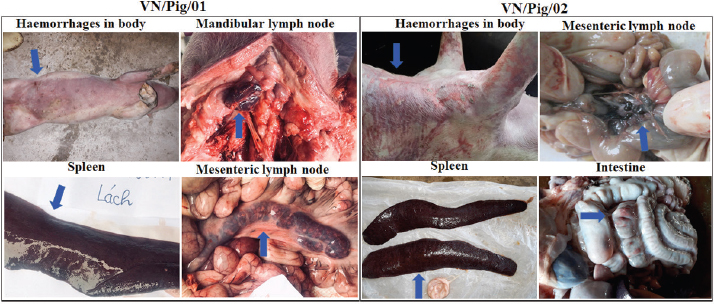
AbstractBackground: The first confirmed case of African swine fever (ASF) in Vietnam was reported officially in February 2019. To date, ASF virus (ASFV) have been detected in 63/63 provinces in Vietnam. Currently, real-time polymerase chain reaction (PCR) is considered to be a powerful tool for viral detection in field samples, including ASFV. However, some recent reports have suggested that mismatches in primer and probe binding regions may directly affect real-time PCR qualification, leading a false-negative result. Aim: This study aims to further examine a conflicting result obtained from two OIE recommended methods, conventional PCR and real-time PCR, for ASFV detection. Methods: Two ASF suspected pigs from different provinces in the north of Vietnam were selected for this study based on clinical signs and postmortem lesions. The different results obtained by OIE-recommended conventional PCR and real-time PCR were further analyzed by the Sanger sequencing method and virus isolation in combination with hemadsorption (HAD) test using porcine alveolar macrophages cells. Results: The results showed that when the primer sequence matched perfectly with the sequences of field isolates, a mutation in probe binding region was found, indicating that a single mismatch in the probe binding site may cause a false-negative result by real-time PCR in detecting ASFV in clinical samples in Vietnam. An agreement between conventional PCR, using PPA1/PPA2 primers and two golden standard methods, virus isolation in combination with HAD assay, and sequencing method was observed in this study. Conclusion: A single mismatch in the probe binding site caused a failse-negative result by realtime PCR method in field diagnosis of ASFV. The needs consideration when selecting the appropriate molecular diagnostic methods is based on the current databases of ASFV sequences, particularly for epidemiological surveillance of ASF. Keywords: African swine fever, PCR, Pigs, Real-time PCR, Vietnam. IntroductionAfrican swine fever (ASF) is a complex viral disease of swine and has a dramatic socioeconomic impact on both developed and developing countries (Gallardo et al., 2015). ASF virus (ASFV) is the causative agent of ASF, and the virus is a large, enveloped, double-stranded DNA virus that is the sole member of the newly named Asfarviridae family (Gallardo et al., 2015; Galindo and Alonso, 2017). ASFV was first identified in Kenya in 1921 (Galindo and Alonso, 2017). ASFV was confined to Africa until it has rapidly spread to Eurasia in the middle of the last century and to South America and the Caribbean in the later half of the last century (Galindo and Alonso, 2017). Until now, a total of 24 genotypes has been identified in the ASFV (Gallardo et al., 2015; Galindo and Alonso, 2017; Quembo et al., 2018; Simulundu et al., 2018). According to the OIE/FAO report, an outbreak of ASF was reported in China during August 2018 and in Mongolia during January 2019 (Zhou et al., 2018), demonstrating that ASFV has a remarkable capacity for transboundary and transcontinental spread. The first confirmed case of ASF in Vietnam was reported officially in early February 2019. To date, ASFV has been detected in 63/63 provinces/cities in Vietnam. Many molecular methods have been developed to detect ASFV, such as conventional polymerase chain reaction (PCR), real-time PCR, enzyme-linked immunosorbent assay (ELISA), and fluorescent antibody test (FAT or DIFT). Each method possesses advantages and disadvantages. Among these methods, real-time PCR is considered to be a powerful tool for detecting ASFV in blood and tissues, including lymph node, spleen, tonsil, kidney, lung, and liver (OIE, 2012). The first report using real-time PCR was demonstrated by King et al. (2003) to detect ASFV and was currently carried out by the OIE reference laboratories as a rapid, sensitive, and specific method for ASFV diagnosis from field samples. However, it is over 15 years since King et al.’s (2003) method has been developed, and novel GenBank deposited ASFV sequences have shown some mutations in its sequences when compared to ones in the past. An updated method of real-time PCR based on King et al.’s (2003) recommendation should be considered, particularly for probe sequences. Recent studies have suggested that the mismatches in primer and probe binding regions may directly affect real-time PCR qualification, leading to a false-negative result. It was reported that a single mismatch on the primer and probe binding site causes false-negative results, and a quantitation error up to 33% was recognized (Suss et al., 2009). Particularly, a conflicting result between real-time PCR, reference method, and direct immunofluorescence test was reported (Kamau et al., 2017) in which the mismatches in probe binding region resulted in a quantification error up to 60%. These studies have shown a weak point of this method on virus detection in the field samples. A combination of real-time PCR, conventional PCR, ELISA, or DIFT is recommended for ASFV detection by OIE (2012). Another virus assay recommended by FAO and OIE for the detection of ASFV is the virus isolation in combination with the hemadsorption (HAD) test. This method is considered as a golden standard test and is often used to confirm the positive results of conventional PCR, real-time PCR, and ELISA (FAO). The HAD test was first described in 1960 by Malmquist and Hay (Malmquist and Hay, 1960) and a positive HAD test is definitive for the presence of ASFV. Additionally, the identification of HAD is recommended to confirm the primary outbreak of ASF that is detected by other molecular assays. However, the performance of this test requires primary cell systems and takes a week to obtain the result. In this study, a conflicting result obtained from the conventional PCR and real-time PCR was further confirmed by sequencing analysis and virus isolation in combination with the HAD test. Our study provides new insight into the potential impact of a mismatch on probe sequence in the detection of ASFV from clinical samples. Materials and MethodsSample collectionTwo ASF suspected pigs from different provinces in the north of Vietnam were selected for this study based on clinical signs and postmortem lesions. Tissues, including mesenteric lymph nodes, spleen, kidney, small intestine, and liver, were collected according to the guidelines of OIE (2012). Nucleic acid extraction and genomic DNA amplificationTissues were homogenized in 1 × phosphate-buffered saline at a ratio of 1:10 (w/v) according to the guidelines of OIE (2012). Tissue homogenates from each animal were used for laboratory diagnosis and molecular analyses. The DNA of ASFV was extracted from infected pig organ samples using PureLink Viral RNA/DNA Mini Kit (Invitrogen, Carlsbad, CA) as per the manufacturer’s instructions. The conventional PCR and real-time PCR analysis was carried out with specific primers, PPA1/PPA2 (Aguero et al., 2003; OIE, 2012), and TaqMan Probe (King et al., 2003; Fernandez-Pinero et al., 2013), respectively, and according to the recommendation by the World Organization for Animal Health (OIE, 2012). Briefly, ASFV DNA was amplified by PCR using the ASF diagnostic primers PPA1 [5′- AGT TAT GGG AAA CCC GAC CC -3′] and PPA2 [5′- CCC CTG AAT CGG AGC ATC CT -3′] that generate an amplicon of 257 bp within the p72 gene, which help to confirm the presence of ASFV (Aguero et al., 2003). PCR was carried out in Agilent PCR System (Agilent, Santa Clara, CA) using Taq polymerase (Thermo Scientific, Waltham, MA), according to the manufacturer’s instructions. Thermal conditions for carrying out PCR are as follows: an initial incubation at 95°C for 10 minutes, 40 cycles of denaturation at 95°C for 15 seconds, annealing at 60°C for 30 seconds, and extension at 72°C for 30 seconds, and final incubation at 72°C for 7 minutes. Real-time PCR, which is more specific and sensitive than the conventional PCR, was carried out with two sets of primers to amplify the ASFV DNA and to confirm the gel-based PCR results. In the first real-time PCR, primer 1 [5′- CTG CTC ATG GTA TCA ATC TTA TCG A -3′ (positive strand)] and primer 2 [5′- GAT ACC ACA AGA TC(AG) GCC GT -3′ (negative strand)] at a concentration of 50 pmol/μl, and TaqMan probe [5′- 6-carboxy-fluorescein (FAM)-CCA CGG GAG GAA TAC CAA CCC AGT G-6-carboxy-tetramethyl-rhodamine (TAMRA) -3′] at a concentration of 5 pmol/μl were used as previously described (King et al., 2003; Fernandez-Pinero et al., 2013). In the second real-time PCR, primer 1 [5′- GAT GAT GAT TAC CTT YGC TTT GAA -3′] and primer 2 [5′- TCT CTT GCT CTR GAT ACR TTA ATA TGA -3′] at a concentration of 50 pmol/μl, and TaqMan probe [5′- CY5-CCA CGG GAG GAA TAC CAA CCC AGT G-CY5 -3′] at a concentration 5 pmol/μl were used as previously described (Haines et al., 2013). Positive amplification control consisted of known DNA of positive ASFV, and negative amplification control consisted of nuclease-free sterile water. Genomic characterizationThe amplification products were electrophoresed on a 1.5% agarose gel against a 50 bp DNA leader marker (Thermo Scientific) and visualized by UV irradiation and ethidium bromide staining (Sigma-Aldrich, Louis, MO). Amplicons of the correct size were excised from the agarose gel and purified using the QIAQuick gel extraction kit (QIAgen, Valencia, CA) according to the manufacturer’s specifications, and sequenced at Gentis (Gentis Jsc, Tay Ho, Hanoi, Vietnam). The percentage of identities and similarity scores of the viral gene sequences were calculated using DNASTAR Lasergene (DNASTAR, Inc., Madison, WI). The gene identities and similarities of two ASFV strains from Vietnam were compared with the reference sequences of the ASFV strain from the National Center for Biotechnology Information (NCBI), including China/2018/Anhui (GenBank accession no. MK128995), China/ASFV-SY18/I (GenBank accession no. MH713612), Russia/Krasnodar/2012 (GenBank accession no. KJ195685), China/Jilin/2018/boar (GenBank accession no. MK189456), Kenya/Ken05 (GenBank accession no. KM111294), and Russia/Irkutsk 2017 (GenBank accession no. KY963545). Virus isolationThe porcine alveolar macrophages (PAM) cultures used for the isolation of the virus were derived from domestic pigs that were tested negative for ASFV upon diagnosis with conventional PCR as previously described (Carrascosa et al., 2011), and according to the recommendation by the World Organization for Animal Health (OIE, 2012). Briefly, these cells were obtained and resuspended in Dulbecco’s modified Eagle’s medium (DMEM, Thermo Fisher Scientific, Waltham MA) supplemented with streptomycin (Sigma-Aldrich, St. Louis, MO), ampicillin (Sigma-Aldrich, St. Louis, MO), and 5% fetal calf serum (FCS, Sigma-Aldrich, St. Louis, MO) to a final concentration of 1 × 106 cells/ml. Six-well culture plates (Thermo Scientific, Waltham MA), containing 2 ml of the cell suspension, were inoculated with 50 μl of a clarified 10% spleen or lymph node suspension. After three days incubation at 37°C, in an atmosphere with 5% CO2, the first passage virus was harvested by freezing and thawing. For the second passage, 50 μl of this virus was harvested and added to 2 ml fresh PAMs, then incubated and harvested as described for the first passage. For the third passage, 50 μl of this virus was harvested and added to 2 ml fresh PAMs, then incubated at 37°C, in an atmosphere with 5% CO2. After one-day incubation, a 20-μl preparation of 1% homologous red blood cells in buffered saline was added to each well. The plates were examined for HAD and cytopathic effects over a 4-day period in PAM cells according to the recommendation by the World Organization for Animal Health (OIE, 2012). Ethical approvalThis study was conducted in compliance with the institutional rules for the care and use of laboratory animals and using a protocol approved by the Ministry of Agriculture and Rural Development (MARD), Vietnam (TCVN 8402:2010). ResultsTypical clinical lesions of the ASFV-infected Vietnam pigsThe identification of clinical signs and pathological lesions are critical for the diagnosis of ASFV. As shown in Figure 1, two dead pigs from different provinces of Vietnam were examined and typical clinical signs, including high fever and hemorrhages, in the skin were found. Importantly, the lymph nodes were hemorrhagic and the spleen was greatly enlarged, hemorrhagic and turned blackish colour. According to the OIE, hemorrhages are frequently present on the body, lymph nodes, liver, spleen, lungs, kidney, heart and bladder, and the hepatic lymph nodes are hemorrhagic in all cases. Moreover, the gall bladder is edematous. In chronic cases, there may be pneumonia, arthritis, pleuritic, and skin ulcers. There are hemorrhages on the skin of aborted fetuses and on the placenta, if present (OIE, 2012). These findings suggest the presence of one of the ASFs in the dead pigs from different provinces of Vietnam.
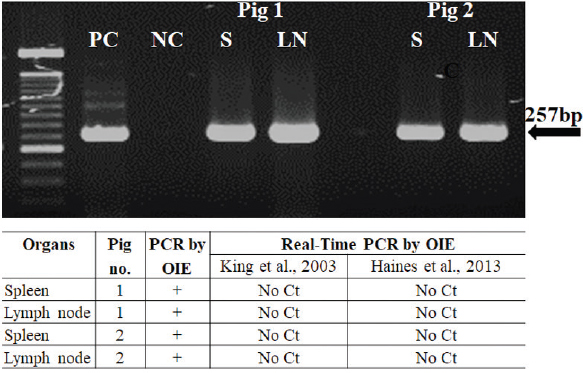
Fig. 1. Clinical signs and pathological findings in organs of two domestic pigs (VN/Pig/01 and VN/Pig/02) infected with ASFV in Vietnam. Confirmation by sequence analysis and virus isolation in combined with HAD testFirst, the spleen and lymph nodes from the two dead pigs was examined by conventional PCR and real-time PCR with specific primers, PPA1/PPA2 (OIE, 2012), and TaqMan probe (King et al., 2003; Fernandez-Pinero et al., 2013; Haines et al., 2013) for ASFV, respectively. We found a conflicting result between conventional PCR and real-time PCR in which conventional PCR showed a positive result, while no Ct values were detected by real-time PCR (Fig. 2). Further confirmation using sequencing method and virus isolation in combination with HAD had been carried out with golden standard tests to verify these data. ASFV-positive lymph nodes were employed for referring the two tests, and the results are shown in Figures 3 and 4.
Fig. 2. Amplification of ASFV DNA in the spleen and lymph nodes of two domestic pigs infected with ASFV in Vietnam by conventional PCR as described by OIE (top). The result of conventional PCR and real-time PCR carried out with two sets of primers as previously described to detect ASFV in organs of two domestic pigs (bottom). PC=Positive control; NC=Negative Control; S=Spleen; LN=Mesenteric lymph node.
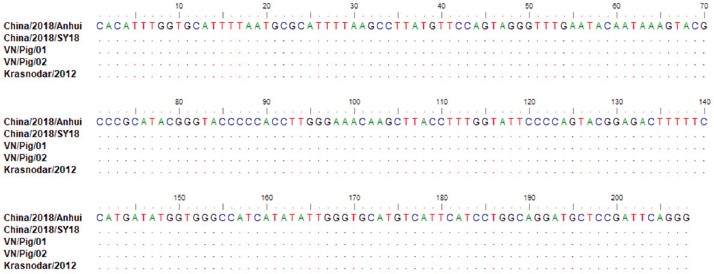
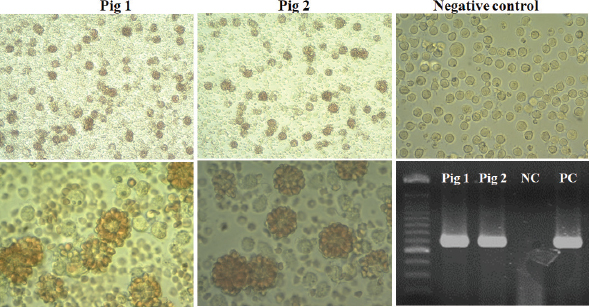
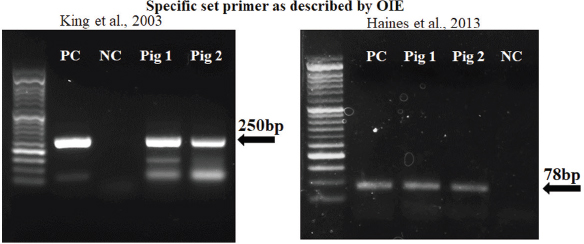
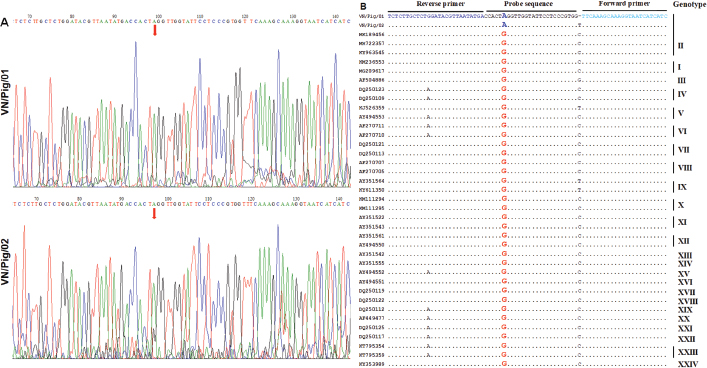
Fig. 3. Multiple sequence alignment of p72 gene amplified by PPA1/PPA2 primers in Vietnam ASFV strains with reference ASFV strains, including China/2018/Anhui (MK128995), China/2018/SY18 (MH713612), and Krasnodar/2012 (KJ195685). Dots indicate similarity with ASFV references sequences. As shown in Figure 3, the nucleotide sequences of ASF isolates from two Vietnam pigs shared 100% identities, with the following strains: ASFV-SY18 and ASFV-Anhui, isolated in China in 2018 (Zhou et al., 2018) and from Krasnodar, Russia, in 2012 (Kolbasov et al., 2017) based on the fragment of the p72 gene amplified by specific PPA1/PPA2 primers (Fig. 3). These reference isolates have been reported previously as genotype II. A further confirmation by virus isolation in combination with HAD test is shown in Figure 4, in which a positive HAD was observed in cultures. This positive HAD test was subsequently checked by conventional PCR using PPA1/PPA2 primers as described in the OIE manual (OIE, 2012), to ensure that it is true-positive. A positive PCR was observed, as shown in Figure 4, and further sequencing of this positive PCR product indicated true-positive result (data not shown). These results indicate the presence of ASFV in our field samples. Additionally, the data obtained from sequencing and virus isolation in combination with HAD indicated a false-negative caused by real-time PCR diagnosis of ASFV in clinical samples. In order to get a better understanding of the molecular mechanisms underlying this event, we carried out conventional PCR using two sets of primers that were used for the real-time PCR. As shown in Figure 5, two PCR products with expected size of 250 bp and 78 bp were observed in gel, indicating that a problem might occur in probe binding regions when we carry out real-time PCR with these primers. Further sequencing data (Fig. 6) indicated that there was no mismatch between primer binding sites and sequences of two isolates in the fragment of 78 nucleotides amplified by OIE primer set (Fernandez-Pinero et al., 2013; Haines et al., 2013). However, a mismatch in probe binding sites (Fig. 6) was observed, suggesting that this mismatch may cause a false-negative result by real-time PCR on ASFV diagnosis in the clinical samples from Vietnam.
Fig. 4. Hemadsorption in the culture of PAM cells from two domestic pigs infected with ASFV in Vietnam (Original magnification, ×200 and ×400), and conventional PCR to detect ASFV after three passages of virus-infected cell culture using PPA1/PPA2 primers as described by OIE.
Fig. 5. The conventional PCR to detect ASFV strains in Vietnam using two sets of real-time PCR primers recommended by OIE.
Fig. 6. (A) The sequencing results of ASFV strains in Vietnam after amplification with a specific set of primers described by Haines et al. (2013). (B) Multiple sequence alignment of the p72 gene fragment of Vietnam ASFV strains amplified by primers described by Haines et al. (2013), with reference ASFV strains from the NCBI. Dots indicate similarity with primer and probe sequences. Mismatching nucleotides at the primer and probe binding sites are highlighted. DiscussionASF is a highly contagious viral disease of swine which causes high mortality, up to 100%, in domestic pigs (Zsak et al., 2005). ASF is a very complex and lethal viral disease for which no vaccine is available to prevent the infection. From early February 2019, an ASF outbreak in Vietnam was reported officially, and to date, the disease has been reported in 63 provinces of Vietnam. Millions of pigs have been culled in infected farms, suggesting the risk of spread of this virus. Many active measures were conducted by Vietnamese Government to prevent and control the spread of this disease, and the improvement of ASF diagnostic strategy is one of the most important steps in controlling and preventing ASF in Vietnam. The clinical signs, including widespread hemorrhages on the body and various postmortem tissues of two died domestic pigs in Vietnam, are consistent with those of the reported ASF cases in Africa, China, and Russia (OIE), suggesting that the outbreaks of a highly fatal hemorrhagic disease are afflicting domestic pigs in Vietnam. Rapid and reliable diagnosis of ASFV is a critical step to control and prevent the spread of this disease worldwide. Many molecular methods were developed to detect the antigen and genome of ASFV in clinical samples, including conventional PCR, real-time PCR, ELISA, DIFT, loop-mediated isothermal amplification, immunofluorescence assays (IFA), and immunoperoxidase test (IPT). Among these, ELISA, IFA, and IPT detect ASFV using antibodies, and the conventional PCR and real-time PCR detect ASFV by amplification of its genome. The virus isolation, followed by HAD test, is one of the golden standard tests recommended by OIE and FAO. Each method possesses advantages and disadvantages. Further improvements should be carried out to increase their sensitive as well as specific value. In this study, a conflicting result between conventional PCR and real-time PCR was further examined in order to bring new insight into the potential effects of mismatches in probe binding regions to detect ASFV in field samples. The different organ samples, including spleen and lymph nodes from two pigs that died due to ASFV infection in Vietnam, were collected according to the guidelines of OIE (2012). The spleen and lymph node samples were subjected for conventional PCR using specific primers (PPA1/PPA2), and two sets of real-time PCR using TaqMan Probe as described by previous studies (King et al., 2003; Fernandez-Pinero et al., 2013; Haines et al., 2013). While conventional PCR using PPA1/PPA2 showed a positive result, no Ct values were detected by real-time PCRs established by OIE (King et al., 2003; Fernandez-Pinero et al., 2013; Haines et al., 2013). In the last decade, the PCRs, including conventional PCR and real-time PCR, have been applied to detect ASFV in clinical samples. Of these PCR methods, real-time PCR is considered to be a powerful tool for ASFV detection in blood and various organ tissues. This method has given accurate quantitative results for virus detection (Moody et al., 2000; Ludwig et al., 2008) in different types of field samples. However, the mismatches on primer and probe binding regions may impair the quantification of this method (Whiley and Sloots, 2005a, 2005b, 2006). It has been reported that conventional PCR using PPA1/PPA2 may also cause false-positives, and therefore, a subsequent sequencing of the PCR product is required to verify the positive results obtained by the OIE approved PCR (Vlad et al., 2015). In this study, we employed two golden standard tests to confirm OIE postive PCR. As a result, a good correlation was observed between conventional PCR, sequence analysis, and virus isolation in the PAM culture from the infected pigs in combination with HAD test, indicating the presence of ASFV in these clinical samples. In the last decade, the PCRs, including convetinal PCR and real-time PCR, have been applied to detect ASFV in clinical samples. It has been indicated that real-time PCR is considered to be a powerful tool for ASFV detection in blood and organ tissues, including lymph node, spleen and tonsil, kidney, lung, and liver. This method has given an accurate quantitative result for virus detection (Moody et al., 2000; Ludwig et al., 2008) in different types of field samples. However, the mismatches on primer and probe binding regions may impair the quantification of this method (Whiley and Sloots, 2005a, 2005b, 2006; Suss et al., 2009). Recent studies have shown that the same region of the p72 gene has been targeted by the method previously described (King et al., 2003), but the efficiency of real-time PCR was improved remarkably due to issues, such as the modification of primers, length of the target size, and modification of probe, PCR reagents, or the use of an internal control that is an important point of quality control (Wong and Medrano, 2005). It provided the assurance of efficient DNA extraction, and the samples contained PCR inhibitors in each sample, therefore leading to false-negative results (McKillen et al., 2010; Tignon et al., 2011). New real-time PCRs developed with modification of primers and shorted probes have been seen to provide greater sensitivity over 10% with positive samples than the real-time TaqMan PCR test recommended in the OIE manual. In particular, the 78 nucleotides of two ASFV isolates in this study shared 97% identity with the following strains: China/ASFV-SY18, China/Jilin/2018/boar, and Russia/Irkutsk 2017 (Kolbasov et al., 2017; Li et al., 2018; Zhou et al., 2018). A single mismatch in the probe binding sites was only noted in our ASFV isolates, i.e., nucleotide A in our ASFV isolates, but nucleotide G in the reference ASFV strains. The strong signals obtained from peak “A” were regconized in both field samples, suggesting that a modified probe sequence should be considered to improve ASFV diagnostic stratagies in Vietnam. It has been recommended by OIE that at least two antigen and genome detection tests, incluling both conventional PCR and real-time PCR, should be used on the samples taken from the same suspected pigs (Oura et al., 2013). Then, positive results from PCRs would be subsequently confirmed by virus isolation and HAD test, particularly for primary outbreak of ASFV, and a positive HAD test is definitive for ASFV diagnosis. In conclusion, real-time PCR is the most sensitive and specific method to detect ASFV in tissue samples but not a perfect method for the field diagnosis of ASFV in Vietnam. The mutation in probe binding site may cause a false-negative result. The selection of the appropriate molecular tests based on the current databases of ASFV seuqences is very essential to obtain an accurate ASF diagnosis in field samples in Vietnam. Additionally, further virus assays, including virus isolation confirmed by HAD, are critical for determining primary outbreak or case of ASF as recommended by OIE. AcknowledgmentsThis work was supported by the Ministry of Science and Technology to Hoang Vu Dang (Project code: ĐTĐL.CN-75/19). The author would like to thank Dr. Deivendran Rengaraj, at the Department of Agricultural Biotechnology, and Research Institute of Agriculture and Life Sciences, College of Agriculture and Life Sciences, Seoul National University, Republic of Korea, for critical review and English revision. Conflict of interestThe authors declare that they have no competing interests. Authors contributionADT, HTTT, and HVD planned and designed the experiments. ADT, DVL, THV, VTH, TCN, TNC, HTN, TVN, and HTTT carried out the experiments. ADT, HTTT, and HVD analyzed the data. HVD contributed the reagents, materials, and analytical tools. ADT, HTTT, NTP, and HVD wrote the manuscript. All authors read and approved the final manuscript. ReferencesAguero, M., Fernandez, J., Romero, L., Sanchez Mascaraque, C., Arias, M. and Sanchez-Vizcaino, J.M. 2003. Highly sensitive PCR assay for routine diagnosis of African swine fever virus in clinical samples. J. Clin. Microbiol. 41(9), 4431–4434. Carrascosa, A.L., Bustos, M.J. and de Leon, P. 2011. Methods for growing and titrating African swine fever virus: field and laboratory samples. Curr. Protoc. Cell Biol. 53(1), 26.14.1–26.14.25. Fernandez-Pinero, J., Gallardo, C., Elizalde, M., Robles, A., Gomez, C., Bishop, R., Heath, L., Couacy-Hymann, E., Fasina, F.O., Pelayo, V., Soler, A. and Arias, M. 2013. Molecular diagnosis of African swine fever by a new real-time PCR using universal probe library. Transbound. Emerg. Dis. 60(1), 48–58. Galindo, I. and Alonso, C. 2017. African swine fever virus: a review. Viruses 9(5), 103. doi:10.3390/v9050103. Gallardo, C., Nieto, R., Soler, A., Pelayo, V., Fernandez-Pinero, J., Markowska-Daniel, I., Pridotkas, G., Nurmoja, I., Granta, R., Simon, A., Perez, C., Martin, E., Fernandez-Pacheco, P. and Arias, M. 2015. Assessment of African swine fever diagnostic techniques as a response to the epidemic outbreaks in Eastern European Union Countries: how to improve surveillance and control programs. J. Clin. Microbiol. 53(8), 2555–2565. Haines, F.J., Hofmann, M.A., King, D.P., Drew, T.W. and Crooke, H.R. 2013. Development and validation of a multiplex, real-time RT PCR assay for the simultaneous detection of classical and African swine fever viruses. PLoS One 8(7), e71019. Kamau, E., Agoti, C.N., Lewa, C.S., Oketch, J., Owor, B.E., Otieno, G.P., Bett, A., Cane, P.A. and Nokes, D.J. 2017. Recent sequence variation in probe binding site affected detection of respiratory syncytial virus group B by real-time RT-PCR. J. Clin. Virol. 88, 21–25. King, D.P., Reid, S.M., Hutchings, G.H., Grierson, S.S., Wilkinson, P.J., Dixon, L.K., Bastos, A.D. and Drew, T.W. 2003. Development of a TaqMan PCR assay with internal amplification control for the detection of African swine fever virus. J. Virol. Methods 107(1), 53–61. Kolbasov, D., Titov, I., Tsybanov, S., Gogin, A. and Malogolovkin, A. 2018. African swine fever virus, Siberia, Russia, 2017. Emerg. Infect. Dis. 24(4), 796–798. Li, L., Wang, Q., Ge, S., Liu, Y., Liu, C., Liu, F., Hu, Y., Li, J., Bao, J., Ren, W., Zhang, Y., Xu, T., Sun, C., Li, L., Wang, S., Fan, X., Huang, B., Wu, X. and Wang, Z. 2018. Infection of African swine fever in wild boar, China, 2018. Transbound. Emerg. Dis. 66(3), 1395–1398. Ludwig, A., Adams, O., Laws, H.J., Schroten, H. and Tenenbaum, T. 2008. Quantitative detection of norovirus excretion in pediatric patients with cancer and prolonged gastroenteritis and shedding of norovirus. J. Med. Virol. 80(8), 1461–1467. Malmquist, W.A. and Hay, D. 1960. Hemadsorption and cytopathic effect produced by African Swine Fever virus in swine bone marrow and buffy coat cultures. Am. J. Vet. Res. 21, 104–108. McKillen, J., McMenamy, M., Hjertner, B., McNeilly, F., Uttenthal, A., Gallardo, C., Adair, B. and Allan, G. 2010. Sensitive detection of African swine fever virus using real-time PCR with a 5' conjugated minor groove binder probe. J. Virol. Methods 168(1–2), 141–146. Moody, A., Sellers, S. and Bumstead, N. 2000. Measuring infectious bursal disease virus RNA in blood by multiplex real-time quantitative RT-PCR. J. Virol. Methods 85(1–2), 55–64. OIE. 2012. African swine fever. Manual of diagnostic tests and vaccines for terrestrial animals 2012, 8th ed. Chapter 2.8.1. Oura, C.A., Edwards, L. and Batten, C.A. 2013. Virological diagnosis of African swine fever–comparative study of available tests. Virus Res. 173(1), 150–158. Quembo, C.J., Jori, F., Vosloo, W. and Heath, L. 2018. Genetic characterization of African swine fever virus isolates from soft ticks at the wildlife/domestic interface in Mozambique and identification of a novel genotype. Transbound. Emerg. Dis. 65(2), 420–431. Simulundu, E., Sinkala, Y., Chambaro, H.M., Chinyemba, A., Banda, F., Mooya, L.E., Ndebe, J., Chitanga, S., Makungu, C., Munthali, G., Fandamu, P., Takada, A. and Mweene, A.S. 2018. Genetic characterisation of African swine fever virus from 2017 outbreaks in Zambia: identification of p72 genotype II variants in domestic pigs. Onderstepoort. J. Vet. Res. 85(1), e1–e5. Suss, B., Flekna, G., Wagner, M. and Hein, I. 2009. Studying the effect of single mismatches in primer and probe binding regions on amplification curves and quantification in real-time PCR. J. Microbiol. Methods 76(3), 316–319. Tignon, M., Gallardo, C., Iscaro, C., Hutet, E., Van der Stede, Y., Kolbasov, D., De Mia, G.M., Le Potier, M.F., Bishop, R.P., Arias, M. and Koenen, F. 2011. Development and inter-laboratory validation study of an improved new real-time PCR assay with internal control for detection and laboratory diagnosis of African swine fever virus. J. Virol. Methods 178(1–2), 161–170. Vlad, P., Laura, B. and Mihaela, Z. 2015. False positive results using PCR detection method for African swine fever virus in wild boars from northern Romanian hunting zones. Turkish J. Vet. Anim. Sci. 39, 1–8. Whiley, D.M. and Sloots, T.P. 2005a. Sequence variation in primer targets affects the accuracy of viral quantitative PCR. J. Clin. Virol. 34(2), 104–107. Whiley, D.M. and Sloots, T.P. 2005b. Comparison of three in-house multiplex PCR assays for the detection of Neisseria gonorrhoeae and Chlamydia trachomatis using real-time and conventional detection methodologies. Pathology 37(5), 364–370. Whiley, D.M. and Sloots, T.P. 2006. Sequence variation can affect the performance of minor groove binder TaqMan probes in viral diagnostic assays. J. Clin. Virol. 35(1), 81–83. Wong, M.L. and Medrano, J.F. 2005. Real-time PCR for mRNA quantitation. Biotechniques 39(1), 75–85. Zhou, X., Li, N., Luo, Y., Liu, Y., Miao, F., Chen, T., Zhang, S., Cao, P., Li, X., Tian, K., Qiu, H.J. and Hu, R. 2018. Emergence of African swine fever in China, 2018. Transbound. Emerg. Dis. 65(6), 1482–1486. Zsak, L., Borca, M.V., Risatti, G.R., Zsak, A., French, R.A., Lu, Z., Kutish, G.F., Neilan, J.G., Callahan, J.D., Nelson, W.M. and Rock, D.L. 2005. Preclinical diagnosis of African swine fever in contact-exposed swine by a real-time PCR assay. J. Clin. Microbiol. 43(1), 112–119. | ||
| How to Cite this Article |
| Pubmed Style Truong AD, Ly DV, Vu TH, Hoang VT, Nguyen TC, Chu TN, Nguyen HT, Nguyen TV, Pham NT, . Unexpected cases in field diagnosis of African swine fever virus in Vietnam: The needs consideration when performing molecular diagnostic tests. Open Vet. J.. 2020; 10(2): 189-197. doi:10.4314/ovj.v10i2.8 Web Style Truong AD, Ly DV, Vu TH, Hoang VT, Nguyen TC, Chu TN, Nguyen HT, Nguyen TV, Pham NT, . Unexpected cases in field diagnosis of African swine fever virus in Vietnam: The needs consideration when performing molecular diagnostic tests. https://www.openveterinaryjournal.com/?mno=76785 [Access: January 25, 2026]. doi:10.4314/ovj.v10i2.8 AMA (American Medical Association) Style Truong AD, Ly DV, Vu TH, Hoang VT, Nguyen TC, Chu TN, Nguyen HT, Nguyen TV, Pham NT, . Unexpected cases in field diagnosis of African swine fever virus in Vietnam: The needs consideration when performing molecular diagnostic tests. Open Vet. J.. 2020; 10(2): 189-197. doi:10.4314/ovj.v10i2.8 Vancouver/ICMJE Style Truong AD, Ly DV, Vu TH, Hoang VT, Nguyen TC, Chu TN, Nguyen HT, Nguyen TV, Pham NT, . Unexpected cases in field diagnosis of African swine fever virus in Vietnam: The needs consideration when performing molecular diagnostic tests. Open Vet. J.. (2020), [cited January 25, 2026]; 10(2): 189-197. doi:10.4314/ovj.v10i2.8 Harvard Style Truong, A. D., Ly, . D. V., Vu, . T. H., Hoang, . V. T., Nguyen, . T. C., Chu, . T. N., Nguyen, . H. T., Nguyen, . T. V., Pham, . N. T. & (2020) Unexpected cases in field diagnosis of African swine fever virus in Vietnam: The needs consideration when performing molecular diagnostic tests. Open Vet. J., 10 (2), 189-197. doi:10.4314/ovj.v10i2.8 Turabian Style Truong, Anh Duc, Duc Viet Ly, Thi Hao Vu, Van Tuan Hoang, Thi Chinh Nguyen, Thi Nhu Chu, Huyen Thi Nguyen, The Vinh Nguyen, Ngoc Thi Pham, and Ha Thi Thanh Tran And Hoang Vu Dang. 2020. Unexpected cases in field diagnosis of African swine fever virus in Vietnam: The needs consideration when performing molecular diagnostic tests. Open Veterinary Journal, 10 (2), 189-197. doi:10.4314/ovj.v10i2.8 Chicago Style Truong, Anh Duc, Duc Viet Ly, Thi Hao Vu, Van Tuan Hoang, Thi Chinh Nguyen, Thi Nhu Chu, Huyen Thi Nguyen, The Vinh Nguyen, Ngoc Thi Pham, and Ha Thi Thanh Tran And Hoang Vu Dang. "Unexpected cases in field diagnosis of African swine fever virus in Vietnam: The needs consideration when performing molecular diagnostic tests." Open Veterinary Journal 10 (2020), 189-197. doi:10.4314/ovj.v10i2.8 MLA (The Modern Language Association) Style Truong, Anh Duc, Duc Viet Ly, Thi Hao Vu, Van Tuan Hoang, Thi Chinh Nguyen, Thi Nhu Chu, Huyen Thi Nguyen, The Vinh Nguyen, Ngoc Thi Pham, and Ha Thi Thanh Tran And Hoang Vu Dang. "Unexpected cases in field diagnosis of African swine fever virus in Vietnam: The needs consideration when performing molecular diagnostic tests." Open Veterinary Journal 10.2 (2020), 189-197. Print. doi:10.4314/ovj.v10i2.8 APA (American Psychological Association) Style Truong, A. D., Ly, . D. V., Vu, . T. H., Hoang, . V. T., Nguyen, . T. C., Chu, . T. N., Nguyen, . H. T., Nguyen, . T. V., Pham, . N. T. & (2020) Unexpected cases in field diagnosis of African swine fever virus in Vietnam: The needs consideration when performing molecular diagnostic tests. Open Veterinary Journal, 10 (2), 189-197. doi:10.4314/ovj.v10i2.8 |