| Original Article | ||
Open Vet J. 2023; 13(1): 26-41 Open Veterinary Journal, (2023), Vol. 13(1): 26–41 Original Research Molecular genotyping, histopathological and immunohistochemical studies of bovine papillomatosisHasanain A. J. Gharban1, Sattar J. J. AL-Shaeli2* and Talal Jabal Hussen21Department of Internal and Preventive Veterinary Medicine, College of Veterinary Medicine, University of Wasit, Kut, Iraq 2Department of Medical Basic Sciences, College of Dentistry, University of Wasit, Kut, Iraq Submitted: 19/09/2022 Accepted: 10/12/2022 Published: 07/01/2023 *Corresponding Author: Sattar J. J. AL-Shaeli. Department of Medical Basic Sciences, College of Dentistry, University of Wasit, Kut, Iraq. Email: salshaeli [at] uowasit.edu.iq © 2023 Open Veterinary Journal
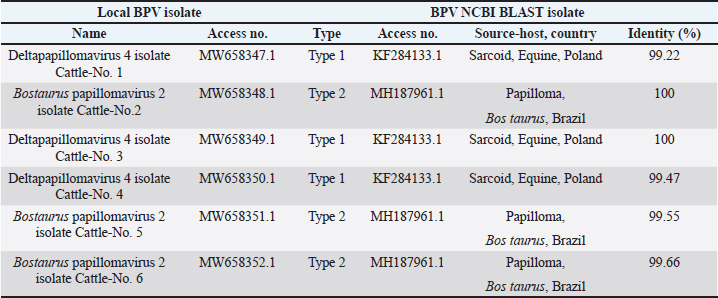
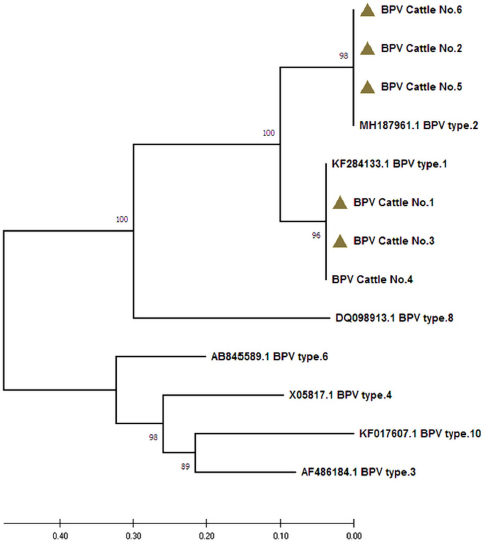
AbstractBackground: Bovine papillomatosis (BP) is considered the most common health problem in large cattle farms. Aim: This study attempts to confirm clinically suspected BP in cattle by polymerase chain reaction (PCR) assay, histopathology, immunohistochemistry (IHC), and genotyping analysis of local isolates. Methods: According to morphological appearance and lesion features, a cross sectional study of 54 clinically diagnosed BP cattle was assigned to this current investigation from May to August (2021) in Al-Kut district (Wasit Province, Iraq) private veterinary clinics using purposive sampling technique based on set criteria. The cattle were diagnosed clinically, and the tissues were collected and some fixed in 10% neutral buffered formalin and other stored frozen and examined by histopathological technique, IHC, and PCR assays. Results: Using PCR assay, all cattle were positive for the BPV L1 gene. According to detect the L1 gene, analysis of the phylogenetic tree showed that local BPV cattle isolates were closely related to the NCBI-BLAST BPV type-1 and type-2 of the Polish equine isolate (KF284133.1) and BPV Brazilian Bostaurus isolate (MH187961.1), respectively. Histological detection showed there were acanthosis, hyperkeratosis, epidermal thickening, severe infiltration of mononuclear cells, massive hemorrhage, dermal fibroplasias, multifocal spongiosis, moderate neovascularization, moderate to severe elongation of the retention ridge towards the dermis, parakeratosis, rings of calcification, and necrosis with nuclear pyknosis of some spinosum cells. Immunohistochemical findings of tumor necrosis factor-alpha, epidermal growth factor receptor and Fascin showed a significant variation in values of immunoreaction in the dermis and epidermis. These results ranged from negative (0) to mild positive (+1) to moderate positive (+2) reactions. Conclusion: The study provided essential molecular and genotyping data to improve our knowledge by emphasizing the crucial of IHC as an elegant diagnostic method to detect cellular alterations. Keywords: Cattle warts, Iraq, Papillomavirus, PCR, Sequence. IntroductionBovine papillomatosis (BP) is a common skin-specific disease of cattle that is promoted by a paraphyletic group of circular double-stranded DNA viruses, bovine papillomaviruses (BPV). The latter belongs to the Papillomaviridae family which represents the oldest and widest family of viruses (Abouelkhair and Kennedy, 2022). There are several distinct BPVs in cattle that are classified based on their site and type of lesion including BPV-1 (Mathewos et al., 2021), BPV-2 (Mathewos et al., 2021), BPV-3 (Pfister et al., 1979), BPV-4 (Campo et al., 1980), BPV-5 (Campo et al., 1981), BPV6 (Jarrett et al., 1984) and BPV-7 (Ogawa et al., 2007). The information of transmission method of BP between animals is limited due to unclear transmission mechanism (Pang et al., 2019). However, the animal populations in restricted spaces are more vulnerable and susceptible to infection due to the direct and indirect virus-spreading behavior (Ugochukwu et al., 2019). The possible mechanisms of transmission are vertical spreading, arthropod vectors, and direct skin contact (Roperto et al., 2019; Ata et al., 2021). The method of BPV spreads through the blood is attracting attention to extensively study the possible methods of transmission which can occur via non-epithelium tissues (Savini et al., 2019) and fluids (Meng et al., 2021). In adults, BPVs are unable to penetrate the host skin; thus, minimum abrasions in the skin are required to initiate the infection. Exposure of skin lesions to BPV leads to the initiation of infection followed by transformation and proliferation of infected epithelium’s basal cells, which develop into benign papilloma or fibropapilloma (Mathewos et al., 2021). In the field, there are different types of papillomatosis. Cutaneous papilloma is the most prevalent type among cattle and displays distinct morphologies, such as the atypical filiform and typical pedunculate (cauliflower) forms that present the verrucous aspect (Constable et al., 2017; Daudt et al., 2018). Although the clinical diagnosis of BP is usually performed when alterations are well characterized in the epidermis, histology and immunohistochemistry (IHC) for stained papilloma tissues can allow an identification of epidermal pathogenic alterations and reveal viral proteins, respectively (Russo et al., 2020; Hassanien et al., 2021). Molecular techniques, in particular polymerase chain reaction (PCR), are important and essential diagnostic tools that are usually applied to confirm infection through the detection of specific DNA of BPV (Emin et al., 2022). Due to the validity and accuracy of PCR test, applying this method can lead to exploring valuable insights into the characteristics of genomic data (Kiselev et al., 2020; Kubacki et al., 2021). Worldwide, the disease is undoubtedly one of the most widely studied due to its significant effects on the veterinary sector. In Iraq, available data remain limited and need to be elucidated because the prevalence of disease has still developed greatly in the last 10 years (Hamad et al., 2017; Mansour et al., 2019; Al-Salihi et al., 2020). Therefore, the current study aimed to confirm clinically suspected BP cattle by using PCR assays, histology, and IHC, with genotyping of local isolates to be documented in the National Center for Biotechnology Information (NCBI). Materials and MethodsStudy animals, design, and topographyA gross sectional study was performed using 54 cattle arrived at the private veterinary clinics in Al-Kut district (Wasit province, Iraq) from May to August (2021). The topography of the current study is including all regions around Al-Kut district in addition to Al-Kut center. The animals were diagnosed clinically to be infected with papillomatosis based on morphological appearance and features of lesions. The set criteria that allowed only the cattle with obvious lesions and clinically diagnosed to employ in this study using purposive sampling technique. Sample collectionAfter injection of lidocaine 5% (Cat No.# N01BB02, ADVANZ, UK) around each lesion, surgical removal of papillomas using a scalpel was performed under aseptic conditions. The collected samples of each animal were divided into two parts; one was kept into a plastic container containing 10% neutral buffered formalin for histology/IHC examination, while the second part was kept into a plastic tube and kept frozen for molecular examination. Molecular genotypingAccording to protocol (B) of G-spinTM, DNAs were extracted from tissue samples using the total DNA Extraction Kit (Cat.No.#IBT-QMS-GT1704, Intron Biotechnology, South Korea). The DNA purity and concentration were measured using a Nanodrop system (Thermo-Scientific, UK). Targeting the conserved region of BPV L1 gene (nt 7250 to 3225) with GenBank access number (KF284133.1), one set of primers was designed to amplify the region [F (5´-CAGTGTCTATCGGGGCCAAA-3´) and R (5´-AATTCAAGAGGAGGGCAAGGC-3´)] with 53.8°C annealing temperature, manufactured by Scientific Researcher Co. (Iraq). The PCR Premix Kit (Cat No.# 162770, Bioneer, South Korea) was used to prepare the mastermix with forwards and reverse gene sat a 20 μl final volume. PCR reaction was performed in thermocycler (Bio-Rad, USA) under the following conditions of: 1 cycle predenaturation (95°C/5 minutes), 35 cycles of denaturation (95°C/30 seconds), annealing (58°C/30 seconds) and extension (72°C/1 minute), and 1 cycle final extension (72°C/5 minutes). The electrophoresis of agarose-gel (1.5%) stained with Ethidium bromide was applied for the resulting PCR products, and then examined under a UV-transilluminator (Wised, South Korea). The samples were considered positive at 409 bp. For genotyping, six positive PCR products were sent to the Macrogen Company (Seoul, South Korea) to be sequenced by the modified Sanger method. Fasta data of DNA sequences were subjected to MEGA software and the UPGMA program for phylogenetic tree analysis and multiple sequence alignment analysis. Finally, all the analyzed local isolates were named and documented in NCBI GenBank to obtain specific access numbers. HistologyFollowing formalin fixation, tissues were exposed to ascending grades of ethanol for dehydration, followed by xylene for clearing, and exposure to paraffin for infiltration, embedding, and blocking. The block was sectioned using the Ultra-Thin Semiautomatic Rotary microtome (MRS3500, Histo-line, Italy) at a thickness of 4–5 μm and mounted on a slide. All prepared slides were stained with the hematoxylin and eosin (Cat. No.# ab245880, Abcam, India), and examined by a trinocular light microscope (MEIJI, Japan) at X10 and X40. IHCEnvision FLEX IHC kits (Cat. No.# 126522-001, Dako, Denmark) were used to detect tumor necrosis factor-alpha (TNF-α), epidermal growth factor receptor (EGFR), and Fascin. Following the manufacturers’ instructions, paraffin-embedded tissues were mounted on positively charged glass, deparaffinized by xylene, rinsed with distilled water and TBS, incubated with antigen retrieval solution at 60°C and then incubated in a water bath at 97°C for 25 minutes. The tissues were flooded with peroxidase block solution for 10 minutes and then with the anti- (TNF-α, EGFR and FASCIN) primary and secondary antibodies, followed by incubation with freshly prepared chromogen for 10 minutes. Then, the tissues were exposed to a counter stain (Mayer’s hematoxylin) for 3 minutes. After dehydration with three ascending ethanol concentrations, the slides were immersed in xylene, mounted with DPX, covered with cover slips and examined under a light microscope at ×10 and ×40. The results of IHC were classified based on their reactions as negative (−), mild positive (+1), moderate positive (+2), and strong positive (+3). Statistical analysisThe obtained data was documented and managed using excel sheet (Microsoft excel, 2016). One-way analysis of variance was applied to analyze the IHC data by using GraphPad Prism (version 6.0.1) Software (GraphPad Software Inc., USA). The differences at p < 0.05 were considered significant among their values. Ethical approvalThe current study was approved by the Scientific Committees of both Colleges of Veterinary Medicine and Dentistry at the University of Wasit (Wasit Province, Iraq) as the work is under their guidelines. ResultsPCR identification and phylogenic tree recordingMolecular assessment by conventional PCR of all clinically suspected BP lesions (Fig. 1) revealed that all the studied cattle (n=54) were positive for the BPV L1 gene (Fig. 2). Analysis of the phylogenetic tree according to derive the L1 gene alignments identified that the local BPV cattle isolates; Deltapapillomavirus 4 isolate Cattle-No. 1, Deltapapillomavirus 4 isolate Cattle-No. 3 and Deltapapillomavirus 4 isolate Cattle-No. 4 were closely related with NCBI-BLAST BPV type-1 (KF284133.1). Local BPV cattle isolates Bostaurus papillomavirus 2 isolate Cattle-No. 2, Bostaurus papillomavirus 2 isolate Cattle-No. 5, and Bostaurus papillomavirus 2 isolate Cattle-No. 6 were closely related with NCBI-BLAST BPV type-2 (MH187961.1) (Illustrations 1–6). NCBI-BLAST homology sequence analysis recorded a highly identity of the local Deltapapillomavirus 4 isolate Cattle-No. 1, No. 3 and No. 4 with the GenBank-NCBI BPV Polish equine isolate (KF284133.1) with 99.22%, 100% and 99.47% identity, respectively. However, the local Bostaurus papillomavirus 2 isolate Cattle-No. 2, No. 5 and No. 6 were more identical to the GenBank-NCBI BPV Brazilian Bostaurus isolate (MH187961.1) with 100%, 99.55% and 99.66% identity, respectively (Table 1, Fig. 3).
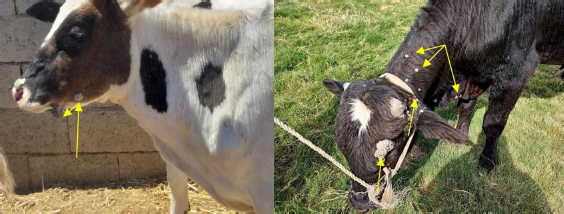
Fig. 1. Left lateral view of studied cattle clinically diagnosed with BP.
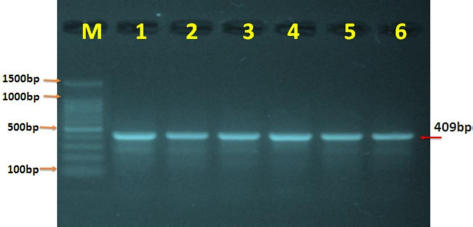
Fig. 2. PCR expression of the BPV L1 gene in agarose gel electrophoresis. M: ladder marker (1,500–100 bp); lanes 1–6: representative positive PCR samples at 409 bp.
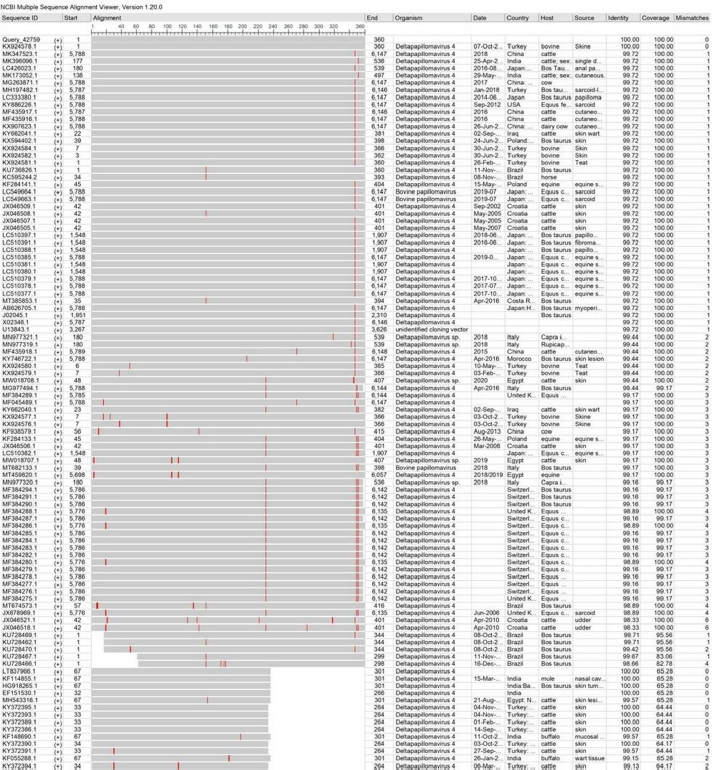
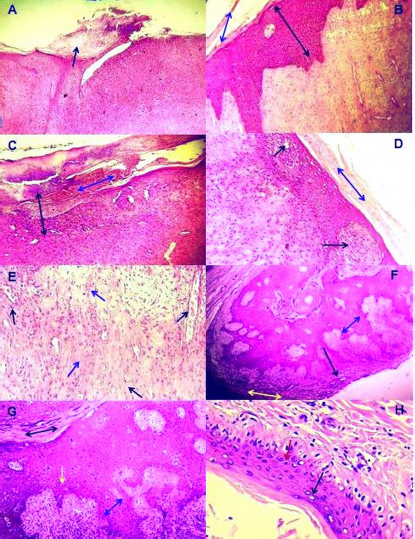
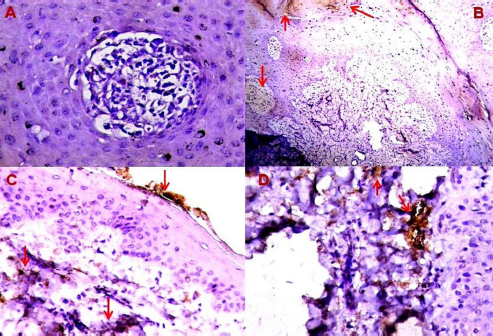
Illustration 1. NCBI multiple sequence alignment for local isolate 1 (MW658347.1). Histological determinationHistological investigation of papilloma under light microscopy showed mature finger-like projection papillae with grown rete pegs expressed in the stratum corneum of the skin epidermis (Fig. 4A). Thickening of the epidermis due to diffuse hyperplasia in stratum spinosum layer (acanthosis) with hyperkeratosis was observed (Fig. 4B). Furthermore, severe infiltration of mononuclear cells (MNCs) mainly macrophages, lymphocytes and fibroblasts, was observed in epidermis and dermis with massive hemorrhage in the epidermal layer in addition to dermal fibroplasias (Fig. 4C). Moreover, multifocal spongiosis and hyperkeratosis (Fig. 4D), with moderate neovascularization and fibroplasias due to fibrous connective proliferation (Fig. 4E), were observed. In addition, there was marked acanthosis with moderate elongation of the retention ridge towards the dermis that showed dense fibrous tissue with infiltration of inflammatory cells (Fig. 4F). Hyperkeratosis, parakeratosis, severe elongation of the retention ridge towards the dermis and signs of calcification observed on the epidermal surface (Fig. 4G). Finally, necrosis in the epidermal layer, mainly in basal cells with nuclear pyknosis of some spinosum cells, was observed (Fig. 4H).
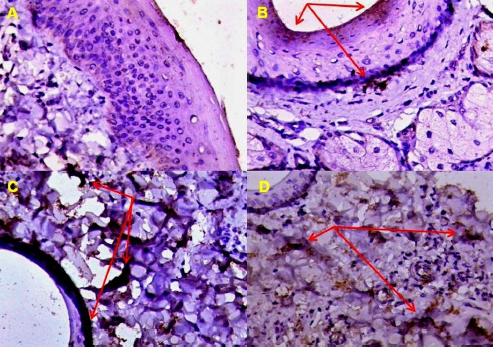
Illustration 2. NCBI multiple sequence alignment for local isolate 2 (MW658348.1). Immunohistochemical detectionIn the present study, the expression of TNF-α, EGFR and FASCIN markers was targeted using IHC. The results of the immunoreactions interestingly displayed significant variation among the examined skin layers (dermis and epidermis), which ranged from negative (0) to mild positive (+1) to moderate positive (+2) reactions (Figs. 5–7).
Illustration 3. NCBI multiple sequence alignment for local isolate 3 (MW658349.1). DiscussionBP is a highly prevalent skin disease of cattle in many countries worldwide including Iraq. However, the global distribution of other types of BPVs still needs to be established. The molecular detection in the current study revealed that all clinically suspected cattle were diagnosed with BV; while genotyping analysis confirmed that the local isolates were closely related to Type 1 and Type 2 BPV. In India, a specific molecular study was performed on normal and BP skin lesions in cattle. The results identified that the normal and BP skin lesions were infected with diagnosed type 1 and 2 BPV, which suggested a wide distribution of the virus that caused a high incidence of the disease (Pangty et al., 2010). Several studies conducted on cattle in the same field showed various spreading of papillomaviruses (PVs) in cattle with clear symptoms and asymptomatic cattle. These results suggested that the cattle without symptoms have a crucial role in spreading the disease, as the virus is endemic (Ataseven et al., 2016; Wassilew, 2018; Kwok et al., 2020).
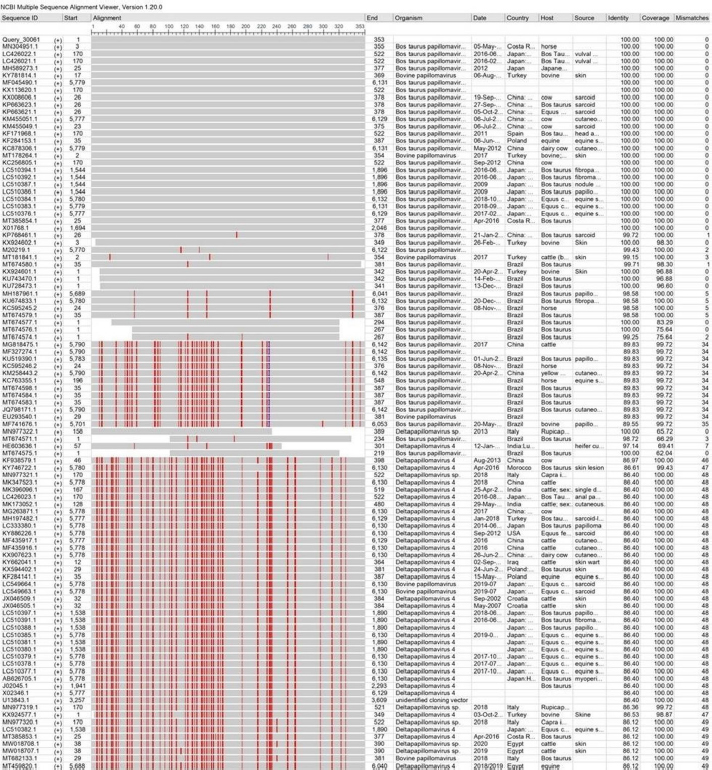
Illustration 4. NCBI multiple sequence alignment for local isolate 4 (MW658350.1). PCR assays, which are important and successful diagnostic methods, have high specificity and sensitivity for detecting agents and pathogens (MacLachlan et al., 2017; Dörttaş and Dağalp, 2020). The conserved region of the L1 gene of PV was used in this study to detect BPVs by PCR (Melo et al., 2014; Ataseven et al., 2016). PVs are classically used in the classification and construction of phylogenetic trees according to biological and medical characterization and sequence alignment of L1 nucleotides (Bocaneti et al., 2016). Our genotyping findings showed that the local isolates were closely related to Type 1 and Type 2 BPV, which is in line with the results of several worldwide studies (Pangty et al., 2010; Bam et al., 2013; Grindatto et al., 2015). Other studies reported that BPV- 1 and BPV2 have the ability to infect buffaloes, as well as other species, such as equines inducing fibropapillomas and fibroplastic tumors (Silvestre et al., 2009). In this study, although Type 2 BPV was detected genotypically in some local isolates, no one of these infected animals showed bladder cancer clinical signs, such as enzootic hematuria. This could be due to a lack of Pteridium aquilinum bracken fernsin in the fields of the study animals.
Illustration 5. NCBI multiple sequence alignment for local isolate 5 (MW658351.1). Histological analysis of stained papillomas indicated the presence of tissue changes suggesting cytopathic action of BP. Acanthosis was the most microscopic skin alteration observed in stained biopsies, which might be due to the mitogenic role of BPVs. Araldi et al. (2015) determined that the epithelium and connective tissues are directly affected by BPV, and chromosomal alterations and DNA breaks were seen incomet assay, suggesting cellular hyperactivity of the virus. However, Özsoy et al. (2011) identified epidermal hyperplasia and dermal irregular papillary projection to various degrees in animals infected with BP. Grindatto et al. (2015) demonstrated that there were no connections between the involved viral types and histopathological findings.
Illustration 6. NCBI multiple sequence alignment for local isolate 6 (MW658352.1). In the current study, mild to moderate positive immunoreactions were observed in the tissues assigned to specific immunohistochemical staining. El-Mandrawy and Alam (2018) identified that infected cells were more susceptible to TNF stimulation by releasing various cytokines and inducing inflammation in various tissues at the viral stage of the disease. Başbuğ et al. (2016) suggested that high levels of TNF-α may be involved in the assessment of the clinical course of the disease. Goodman et al. (2011) determined that the combination of TNF and recombinant vaccine based on virus protein rapidly reduced infection; however, the overall picture obtained during anti-TNF treatment of infected animals is unclear and may reveal some differences. It has been suggested that the convenient support for TNF antiviral effects comes from the fact that the family of viruses contains a variety of factors that regulate TNF-related activities. Many poxviruses encode soluble versions of TNF receptors and, more importantly, disrupt the viral TNFR gene in a virus, leading to a reduction in virus virulence (Kerr et al., 2015; Alvarez-de Miranda et al., 2021). Table 1. Identity (%) sequencing between local BPV isolates and NCBI-BLAST BPV isolates based on NCBI-BLAST homology.
Fig. 3. Analysis of phylogenetic tree according to the L1 gene in local BPV isolates. A UPGMA tree in MEGA (version 6.0) was applied. The local BPV isolates exhibited a close relationship to NCBI-BLAST isolates in terms of total genetic changes (0.4%–0.10%).
Fig. 4. Histopathological changes in skin lesions infected with BP. (A): Mature finger like projection papillae with rete pegs which grown in the stratum corneum in the epidermis of skin (×400); (B): Thickening of epidermis due to diffuse hyperplasia in the stratum spinosum layer (Black dual arrow) with hyperkeratosis (Blue dual arrow) (×100); (C): Severe infiltration of MNCs in epidermis and dermis of skin (Black dual arrow) with massive hemorrhage in epidermal layer (Blue dual arrow) in addition to dermal fibroplasia (×100); (D): Multifocal spongiosis (Black arrow) with hyperkeratosis (Blue dual arrow), (×200); (E): Dermis with moderate neovascularization (Black arrow) and fibroplasia due to fibrous connective proliferation (Blue arrow), (×200); (F): Marked acanthosis (Yellow dual arrow) with moderate elongation of retention ridge towards dermis (Blue dual arrow) and rings of calcification observed in epidermal surface (Black arrow), (×100); (G): Hyperkeratosis, parakeratosis, severe elongation of retention ridge towards dermis, acanthosis (Blue dual arrow) and infiltration of inflammatory cells (Yellow arrow), (×200); (H): Necrotic finding in epidermal layer mainly in basal cells (Black arrow) with nuclear pyknosis of some spinosum cells (Blue arrow), (×400). All sections were stained with H&E, and the images represent all study animals. Growth factors are crucial for the growth, development and homeostasis of almost all organisms and are required for tissue specialization, apoptosis, cell survival and viability, and fate determination (Mukai et al., 2018; Koons and Mikos, 2019). Growth factor receptors transmit extracellular signals by activating intracellular messengers or by transporting the receptors directly to the nucleus (Liu et al., 2020). EGFR is a specific factor receptor of transmembrane proteins that share structural and functional similarities (Nagano et al., 2018). Although, EGFR is a key indicator involved in predicting the diseases and their prognostic value, however, the EGFR-IHC role is still unclear. Several studies have reported that reduced signals of EGFR and other tyrosine kinase receptors are associated with disease, while increased activity is associated with the development of various tumors (Lian et al., 2019; Xia et al., 2020; Hong et al., 2022). Suppression of binding sites of EGFR signaling at the level of extracellular receptor or tyrosine kinase intracellular activity may inhibit EGFR expression networks and thus improve patient outcomes (Kitamura et al., 2020; Wingert et al., 2021). Several authors have suggested that the increased expression of EGFR has negative impacts (Kwon et al., 2018; Hashmi et al., 2019; London and Gallo, 2020), while others maintain the opposite (Pidugu et al., 2019; Feng et al., 2020; Lamtha et al., 2022). However, meta-analysis data summarizing the results of various studies show that exposure to EGFR is not an independent predictor of survival in various diseases (Yang et al., 2019; Nastały et al., 2020; Atef et al., 2021).
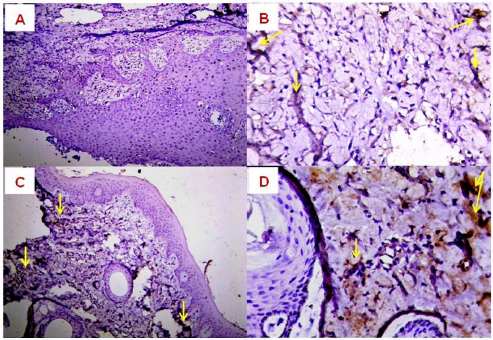
Fig. 5. Expression of TNF-α in BP skin lesions using IHC with Mayer’s hematoxylin. (A): 0 (×100); (B): +1 (×400); (C): +2 (×200); (D): +2 (×400) reactions.
Fig. 6. Expression of EGFR in BP skin lesions using IHC with Mayer’s hematoxylin. (A): 0 (×100); (B): +1 (×200); (C): +1 (×400); (D): +2 (×200) reactions. Fascin represents a family of active binding proteins that are localized in stress fibers and throughout the proliferation of various cells, such as myoblasts and glioma cells (Ogunlade et al., 2020). This protein is unique to muscle and cell types, such as glial cells, neurons, antigen-presenting dendritic cells, and endothelial cells (Zhang et al., 2022). This observation suggested that Fascin plays a role in cell movement or interaction with other cell types (Chung et al., 2022).
Fig. 7. Expression of Fascin in BP skin lesions using IHC with Mayer’s hematoxylin. (A): 0 (×400); (B): +1 (×100); (C): +1 (×400); (D): +2 (×400) reactions. ConclusionInterestingly, the BPVs exhibited a cytopathic role that interacted with cellular structure and function and promoted cytological and histomorphological changes; however, the number of available studies remains low. Thus, the current study highlights the interesting essential data that provide insight into mechanisms to acknowledge the possible oncogenesis and pathogenesis of BPV. Furthermore, the study emphasized that IHC is a specific crucial diagnostic tool that provides precise detection of the cellular and intracellular distribution of viral and antigenic proteins using specific antibodies. Moreover, targeting other PV types in cattle and other animal species, disease prevalence, animal breeding biosafety evaluation and fundamental vaccine roles and production are of great interest. AcknowledgementsThe authors gratefully acknowledge Lect. Dr. Ali H.M. Al-Shraify for active help throughout the collection of required information about study animals. Authors’ contributionHAJG: Clinical examination of study animals, collection of papilloma samples, molecular examination and documentation of local isolates in the NCBI. SJJA: Histological and immunohistochemical processing of papilloma tissue samples, and created stained slides. TJH: Histopathological and immunohistochemical examination and reading of the slides by light microscopy. Data analysis was performed by both HAJG and SJJA. The authors have read and approved the final version of this manuscript as they participated in the writing of the manuscript. FundingThe authors provided fund from themselves with no external funds. Conflict of interestThe authors declare that there is no conflict of interest. ReferencesAbouelkhair, M.A. and Kennedy, M. 2022. Papillomaviridae and Polyomaviridae. Vet. Microbiol. 4, 484–488. Al-Salihi, K.A., Al-Dabhawi, A.H., Ajeel, A.A., Erzuki, I.A. and Ali, T.A.H. 2020. Clinico-histopathological and immunohistochemical study of ruminant’s cutaneous papillomavirus in Iraq. Vet. Med. Int. 2020, 1–11. Alvarez-de Miranda, F.J., Alonso-Sánchez, I., Alcamí, A. and Hernaez, B. 2021. TNF decoy receptors encoded by poxviruses. Pathogens. 10, 1065–1079. Araldi, R.P., Melo, T.C., Neves, A.C., Spadacci-Morena, D.D., Magnelli, R.F., Modolo, D.G., de-Sá-Júnior, P.L., Mazucchelli-de-Souza, J., Carvalho, R.F., Beçak, W. and Stocco, RC. 2015. Hyperproliferative action of bovine papillomavirus: genetic and histopathological aspects. Genet. Mol. Res. 14, 12942–12954. Ata, E.B., Allam, A.M., Elbayoumy, M.K. and Mahmoud, M.A.E.F. 2021. Electron microscopy and phylogenetic analysis of bovine papillomavirus infection in cattle from four Egyptian governorates. Trop. Anim. Health Prod. 53, 1–7. Ataseven, V.S., Kanat, Ö. And Ergün, Y. 2016. Molecular identification of bovine papillomaviruses in dairy and beef cattle: first description of Xi-and Epsilonpapillomavirus in Turkey. Turk. J. Vet. Anim. Sci. 40, 757–763. Atef, M.M., Amer, A.I., Hafez, Y.M., Elsebaey, M.A., Saber, S.A. and Abd El-Khalik, S.R. 2021. Long non-coding RNA EGFR-AS1 in colorectal cancer: potential role in tumorigenesis and survival via miRNA-133b sponge and EGFR/STAT3 axis regulation. Br. J. Biomed. Sci. 78(3), 122–129. Bam, J., Kumar, P., Leishangthem, G. D., Saikia, A. and Somvanshi, R. 2013. Spontaneous cutaneous papillomatosis in yaks and detection and quantification of bovine papillomavirus-1 and-2. Transbound. Emerg. Dis. 60, 475–480. Başbuğ, O., Zahid, T.A. and Tuzcu, N. 2016. Tumour necrosis factor-alpha, haptoglobin, serum amyloid A and neopterin levels in cattle with lumpy skin disease. Kafkas. Üniv. Vet. Fak. Derg. 22, 417–424. Bocaneti, F., Altamura, G., Corteggio, A., Velescu, E., Roperto, F. and Borzacchiello, G. 2016. Bovine papillomavirus: new insights into an old disease. Transbound. Emerg. Dis. 63, 14–23. Campo, M.S., Moar, M.H., Jarrett, W.F. and Laird, H.M. 1980. A new papillomavirus associated with alimentary cancer in cattle. Nature 286, 180–182. Campo, M.S., Moar, M.H., Laird, H.M. and Jarrett, W.F. 1981. Molecular heterogeneity and lesion site specificity of cutaneous bovine papillomaviruses. Virology 113, 323–335. Chung, J.M., Sato, O., Ikebe, R., Lee, S., Ikebe, M. and Jung, H.S. 2022. Structural analysis of human fascin-1: essential protein for actin filaments bundling. Life. 12, 843–855. Constable, P.D., Hinchcliff, K.W., Done, S.H. and Gruenberg, W. 2017. A textbook of the diseases of cattle, horses, sheep, pigs, and goats, 11th ed. Elsevier, NY: Saunders, p: 1580. Daudt, C., Da Silva, F.R.C., Lunardi, M., Alves, C.B.D. T., Weber, M.N., Cibulski, S.P., Alfieri, A.F., Alfieri, A.A. and Canal, C.W. 2018. Papillomaviruses in ruminants: an update. Transbound. Emerg. Dis. 65 (5), 1381–1395. Dörttaş, S.D. and Dağalp, S.B. 2020. Veteriner hekimlikte papillomaviruslar veönemi. Atatürk Üniv. Vet. Bilim. Derg. 15, 91–99. El-Mandrawy, S.A. and Alam, R.T. 2018. Hematological, biochemical and oxidative stress studies of lumpy skin disease virus infection in cattle. J. Appl. Anim. Res. 46, 1073–1077. Emin, K., Nuvit, C., Serpil, D., Enver, B., Ataseven, VS., Volkan, Y., Fırat, D., Hilmi, N., Celal Sahin, E., Ugur, A., Mushap, K. and Ayfer, Y. 2022. Molecular detection of Papillomavirus and immunohistochemical investigation of p53 gene expressions in bovine papillomas and fibropapillomas. Arch. Microbiol. 204, 1–12. Feng, Z., Li, X., Qiu, M., Luo, R., Lin, J. and Liu, B. 2020. LncRNA EGFR-AS1 upregulates ROCK1 by sponging miR-145 to promote esophageal squamous cell carcinoma cell invasion and migration. Cancer Biother. Radiopharm. 35, 66–71. Goodman, A.G., Heinen, P.P., Guerra, S., Vijayan, A., Sorzano, C.O.S., Gomez, C.E. and Esteban, M. 2011. A human multi-epitope recombinant vaccinia virus as a universal T cell vaccine candidate against influenza virus. PLoS. One. 6, e25938. Grindatto, A., Ferraro, G., Varello, K., Crescio, M.I., Miceli, I., Bozzetta, E., Goria, M. and Nappi, R. 2015. Molecular and histological characterization of bovine papillomavirus in North West Italy. Vet. Microbiol. 180, 113–117. Hamad, M.A., Al-Shammari, A.M., Odisho, S.M. and Yaseen, N.Y. 2017. Molecular epidemiology of bovine papillomatosis and identification of three genotypes in central Iraq. Intervirology 60, 156–164. Hashmi, A.A., Naz, S., Hashmi, S.K., Irfan, M., Hussain, Z.F., Khan, E.Y., Asif, H. and Faridi, N. 2019. Epidermal growth factor receptor (EGFR) overexpression in triple-negative breast cancer: association with clinicopathologic features and prognostic parameters. Surg. Exp. Pathol. 2, 1–7. Hassanien, R.T., Hamdy, M.E., Elnomrosy, S.M., Hussein, H.A., Afify, A.F., Darwish, F.M., Shehab, G., Emran, R., Abd-El-Moniem, M.I.I., Habashi, AR., Fahmy, HA., Ibraheem, E.M., Shahein, M.A., Attya, M., Abdelhakim, A.M.M. and Hagag, N.M. 2021. Molecular characterization and pathological identification of a novel strain of delta papillomavirus-4 (bovine papillomavirus-2) in Egypt. Vet. World. 14, 2296–2305. Hong, D., Zhou, B., Zhang, B., Ren, H., Zhu, L., Zheng, G. and Ge, J. 2022. Recent advances in the development of EGFR degraders: PROTACs and LYTACs. Eur. J. Med. Chem. 12, 1257–1272. Jarrett, W.F.H., Campo, M.S., Oweil, B.W., Laird, H.M. and Coggins, L.W. 1984. A novel bovine papillomavirus (BPV-6) causing true epithelial papillomas of the mammary gland skin: a member of a proposed new BPV subgroup. Virology 136, 255–264. Kerr, P.J., Liu, J., Cattadori, I., Ghedin, E., Read, A.F. and Holmes, E.C. 2015. Myxoma virus and the Leporipoxviruses: an evolutionary paradigm. Viruses 7, 1020–1061. Kiselev, D., Matsvay, A., Abramov, I., Dedkov, V., Shipulin, G. and Khafizov, K. 2020. Current trends in diagnostics of viral infections of unknown etiology. Viruses 12, 211–239. Kitamura, S., Maeda, T. and Yanagi, T. 2020. Vandetanib inhibits cell growth in EGFR-expressing cutaneous squamous cell carcinoma. Biochem. Biophys. Res. Commun. 531, 396–401. Koons, G.L. and Mikos, A.G. 2019. Progress in three-dimensional printing with growth factors. J. Control. Release. 295, 50–59. Kubacki, J., Fraefel, C. and Bachofen, C. 2021. Implementation of next-generation sequencing for virus identification in veterinary diagnostic laboratories. J. Vet. Diagn. Invest. 33, 235–247. Kwok, K.T., Nieuwenhuijse, D.F., Phan, M.V. and Koopmans, M.P. 2020. Virus metagenomics in farm animals: a systematic review. Viruses 12, 107–129. Kwon, D., Koh, J., Kim, S., Go, H., Min, H.S., Kim, Y.A. and Chung, D.H. 2018. Overexpression of endoplasmic reticulum stress-related proteins, XBP1s and GRP78, predicts poor prognosis in pulmonary adenocarcinoma. Lung Cancer 122, 131–137. Lamtha, T., Krobthong, S., Yingchutrakul, Y., Samutrtai, P., Gerner, C., Tabtimmai, L. and Choowongkomon, K. 2022. A novel nanobody as therapeutics target for EGFR-positive colorectal cancer therapy: exploring the effects of the nanobody on SW480 cells using proteomics approach. Proteome Sci. 20, 1–11. Lian, G., Chen, S., Ouyang, M., Li, F., Chen, L. and Yang, J. 2019. Colon cancer cell secretes EGF to promote M2 polarization of TAM through EGFR/PI3K/AKT/mTOR pathway. Technol. Cancer Res. Treat. 18, 1–9. Liu, P.C., Koblish, H., Wu, L., Bowman, K., Diamond, S., DiMatteo, D. and Hollis, G. 2020. INCB054828 (pemigatinib), a potent and selective inhibitor of fibroblast growth factor receptors 1, 2, and 3, displays activity against genetically defined tumor models. PLoS. One. 15, e0231877. London, M. and Gallo, E. 2020. Epidermal growth factor receptor (EGFR) involvement in epithelial-derived cancers and its current antibody-based immunotherapies. Cell Biol. Int. 44, 1267–1282. MacLachlan, N.J., Dubovi, E.J., Barthold, S.W., Swayne, D.E. and Winton, J.R. 2017. Fenner’s Veterinary Virology, 5th edition. In Papillomaviridae and Polyomaviridae. Eds., Maclachlan, N.J. and Dubovi, E.J. San Diego, CA: Elsevier Inc, pp: 327–356. Mansour, K.A., Naser, H.H. and Hussain, M.H. 2019. Phylogenetic tree analysis study of bovine papillomaviruses type 1 based on L1 gene in Al-Qadisiyah governorate, Iraq. Iraqi J. Vet. Sci. 33, 151–155. Mathewos, M., Teshome, T., Fesseha, H. and Yirgalem, M. 2021. Cytopathogical characterization of papillomatosis in cattle of Wolaita Sodo district, Southern Ethiopia. Sci. Afr. 13, 1–9. Melo, T.C., Carvalho, R.F., Mazzucchelli-de-Souza, J., Diniz, N., Vasconcelos, S., Assaf, SL., Araldi, R.P., Ruiz, R.M., Kerkis, I., Beçak, W. and Stocco, R.C. 2014. Phylogenetic classification and clinical aspects of a new putative Deltapapillomavirus associated with skin lesions in cattle. Genet. Mol. Res. 13, 2458–2469. Meng, Q., Ning, C., Wang, L., Ren, Y., Li, J., Xiao, C., Li, Y., Li, Z., He, Z., Cai, X. and Qiao, J. 2021. Molecular detection and genetic diversity of bovine papillomavirus in dairy cows in Xinjiang, China. J. Vet. Sci. 22, e50–e59. Mukai, K., Tsai, M., Saito, H. and Galli, S.J. 2018. Mast cells as sources of cytokines, chemokines, and growth factors. Immunol. Rev. 282, 121–150. Nagano, T., Tachihara, M. and Nishimura, Y. 2018. Mechanism of resistance to epidermal growth factor receptor-tyrosine kinase inhibitors and a potential treatment strategy. Cells 7, 212–228. Nastały, P., Stoupiec, S., Popęda, M., Smentoch, J., Schlomm, T., Morrissey, C., Żaczek, AJ., Beyer, B., Tennstedt, P., Graefen, M., Eltze, E., Maiuri, P., Semjonow, A., Pantel, K., Brandt, B. and Bednarz-Knoll, N. 2020. EGFR as a stable marker of prostate cancer dissemination to bones. Br. J. Cancer 123, 1767–1774. Ogawa, T., Tomita, Y., Okada, M. and Shirasawa, H. 2007. Complete genome and phylogenetic position of bovine papillomavirus type 7. J. Gen. Virol. 88, 1934–1938. Ogunlade, B., Guidry, J.J., Mukerjee, S., Sriramula, S., Lazartigues, E. and Filipeanu, C.M. 2020. The actin bundling protein Fascin-1 as an ACE2-accessory protein. Cell. Mol. Neurobiol. 42, 255–263. Özsoy, Ş.Y., Özyildiz, Z. and Güzel, M. 2011. Clinical, pathological and immunohistochemical findings of bovine cutaneous papillomatosis. Ankara Üniv. Vet. Fakül. Derg. 58, 161–165. Pang, F., Zhang, M., Li, G., Zhang, Z., Huang, H., Li, B. and Wang, F. 2019. Integrated mRNA and miRNA profiling in NIH/3T3 cells in response to bovine papillomavirus E6 gene expression. Peer J. 7, e7442. Pangty, K., Singh, S., Goswami, R., Saikumar, G. and Somvanshi, R. 2010. Detection of BPV-1 and-2 and quantification of BPV-1 by real-time PCR in cutaneous warts in cattle and buffaloes. Transbound. Emerg. Dis. 57, 185–196. Pfister, H., Linz, U., Gissmann, L., Huchthausen, B., Hoffmann, D. and ZurHausen, H. 1979. Partial characterization of a new type of bovine papilloma viruses. Virology 96, 1–8. Pidugu, V.K., Wu, M.M., Yen, A.H., Pidugu, H.B., Chang, K.W., Liu, C.J. and Lee, T.C. 2019. IFIT1 and IFIT3 promote oral squamous cell carcinoma metastasis and contribute to the anti-tumor effect of gefitinib via enhancing p-EGFR recycling. Oncogene 38, 3232–3247. Roperto, S., Russo, V., De Falco, F., Taulescu, M. and Roperto, F. 2019. Congenital papillomavirus infection in cattle: evidence for transplacental transmission. Vet. Microbiol. 230, 95–100. Russo, V., Roperto, F., De Biase, D., Cerino, P., Urraro, C., Munday, J.S. and Roperto, S. 2020. Bovine papillomavirus type 2 infection associated with papillomatosis of the amniotic membrane in water buffaloes (Bubalus bubalis). Pathogens 9, 1–15. Savini, F., Gallina, L., Mazza, F., Mariella, J., Castagnetti, C. and Scagliarini, A. 2019. Molecular detection of bovine papillomavirus DNA in the placenta and blood of healthy mares and respective foals. Vet. Sci. 6, 1–5. Silvestre, O., Borzacchiello, G., Nava, D., Iovane, G., Russo, V., Vecchio, D. and Paciello, O. 2009. Bovine papillomavirus type 1 DNA and E5 oncoprotein expression in water buffalo fibropapillomas. Vet. Pathol. 46, 636–641. Ugochukwu, IC., Aneke, CI., Idoko, IS., Sani, NA., Amoche, AJ., Mshiela, WP., Ede, RE., Ibrahim, ND., Njoku, CI. and Sackey, AK. 2019. Bovine papilloma: Aetiology, pathology, immunology, disease status, diagnosis, control, prevention and treatment: a review. Comp. Clin. Pathol. 28, 737–745. Wassilew, S.W. 2018. Viral infections. In Pigmented ethnic skin and imported dermatoses. Cham, Switzerland: Springer, pp: 69–82. Wingert, S., Reusch, U., Knackmuss, S., Kluge, M., Damrat, M., Pahl, J., Schniegler-Mattox, U., Mueller, T., Fucek, I., Ellwanger, K., Tesar, M., Haneke, T., Koch, J., Treder, M., Fischer, W. and Rajkovic, E. 2021. Preclinical evaluation of AFM24, a novel CD16A-specific innate immune cell engager targeting EGFR-positive tumors. In MAbs. Taylor Francis (London, UK), vol. 13, p 1950264. Xia, H., Xue, X., Ding, H., Ou, Q., Wu, X., Nagasaka, M. and Ou, S.H.I. 2020. Evidence of NTRK1 fusion as resistance mechanism to EGFR TKI in EGFR+ NSCLC: results from a large-scale survey of NTRK1 fusions in Chinese patients with lung cancer. Clin. Lung Cancer 21, 247–254. Yang, C.C., Lin, L.C., Lin, Y.W., Tian, Y.F., Lin, C.Y., Sheu, M.J. and Tai, M.H. 2019. Higher nuclear EGFR expression is a better predictor of survival in rectal cancer patients following neoadjuvant chemoradiotherapy than cytoplasmic EGFR expression. Oncol. Lett. 17, 1551–1558. Zhang, Z.D., Wen, B., Li, D.J., Deng, D.X., Wu, X.D., Cheng, Y.W. and Li, E.M. 2022. AKT serine / threonine kinase 2-mediated phosphorylation of fascin threonine 403 regulates esophageal cancer progression. Int. J. Biochem. Cell Biol. 145, 106188. | ||
| How to Cite this Article |
| Pubmed Style Gharban H, AL-Shaeli SJ, Hussen TJ. Molecular genotyping, histopathological and immunohistochemical studies of bovine papillomatosis. Open Vet J. 2023; 13(1): 26-41. doi:10.5455/OVJ.2023.v13.i1.4 Web Style Gharban H, AL-Shaeli SJ, Hussen TJ. Molecular genotyping, histopathological and immunohistochemical studies of bovine papillomatosis. https://www.openveterinaryjournal.com/?mno=115559 [Access: July 27, 2024]. doi:10.5455/OVJ.2023.v13.i1.4 AMA (American Medical Association) Style Gharban H, AL-Shaeli SJ, Hussen TJ. Molecular genotyping, histopathological and immunohistochemical studies of bovine papillomatosis. Open Vet J. 2023; 13(1): 26-41. doi:10.5455/OVJ.2023.v13.i1.4 Vancouver/ICMJE Style Gharban H, AL-Shaeli SJ, Hussen TJ. Molecular genotyping, histopathological and immunohistochemical studies of bovine papillomatosis. Open Vet J. (2023), [cited July 27, 2024]; 13(1): 26-41. doi:10.5455/OVJ.2023.v13.i1.4 Harvard Style Gharban, H., AL-Shaeli, . S. J. & Hussen, . T. J. (2023) Molecular genotyping, histopathological and immunohistochemical studies of bovine papillomatosis. Open Vet J, 13 (1), 26-41. doi:10.5455/OVJ.2023.v13.i1.4 Turabian Style Gharban, Hasanain, Sattar Jabbar AL-Shaeli, and Talal Jabal Hussen. 2023. Molecular genotyping, histopathological and immunohistochemical studies of bovine papillomatosis. Open Veterinary Journal, 13 (1), 26-41. doi:10.5455/OVJ.2023.v13.i1.4 Chicago Style Gharban, Hasanain, Sattar Jabbar AL-Shaeli, and Talal Jabal Hussen. "Molecular genotyping, histopathological and immunohistochemical studies of bovine papillomatosis." Open Veterinary Journal 13 (2023), 26-41. doi:10.5455/OVJ.2023.v13.i1.4 MLA (The Modern Language Association) Style Gharban, Hasanain, Sattar Jabbar AL-Shaeli, and Talal Jabal Hussen. "Molecular genotyping, histopathological and immunohistochemical studies of bovine papillomatosis." Open Veterinary Journal 13.1 (2023), 26-41. Print. doi:10.5455/OVJ.2023.v13.i1.4 APA (American Psychological Association) Style Gharban, H., AL-Shaeli, . S. J. & Hussen, . T. J. (2023) Molecular genotyping, histopathological and immunohistochemical studies of bovine papillomatosis. Open Veterinary Journal, 13 (1), 26-41. doi:10.5455/OVJ.2023.v13.i1.4 |