| Research Article | ||
Open Vet J. 2023; 13(5): 515-522 Open Veterinary Journal, (2023), Vol. 13(5): 515–522 Original Research Serological and genetic diversity of hepatitis E virus among rabbits population in EgyptAhmed M. El-Adly*Botany and Microbiology Department, Faculty of Science, Al-Azhar University, Assiut, Egypt *Corresponding Author: Ahmed M. El-Adly. Botany and Microbiology Department, Faculty of Science, Al-Azhar University, Assiut, Egypt. Email: ahmedeladly.ast [at] azhar.edu.eg. Submitted: 25/04/2022 Accepted: 03/04/2023 Published: 03/05/2023 © 2023 Open Veterinary Journal
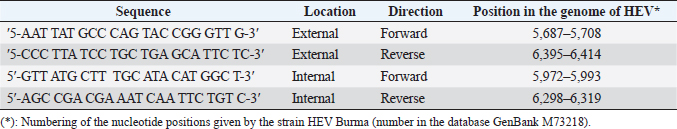
AbstractBackground: Hepatitis E virus (HEV) infection has a worldwide distribution and represents an important cause of acute hepatitis. Data on rabbit HEV prevalence and genetic diversity in hyperendemic regions (Egypt) are limited, per the information on rabbit HEV’s implications for human pathology. Aim: This study aimed to determine the prevalence of HEV infection in farmed rabbits from hyperendemic (Egypt) regions, as well as to examine the genetic relatedness of rabbit strains to human strains isolated in these regions. Methods: Anti-HEV was tested by ELISA from 164 serum samples isolated from rabbits in Egypt. HEV RNA was tested using reverse transcription of a nested polymerase chain reaction with degenerative primers to open reading frames 2 in feces samples from 355 farmed rabbits from Egypt (3 farms from different regions). Results: All of the animals were between the ages of 2 and 24 months. Age groups at various governorates, with the bulk of infections occurring between the ages of 2 and 12 months. HEV RNA prevalence in rabbits at the age between 2 and 12 months was varied in different governorates from 13.40%, 18.20%, and 32.10% in Qena, Luxor, and Assiut, respectively. While at the age between 12 and 24 months, HEV RNA prevalence in rabbits was 0.0%, 3.70%, and 4.30% in Assiut, Qena, and Luxor, respectively. Phylogenetic analysis did not reveal any relatedness of rabbit HEV strains neither to HEV genotype 3 sequences from patients with autochthonous hepatitis E in Egypt. Conclusion: HEV is prevalent in rabbits from Egypt with other rabbit strains belonging to species-specific group which is close to genotype 3. Keywords: HEV, Egypt, Rabbit, ELISA, Phylogenetic tree. IntroductionHepatitis E is a virus (HEV) that causes liver disease is found worldwide. In many developing countries, hepatitis E epidemics is responsible the disease, which is likewise endemic in few industrialized countries (Balayan et al., 1983; Sreenivasan et al., 1984). HEV belongs to the genus Hepevirus in the family Hepeviridae, is a small, icosahedral, spherical particles of 27–34 nm, non-enveloped virus with a single-stranded, positive-sense RNA genome of approximately 7.2 kb containing three open reading frames (ORFs), ORF1, ORF2 and ORF3, where ORF3 partially overlaps ORF2 (Fauquet et al., 2005). HEV divided into four genotypes with at least two putative new genotypes belong to only one serotype that can infect mammals, named after the place of isolation of the reference strains. Two of the genotypes contain strains that only infect people (HEV-1 “Burma,” HEV-2 “Mexico”); the other two genotypes (HEV-3 “USA,” HEV-4 “China”) compries viruses isolated from both human and animal samples (Worm et al., 2002). HEV has been genetically identified in rats, wild boars, domestic swine, mongooses, rabbits, chickens, ferrets, cutthroat trout, bats, and deer (Lhomme et al., 2013). HEV is considered hyperendemic in many developing countries, with a prevalence of up to 25%, acute hepatitis cases transmitted via the fecal-oral route and associated with contamination of drinking water by HEV genotypes 1 and 2. While, HEV is regarded endemic in industrialized countries, where the prevalence of HEV is less than 25% of acute hepatitis cases (Ashbolt, 2004). HEV is transmitted zoonotically by the consumption of contaminated meat and meat products or by contact with infected animals (Petral et al., 2013). Following the discovery of an HEV strain in a rabbit that belongs to genotype 3, rabbits were evaluated as a potential HEV animal model (Wang et al., 2002). Rabbits inoculated with the rabbit HEV shed virus in feces were viremic and exhibited increased serum ALT activity, as well as irregular multifocal lymphohistiocytic in filtrates and local hepatocellular necrosis (Prinja et al., 2008). Liu et al. (2013) reported that two of the nine rabbits inoculated with human HEV genotype 4 developed viremia and fecal virus shredding, whereas none of the rabbits inoculated with human HEV genotype 1 developed viremia or fecal virus shredding. Although rabbits may be of limited utility as a model for studies of human HEV genotypes 1 and 2, the rabbit model of HEV infection may be useful for evaluating some characteristics of HEV infection and replication of the rabbit HEV strain. Given the genetic identification of zoonotic genotype 3 HEV strains in rabbits from China, the United States, and France (Cossaboom et al., 2012a; Izopet et al., 2012; Birke et al., 2014), Rabbits could serve as reservoir hosts for the transfer of HEV to humans. Rabbits are susceptible to human HEV infection of genotype 4, and infected rabbits developed viremia, anti-HEV seroconversion, and fecal virus shredding (Liu et al., 2014). The rabbit HEV is closely related to other mammalian HEV both genetically and antigenically. The capsid protein of the HEV strain genotype 3 rabbit was capable of cross-reacting with antibodies from other HEV strains such as rat, swine, human, and chicken (Ma et al., 2010). Rabbit HEV, like swine HEV in pigs, is widely spread throughout the rabbit population. So, similar to swine HEV, direct contact with infected rabbits, consumption of undercooked or raw rabbit meats, and water contaminated with rabbit HEV could all be sources of human infection (Cossaboom et al., 2012b). A rabbit-HEV was discovered in a patient, implying that rabbit HEV can be transmitted to humans. The contribution of rabbit HEV to the epidemiology of hepatitis E in humans is uncertain (Wang et al., 2013). HEV is endemic in southwest France, with an annual incidence of 3.2% locally acquired HEV infections (Jeblaoui et al., 2013). Consumption of not farmed animal meats, primarily wild boar, deer, and wild rabbits, was revealed to be the only factor independently associated with HEV infection in a case-control investigation (Cheng et al., 2012). Molecular data from several studies carried out in France, on the other hand, show that the majority of HEV strains identified belong to genotypes 3f, 3c, or 3e, which are prevalent in pigs and wild boars (Takahashi and Okamoto, 2014). Materials and MethodsBlood and fecal sample collectionIn this study, 164 and 355 blood and feces samples, respectively, were collected from several governorates in Egypt, including 80 and 156 (Luxor governorate), 36 and 75 (Assiut governorate), 48 and 124 (Qena governorate) blood and fecal samples, respectively. Specimens were collected in January and April 2021. Rabbit serum and feces samples were transported in a sterile tube and container with a lid and a spoon. The samples were accompanied by paperwork indicating their age, collecting location, and date. Individual samples were taken from each animal. Blood samples were collected by cleaning the dorsal surface of the ear with 70%. The vein is normally occluded distally before the needle is inserted. Blood is collected from the tip of the ear using sterile needles and distributed to clean plastic, away from the base of the ear. Blood samples were centrifuged at 4,000 rpm for 10 minutes, and the supernatant was collected as a serum sample. All serum and feces samples were stored at -70°C until tested by an ELISA and polymerase chain reaction (PCR) assay following the manufacturer’s instruction. Serological detectionSerum markers for anti-HEV IgG Anti-HEV IgG was detected using third generation enzyme immunoassay (EIA) according to the manufacturer’s instructions (DIA.PRO, Milano, Italy). Since this set is designed to detecting of human anti-HEV IgG, species-specific conjugate in the formulation instead of using the conjugate of the set of anti-rabbit conjugate (goat antibodies, affinity purified, specific for immunoglobulin IgG, IgA, IgM rabbit horseradish-labeled, Russia) at a dilution of 1: 10,000 in phosphate-buffered saline. In accordance with the manufacturer’s guidelines, the cut-off point was determined by multiplying 0.350 by mean optical density value of the negative control (NC) and samples were considered as positive when the ratio of the test result of sample (od 450 nm) and according to the manufacturer’s instructions, the cut-off value was more than 1 (or ≤1). The EL × 800 universal microplate reader is used to read the results, (Biotek Instruments Inc.). All positive samples were retested in duplicate with the same EIA assay to confirm the initial results. Molecular detection of HEV by reverse transcription of a nested polymerase chain reaction (RT-PCR)RNA extraction For the isolation of nucleic acids from faecal samples, 10%–20% clarified fecal extract was used. Samples of faeces up to 1.0 ml (0.4–1.0 g) were collected with a sterile spatula and placed in a sterile tube for this study. After that, 4.0 ml of phosphate buffer solution was added to form a 10%–20% slurry. To make a slurry, the feces was vortexed vigorously. The resulting suspension was clarified by centrifugation at 3,000 rpm for 30 minutes, and the supernatants were then transferred to new sterile tubes and centrifuged at 2,000 rpm for 20 minutes to remove debris and bacteria. The supernatant was collected and stored in sterile containers at -70°C. Amplification of DNA fragments by PCRHEV RNA was detected using degenerate primers in reverse transcription of a nested polymerase chain reaction. The HEV ORF 2 is located at the position of the HEV ORF 1. (ORF 2) (Table 1). The detection of serum and fecal HEV RNA by nested RT-PCR was carried out in accordance with the manufacturer’s specifications using a QIAquick Gel Extraction kit (QIAGEN) One-Step RT-PCR kit. Following Huang et al. (2002), the primers were adopted (Table 1) after Huang et al. (2002). Sterile distilled water was used as an NC. The human HEV strain was the positive control. Positive and NCs, as well as specific molecular weight markers, were included in each run. Table 1. The nucleotide sequences that used in the amplification of HEV RNA.
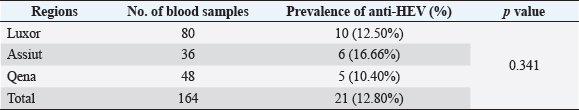
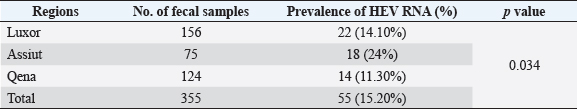
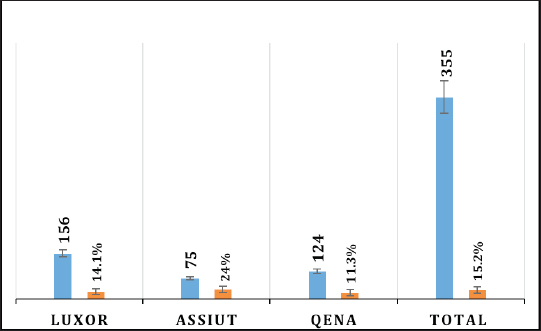
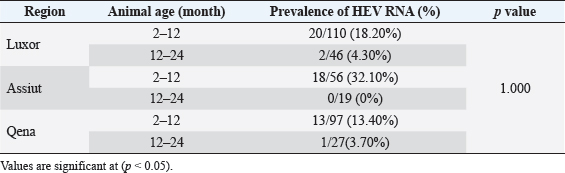
Electrophoresis agarose gel detectionThe amplified PCR-HEV product was identified using 1.5% agarose gel electrophoresis, etidium bromide staining, and an ultraviolet light reaction. 348 bp is the predicted result of universal nesting RT-PCR. Sequencing of DNATo confirm the specificity of the detection of HEV RNA and subsequent phylogenetic analysis revealed variants of the virus performed direct sequencing of amplified genomic fragments HEV. The PCR product size 350 nt was excised from the gel and purified from the agarose using a set QIAGEN. Sequencing was performed on fragment SEQ8800 automatic sequencer (Beckman Coulter). Sequencing was performed using a set of Genome Lab TM methods development kit dye terminator cycle sequencing (“Beckman Coulter,” USA), based on the use of fluorescent labels associated with ddNTP (Dye Terminators) according to the manufacturer’s protocol. Sequencing each PCR fragment was carried out in two ways – with the forward and reverse primers. The resulting chromatograms were collected into finished sequence using Seqman 4.03 software module (software package LASERGENE, DNASTAR, USA). Nucleotide sequence analysisTo determine the specificity and the amplification of sequences that differ by HEV, BLAST search was carried out in the database national center for biotechnology information. Using MEGA version 5.05., the obtained nucleotide sequences were aligned with one another and with their respective portions of whole and partial genomic sequences of other HEV isolates available in GenBank at the time of the survey. The size of ORF2 is placed in 300 nucleotides to analyze the nucleotide sequence of the viral genome of rabbits HEV isolates (after removal of the primer sequences used for amplification). The algorithm neighbour-joining (NJ) was used to create phylogenetic trees using the application Clustal W. To assess the evolutionary distance among sequences of calculation used p-distance. For performance reliability of phylogenetic clustering was analyzed 100 random samples (bootstrap pseudoreplicates). The paper presents the NJ-trees for which the sum of the lengths of the branches correspond to differences among nucleotide sequences of isolates from each other as a percentage. On phylogenetic trees was done confidence index phylogenetic grouping (bootstrap value). According to generally accepted standards, indicators of reliability of the phylogenetic grouping of more than 70% were considered significant. Statistical analysisAll statistical analyses were conducted by SPSS software for Windows, release 18 (SPSS Inc., Chicago, IL). A p-value < 0.05 was considered significant. Ethical approvalThe Ministry of Animal Health reported that all guidelines and analysis steps had been accepted. ResultsSerum samples were collected from 235 different farmed rabbits at different governorates in Egypt, 80 (48.80%) serum samples were collected from Luxor governorate, 36 (21.95%) from Assiut governorate, 48 (29.25%) from Qena governorate (Table 2). Out of the 164 rabbit serum samples, the total rate of anti-HEV IgG positivity was 21 (12.80%) examined by ELISA test. Rabbits with the highest anti-HEV IgG prevalence were found in Assiut governorates 6 (16.66%), followed by Luxor and Qena governorates (12.50% and 10.40%, respectively) (Table 2 and Fig. 1). Fecal samples were collected from 355 different farmed rabbits at different governorates in Egypt, 156 (43.95%) fecal samples were collected from Luxor governorate, 75 (21.15%) from Assiut governorate, 124 (34.90%) from Qena governorate. Out of the 355 fecal samples, the total positive rate HEV RNA was 55 (15.20%). Prevalence of HEV RNA in fecal samples of rabbits varied in different studied governorates, Assiut recorded the highest governorate by 18/75 (24%). While prevalence of HEV RNA was recorded 22 (14.10%) and 14 (11.30%) in Luxor and Qena governorates, respectively (Table 3 and Fig. 2). All animals examined were aged between 2 and 24 months, and among them, the distribution of HEV infection varied from 0% to 32.10%. HEV prevalence peaked in different age groups at different farms, with a majority of infections at age low 12 months. HEV RNA in fecal samples at Assuit governorates was detected only in fecal samples of rabbits age low 12 months as 32.10% and not detectable in fecal samples of rabbits age high 12 months. Luxor and Qena governorates were recorded high positively of HEV RNA in fecal samples at age low 12 months as 18.20%, and 13.40% compared to at age high 12 months 4.30% and 3.70%, respectively (Table 4). Table 2. Prevalence of serum anti-HEV among rabbits at different regions in Egypt.
Fig. 1. Prevalence of serum anti-HEV among rabbits in different regions in Egypt. Table 3. Prevalence of HEV RNA in fecal samples among rabbits at different regions in Egypt.
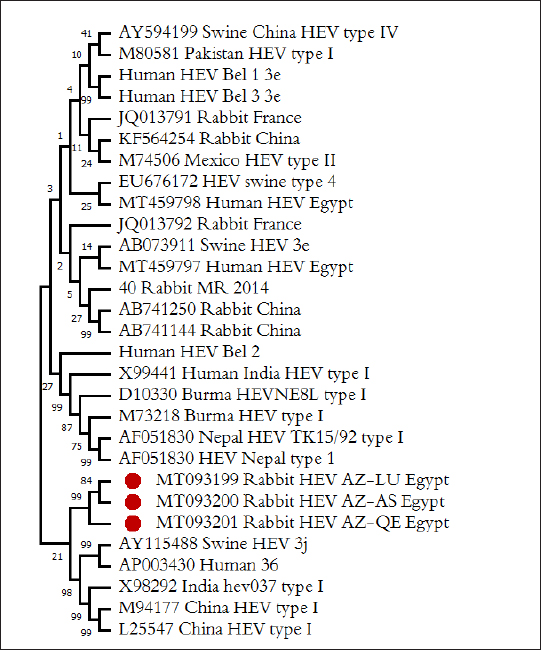
Nucleotide sequence analysis of HEVTotal installed nucleotide sequence of three isolates of rabbit HEV, one isolates from every governorate. After the exclusion of identical sequences in the GenBank database sequences were deposited three fragment ORF2 HEV rabbits, by accession number MT093199–MT093201. For phylogenetic analysis revealed fragments of HEV genome in GenBank database and we have selected the reference sequence of the rabbit HEV isolated from China, Russia, and France, and differ from each other by more than 1%, as well as a large number of sequences from the human HEV and pigs of various genotypes and subtypes circulating in different country of the world. The results of phylogenetic analysis recorded that HEV isolated from rabbits in this study form a single cluster with all known to date HEV sequences from rabbits in other city, forming phylogroups rabbit HEV close genotype 3 HEV. Sequences HEV from rabbits in different governorates in Egypt in this study (indicated in Fig. 3 as the Rabbit HEV AZ) is formed on tree a single phylogenetic group, with the degree of similarity within the group of 84%–99%. Isolated from the Luxor and Assiut region form one group [MT093199 Rabbit HEV AZ-LU Egypt and MT093200 Rabbit HEV AZ-AS Egypt, respectively, (Fig. 3)], the second group—included isolates from Qena region [MT093201 Rabbit HEV AZ-QE Egypt, (Fig. 3)]. The degree of similarity between the sequences of two groups of isolates—84%, the degree of similarity within groups 99%. All three groups of rabbit HEV identified in our work sequencing grouped with Chinese HEV isolates, with them as the group includes only published rabbit HEV isolate from the US. Frence rabbit HEV isolates form a separate group in the tree, the differences between them and identified in our work sequences formed 27%. It should be shown that all the rabbits' HEV sequences, identified in our work, and released earlier, form a separate group, and are different from the genotype 3 HEV sequences isolated from humans and pigs in Egypt and other countries in the 21%–27%.
Fig. 2. Prevalence of HEV RNA in fecal samples among rabbits at different regions in Egypt. Table 4. Age-related frequency of HEV infection at different Egypt governorates among rabbits population.
DiscussionIn Egypt, HEV is most likely endemic and appears to be a common infection. In humans, the reported HEV prevalence was higher than in other countries (El-Adly, 2019). In rural parts of suburban Cairo and the Nile Delta, high seroprevalence rates were recorded (Stoszek et al., 2006). HEV seropositivity was found in 21.6%, 14%, 4.4%, and 11.9% of the animals tested, respectively, in cows, buffaloes, sheep, and rabbits. Infected food animals were useful to HEV-positive individuals who might declare the epidemiological picture of zoonotic HEV (El-Tras et al., 2013; El-Adly, 2020). Given the genetic identification of zoonotic genotype 3 strains of HEV from rabbits in China, the United States, and France, rabbits may act as reservoir hosts for HEV transmission to humans (Zhao et al., 2009; Cossaboom et al., 2012c; Izopet et al., 2012b). Rabbits are susceptible to Genotype 4 human HEV infection, and infected rabbits developed viremia, anti-HEV antibodies, and fecal virus shredding (Cossaboom et al., 2012b). Rabbit HEV is closely related to other mammalian HEV both genetically and antigenically. The capsid protein in the genotype 3 rabbit-HEV strain was capable of cross-reacting with antibodies from rat, swine, human, and chicken HEV strains (Birke et al., 2014). Seroprevalance of anti-HEV among rabbit populations in the present study at different governorates in Egypt was recorded in total 12.80%. The highest seroprevalance of anti-HEV prevalence was demonstrated in Assiut governorates 16.66%, followed by Luxor and Qena governorates 12.50% and 10.40%, respectively. According to previous authors, the rate of seropositive of anti-HEV in blood samples of rabbits ranged from 7% to 36.5% among rabbit populations at different regions in the USA, France, Egypt and Germany (Cossaboom et al., 2012a; Izopet et al., 2012; Eiden et al., 2016; El-Adly 2020). However, Rabbits in Inner Mongolia, China, have the highest anti-HIV prevalence (57%) and viraemia (72%) (Zhao et al., 2009). HEV-infected rabbits were also found in the workplaces of two different American firms; When antibodies against HEV were evaluated, seroprevalence was 40% for supplier A and 50% for supplier B (Caitlin et al., 2011; Birke et al., 2014).
Fig. 3.Phylogenetic relationships of HEV isolated from rabbits and humans and pigs were illustrated in the diagram. Bold marked indicates HEV isolated in this study. HEV isolates which isolated from rabbits labeled rabbit, from pigs—Swine, and from humans—Human. To identify the region of viral shedding following the abbreviations used: Moscow region—MR, Belgorod region—Bel, Qena governorate, Egypt—QE, Luxor governorate, Egypt—LU, Assiut governorate, Egypt—AS. Prototype HEV strains from GenBank listed with their numbers in the database. The overall prevalence of HEV RNA from feces samples among rabbit population in our study was demonstrated in 15.20%. HEV RNA prevalence in fecal samples of rabbits varied in different studied governorates, Assiut governorate demonstrated the highest HEV RNA prevalence by 24%, while were recorded 14.10% and 11.30% in Luxor and Qena governorate respectively. Agreement with other studies, the rate of HEV RNA prevalence in feces sample among rabbit populations ranged from 7% to 16.5% at different regions studied in USA, China, Egypt, and France (Caitlin et al., 2011; Geng et al., 2011; Izopet et al., 2012). Other research, however, found a low prevalence rate of fecal HEV RNA of 1.0% in three Chinese locations, and all rabbits were caged separately, which reduced the risk of fecal oral transmission between cage mates (Xia et al., 2015). All animals examined in our study were aged between 2 and 24 months. HEV prevalence peaked in different age groups at different farms, with a majority of infections at age low 12 months. Thus, to date, no cases of Human HEV associated with HEV of rabbits have been identified in Egypt. Only one isolate is known in the world, isolated in France from a sick person and related to the HEV of rabbits, according to the results of the analysis of the genomic sequence (Lhomme et al., 2013). Therefore, the data for the conclusion about the importance of HEV in rabbits for infectious human pathology are still limited. The posibility of HEV transmission from rabbits to humans is not excluded, similar to the well-documented and proven transmission of HEV from swine to humans, especially since successful cases of experimental infection of primates, as well as pigs with rabbit-HEV (Liu et al., 2013), and rabbits with HEV strains of genotypes 3 and 4 (Mao et al., 2014). However, our data do not support the assumption of any significant role of rabbits as a reservoir of HEV for humans, in contrast to pigs. Similarly, we have not identified cases of infection of pigs with rabbit variants of HEV. ConclusionCirculation of HEV among rabbit population in hyperendemic region (Egypt) is not installed. HEV is widespread among rabbits in Egypt, at this point high frequency of detection of anti-HEV (from 10.40% to 16.66%) and HEV RNA (from 11.30% up to 24%). Rabbit HEV prevalence peaked in different age groups at different regions, with the majority of infections at age low 12 months. Circulating of variants rabbit HEV in the Egypt belong to HEV genotype 3 and all the known sequence of HEV rabbits. Identification of HEV isolates from rabbits are not genetically related to isolates of human and pig HEV isolated in the same and different regions, which is not considered rabbits as a reservoir for pathogenic human strains of HEV. HEV is prevalent among rabbits in Egypt with all rabbit strains belonging to species-specific group which is close to genotype 3. Conflict of interestWith respect to the research, authorship, and publication of this article, the author reported no potential conflicts of interest. FundingFor the research, authoring, and publication of this article, the author received no financial assistance. ReferencesAshbolt, N.J. 2004. Microbial contamination of drinking water and disease outcomes in developing regions. Toxicology 198, 229–238. Balayan, M.S., Andjaparidze, A.G., Savinskaya, S.S., Ketiladze, E.S., Braginsky, D.N., Savinov, A.P. and Poleschuk, V.F. 1983. Evidence for a virus in non-A, non-B hepatitis transmitted via faecal-oral route. Intervirology 20, 23–31. Birke, L., Cormier, S.A., You, D., Stout, R.W., Clement, C., Johnson, M. and Thompson, H. 2014. Hepatitis E antibodies in laboratory rabbits from 2 US vendors. Emerg. Infect. Dis. 20, 693–696. Caitlin, M.C., Barbara, A.D. and Xiang-Jin, M. 2011. Hepatitis E virus in rabbits, Virginia, USA. Emerg. Infect. Dis. 17, 2047–2049. Cheng, X., Wang, S., Dai, X., Shi, C., Wen, Y., Zhu, M., Zhan, S. and Meng, J. 2012. Rabbit as a novel animal model for hepatitis E virus infection and vaccine evaluation. PLoS One 7(12), e51616. Cossaboom, C.M., Cordoba, L., Cao, D., Ni, Y.Y. and Meng, X.J. 2012b. Complete genome sequence of hepatitis E virus from rabbits in the United States. J. Virol. 86, 13124–13125. Cossaboom, C.M., Cordoba, L., Dryman, B.A. and Meng, X.J. 2012a. Hepatitis E virus in rabbits, Virginia, USA. Emerg. Infect. Dis. 17, 2047–2049. Cossaboom, C.M., Cordoba, L., Sanford, B.J., Pineyro, P., Kenney, S.P., Dryman, B.A., Wang, Y. and Meng, X.J. 2012c. Cross-species infection of pigs with a novel rabbit, but not rat, strain of hepatitis E virus isolated in the United States. J. Gen. Virol. 93, 1687–1695. Eiden, M., Vina-Rodriguez, A., Schlosser, J., Schirrmeier, H. and Groschup, M.H. 2016. Detection of hepatitis E virus in archived rabbit serum samples, Germany. Food. Environ. Virol. 1989(8), 105–107. El-Adly, A.M. 2019. Prevalence of HEV markers among healthy and patients with hepatitis B and C in Upper Egypt. Afr. J. Microbiol. Res. 13(28), 552–561. El-Adly, A.M. 2020. Incidence and circulation of hepatitis E virus among farmed rabbits in Egypt. Assiut. Vet. Med. J. 66(166), 1–18. El-Tras, W.F., Tayel, A.A. and El-Kady, N.N. 2013. Seroprevalence of hepatitis E virus in humans and geographically matched food animals in Egypt. Zoonoses. Public. Health. 60, 244–251. Fauquet, C.M., Mayo, M.A., Maniloff, J., Desselberger, U. and Ball, L.A. 2005. VIIIth report of the International Committee on taxonomy of viruses. Cambridge, MA: Academic Press, pp: 583–587. Geng, Y., Zhao, C., Song, A., Wang, J., Zhang, X. and Harrison, T.J. 2011. The serological prevalence and genetic diversity of hepatitis E virus in farmed rabbits in China. Infect. Genet. Evol. 1, 476–482. Huang, F.F., Haqshenas, G., Guenette, D.K., Halbur, P.G., Schommer, S.K., Pierson, F.W., Toth, T.E. and Meng, X.J. 2002. Detection by reverse transcription-PCR and genetic characterization of field isolates of swine hepatitis E virus from pigs in different geographic regions of the United States. J. Clin. Microbiol. 40, 1326–1332. Izopet, J., Dubois, M., Bertagnoli, S., Lhomme, S., Marchandeau, S., Boucher, S., Kamar, N., Abravanel, F. and Guerin, J.L. 2012. Hepatitis E virus strains in rabbits and evidence of a closely related strain in humans, France. Emerg. Infect. Dis. 18, 1274–1281. Jeblaoui, A., Haim-Boukobza, S., Marchadier, E., Mokhtari, C. and Roque-Afonso, A.M. 2013. Genotype 4 hepatitis E virus in France: an autochthonous infection with a more severe presentation. Clin. Infect. Dis. 57, 122–126. Lhomme, S., Dubois, M. and Abravanel, F. 2013. Risk of zoonotic transmission of HEV from rabbits. J. Clin. Virol. 58, 357–362. Liu, P., Bu, Q.N., Wang, L., Han, J., Du, R.J., Lei, Y.X., Ouyang, Y.Q., Li, J., Zhu, Y.H., Lu, F.M. and Zhuang, H. 2013. Transmission of hepatitis E virus from rabbits to cynomolgus macaque. Emerg. Infect. Dis. 19, 559–565. Liu, P., Du, R.J., Wang, L., Han, J., Liu, L., Zhang, Y.L., Xia, J.K., Lu, F.M. and Zhuang, H. 2014. Management of hepatitis E virus (HEV) zoonotic transmission: protection of rabbits against HEV challenge following immunization with HEV 239 vaccine. PLoS One 30, e87600. Ma, H., Zheng, L., Liu, Y., Zhao, C., Harrison, T.J., Ma, Y., Sun, S., Zhang, J. and Wang, Y. 2010. Experimental infection of rabbits with rabbit and genotypes 1 and 4 hepatitis E viruses. PLoS One 5, e9160. Mao, J., Zhao, Y., She, R., Cao, B., Xiao, P., Wu, Q., Guo, Z., Ma, L. and Soomro, M.H. 2014. Detection and localization of rabbit hepatitis E virus and antigen in systemic tissues from experimentally intraperitoneally infected rabbits. PLoS One 9, e88607. Petra1, L.R., Ivan, T. and Andrej, K. 2013. Detection of hepatitis E virus in faeces and liver of pigs collected at two Slovenian slaughter houses. Mac. Vet. Rev. 36, 97–100. Prinja, S., Kumar, S., Reddy, G.M., Ratho, R.K. and Kumar, R. 2008. Investigation of viral hepatitis E outbreak in a town in Haryana. J. Commun. Dis. 40, 249–254. Sreenivasan, M.A., Arankalle, V.A., Sehgal, A. and Pavri, K.M. 1984. Non-A, non-B epidemic hepatitis: visualization of virus-like particles in the stool by immune electron microscopy. J. Gen. Virol. 65, 1005–1007. Stoszek, S.K., Engle, R.E. and Abdel-Hamid, M. 2006. Hepatitis E antibody seroconversion without disease in highly endemic rural Egyptian communities. Trans. R. Soc. Trop. Med. Hyg. 100, 89–94. Takahashi, M. and Okamoto, H. 2014. Features of hepatitis E virus infection in humans and animals in Japan. Hepatol. Res. 44, 43–58. Wang, S., Dong, C., Dai, X., Cheng, X., Liang, J., Dong, M., Purdy, M.A. and Meng, J. 2013. Hepatitis E virus isolated from rabbits is genetically heterogeneous but with very similar antigenicity to human HEV. J. Med. Virol. 85(4), 627–635. Wang, Y.C., Zhang, H.Y., Xia, N.S., Peng, G. and Lan, H.Y. 2002. Prevalence, isolation, and partial sequence analysis of hepatitis E virus from domestic animals in China. J. Med. Virol. 67, 516–521. Worm, H.C., van der Poel, W.H.M. and Brandstätter, G. 2002. Hepatitis E: an overview. Microbes. Infect. 4(6), 657–666. Xia, J., Zeng, H. and Liu, L. 2015. Swine and rabbits are the main reservoirs of hepatitis E virus in China: detection of HEV RNA in feces of farmed and wild animals. Arch. Virol. 160, 2791–2798. Zhao, C., Ma, Z., Harrison, T.J., Feng, R. and Zhang, C. 2009. A novel genotype of hepatitis E virus prevalent among farmed rabbits in China. J. Med. Virol. 81, 1371–1379. | ||
| How to Cite this Article |
| Pubmed Style Ahmed M. El-Adly. Serological and genetic diversity of hepatitis E virus among rabbits population in Egypt. Open Vet J. 2023; 13(5): 515-522. doi:10.5455/OVJ.2023.v13.i5.3 Web Style Ahmed M. El-Adly. Serological and genetic diversity of hepatitis E virus among rabbits population in Egypt. https://www.openveterinaryjournal.com/?mno=13277 [Access: July 05, 2025]. doi:10.5455/OVJ.2023.v13.i5.3 AMA (American Medical Association) Style Ahmed M. El-Adly. Serological and genetic diversity of hepatitis E virus among rabbits population in Egypt. Open Vet J. 2023; 13(5): 515-522. doi:10.5455/OVJ.2023.v13.i5.3 Vancouver/ICMJE Style Ahmed M. El-Adly. Serological and genetic diversity of hepatitis E virus among rabbits population in Egypt. Open Vet J. (2023), [cited July 05, 2025]; 13(5): 515-522. doi:10.5455/OVJ.2023.v13.i5.3 Harvard Style Ahmed M. El-Adly (2023) Serological and genetic diversity of hepatitis E virus among rabbits population in Egypt. Open Vet J, 13 (5), 515-522. doi:10.5455/OVJ.2023.v13.i5.3 Turabian Style Ahmed M. El-Adly. 2023. Serological and genetic diversity of hepatitis E virus among rabbits population in Egypt. Open Veterinary Journal, 13 (5), 515-522. doi:10.5455/OVJ.2023.v13.i5.3 Chicago Style Ahmed M. El-Adly. "Serological and genetic diversity of hepatitis E virus among rabbits population in Egypt." Open Veterinary Journal 13 (2023), 515-522. doi:10.5455/OVJ.2023.v13.i5.3 MLA (The Modern Language Association) Style Ahmed M. El-Adly. "Serological and genetic diversity of hepatitis E virus among rabbits population in Egypt." Open Veterinary Journal 13.5 (2023), 515-522. Print. doi:10.5455/OVJ.2023.v13.i5.3 APA (American Psychological Association) Style Ahmed M. El-Adly (2023) Serological and genetic diversity of hepatitis E virus among rabbits population in Egypt. Open Veterinary Journal, 13 (5), 515-522. doi:10.5455/OVJ.2023.v13.i5.3 |