| Short Communication | ||
Open Vet J. 2023; 13(3): 352-357 Open Veterinary Journal, (2023), Vol. 13(3): 352–357 Short Communication Genotyping of cows by LHCGR, FSHR loci, and determination of the level of ovulation depending on the expression of the studied genesYktiyar Sarybayev1*, Yessengali Ussenbekov1, Orynbassar Turebekov1, Bekbolat Turumbetov2 and Ibragim Tutkyshbay21Department of Obstetrics, Surgery and Biotechnology of Animal Reproduction, Kazakh National Agrarian Research University, Almaty, Republic of Kazakhstan 2Department of Veterinary Medicine, Mukhtar Auezov South Kazakhstan University, Shymkent, Republic of Kazakhsta *Corresponding Author: Yktiyar Sarybayev. Department of Obstetrics, Surgery and Biotechnology of Animal Reproduction, Kazakh National Agrarian Research University, Almaty, Republic of Kazakhstan. Email: yktiyar.sarybayev [at] gmail.com. Submitted: 03/01/2023 Accepted: 13/02/2023 Published: 18/03/2023 © 2023 Open Veterinary Journal
AbstractBackground: Genotyping offers a promising avenue for identifying the healthy reproductive system in cows. The healthy reproductive system in cows is determined by measuring the level of ovulation and by identifying the type polymorphism of specific genes. Aim: The aim of the article is to explore how polymorphism of follicle stimulating hormone Receptor (FSHR) and luteinizing hormone/choriogonadotropin receptor (LHCGR) genes affect the reproduction trait of Holstein cows. Methods: Here we define a reproducible protocol to genotype and identify the polymorphism in specific genes from the extracted DNA of cows. Results: The results of genotyping showed that the only C allele (CC genotype) was observed in 100% of cows at the LHCGR locus, and three genotypes were observed at the FSHR locus (CC—67.74%, CG—9.03%, GG—23.22%). In cows with the CC genotype at the FSHR locus, the hormone concentration during ovulation was 1.1–2.5 ng/ml, which is within the physiological range for healthy reproduction. Conclusion: Cows with the CC genotype at the FSHR locus have a healthy course of the ovulation process, therefore good reproduction. Keywords: Genotyping, Ovulation, LHCGR, FSHR, Polymorphism. IntroductionThe priority basis for the successful management of dairy cattle breeding is animals with a high level of milk productivity and reproductive system. Recent advances in molecular science contribute to the use of the genome and the selection of the necessary genes to improve the genetics of domestic stingrays, in this case, cows (Sharifiyazdi et al., 2018; Schniewind et al., 2021; Abeygunawardana et al., 2022). In other words, modern biotechnological techniques allow us to find single nucleotide polymorphisms (SNP) among the population. A gene polymorphism is a difference in the DNA sequence (mutant and normal) in the genome of representatives of the same species. Single-nucleotide polymorphism is the difference of a single nucleotide in alternative genes (Sharifiyazdi et al., 2018; Guo et al., 2021; Gutiérrez-Reinoso et al., 2022). In the body of animals, there are usually pituitary hormones gonadotropins: luteinizing (LH) and follicle-stimulating (FSH). The latter, entering the general bloodstream, regulate the functions of the testes and ovaries, whereas FSH controls the processes of gamete maturation in the animal body (Mylostyvyi et al., 2021; Gritsienko et al., 2022) time, FSH in the ovaries regulates the growth and maturation phases of the follicle, stimulating the activity of the FSH receptor (FSHR) in the cells of the theca and granulosa. The function of LH is to stimulate the synthesis of sex steroid hormones: estrogens and testosterone. LH hormone participates in follicular development and contributes to the normal course of the ovulation process (Ponsart et al., 2014). FSH and LH are glycoprotein polypeptide hormones, meaning that they cannot enter the cell (Ziba et al., 2017). Their effect on target cells is mediated by special receptors located on the cell membrane of target cells (Younis et al., 2020). Each hormone has special receptors FSHR and LH receptor (LHCGR) (Sharifiyazdi et al., 2018). During ovulation in cows, the concentration of the above-mentioned hormones should be in the region of 1.0–2.0 ng/ml for a healthy course of the reproductive system. Another way to determine cows with a healthy reproductive system is by genotyping by locus, such as LHCGR and FSHR. In other words, the detection of polymorphism in the gene markers of calving. According to studies, three genotypes were observed for each locus: FSHR—CC, CG and GG, LHCGR—CC, TT, and CT (Sharifiyazdi et al., 2018). To determine the cows with which SNPs have a high reproductive function, it is necessary to measure the level of hormones during ovulation in a group of cows with different polymorphisms. Despite numerous studies on this topic, the topic of the level of ovulation depending on the expression of certain genes in cows has not been sufficiently studied. The novelty of the study is that the authors for the first time identified cows with a healthy reproductive function by genotyping at the FSHR and LHCGR loci and determining the level of ovulation. The aim of the article is to explore how polymorphism of FSHR and LHCGR genes affect the reproduction trait of Holstein cows. The relationship between the level of ovulation and polymorphism in cows has been studied. According to the research hypothesis it is expected that heifers with CC genotype at the FSHR and LHCGR loci will have a healthy ovulation period. All experimental procedures were carried out in compliance with ethical standards in the study with animals. The study was conducted with the permission of the owner of the farm, who gave informed consent to the study. Materials and MethodsThe blood samples were obtained from Holstein cows of the dairy farm "Baiserke-Agro" LLP (Almaty region). Bovine LH and FSH concentrations were determined using a competitive ELISA kit according to manufacturer's instructions (Bautista et al., 2019). The working area, pipettes, and equipment were disinfected. Ready Thermo scientific genomic DNA purification kit was used. Procedures only for one sample of blood are given below: A lysis buffer was added to the 5 ml of blood sample and incubated at 65°C for 5 minutes. After, chloroform was added to the tube and shaken by hand for 20 seconds. Then, the mixture was centrifuged at 3,500 rpm for 2 minutes. The tube had two separate phases. The aqueous phase was transferred to a new tube to proceed to the precipitation stage. The precipitation solution was added to the tube aqueous phase, mixed gently, and centrifuged for 2 minutes. The resulting supernatant liquid part on the top was discarded. The pellet was dissolved in NaCl solution and 96% Ethanol was added to the precipitate. Then again, the tube was centrifuged and the supernatant was removed. The resulting pellet was washed with chilled 70% Ethanol and dissolved in water or TE. The extracted DNA was stored at −20°C until the polymerase chain reaction (LH ELISA kit: Bovine Luteinizing Hormone ELISA Kit-AAD14960.1. https://www.mybiosource.com/bovine-ovine-elisa-kits/luteinizing-hormone-lh/2609425). The PCR reaction was done mixing the cocktail (0.25 mM primers, 150 μM deoxynucleotides, 1× reaction buffer, and Taq DNA polymerase) and 100 ng of extracted DNA. The sample tubes were denatured at 96°C for 2 minutes and following 35 cycles of denaturation (96°C for 45 seconds), annealing (65°C for 30 seconds), and extension (72°C for 5 minutes) steps. Below primers from a gene bank were used: Primers for FSHR forward: 5ʹ-CATCACTGTGTCCA AGTCAAAGA-3′ and reverse: 5ʹ-CTCTTCCATGGC ATATTCTTCAA-3' Primers for LHCGR forward: 5-CTTTCAATCCC TGTGAAGACATT-3' and reverse: 5'-GGTCCAGTTGAATAGCATAGGTG-3' After the PCR amplification reaction completion, each target gene is analyzed using RFLP by enzymatic digestion. The samples with LHCGR and FSHR were digested with HhaI and AluI enzymes respectively. The restriction products were visualized by agarose gel electrophoresis. The Infinity VX2 3026, WL/LC/26m XPress, Vilber Lourmat gel documentation imaging system, bromophenol blue dye, and pUC19 / DNAMspI DNA marker were used. 5 ul of gDNA samples with dye were loaded on the gel and visualized using the system mentioned above. All experimental procedures were carried out in compliance with the ethical standards of animal research of the Veterinary faculty, Kazakhstan National Agrarian University. The study was conducted with the permission of the owner of the farm, who gave informed consent to carry out the study (Desjardins and Conklin, 2010).
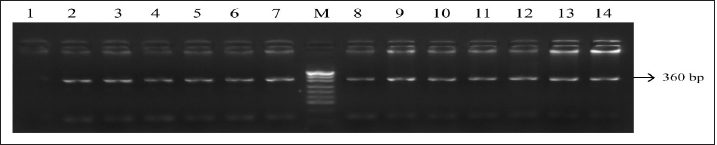
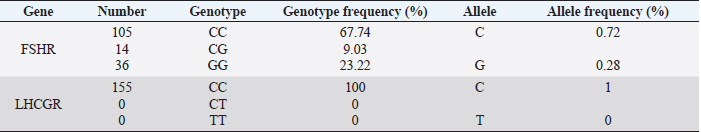
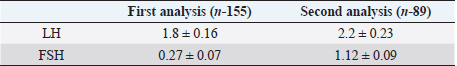
Fig. 1. PCR FSHR gene. Electrophoresis showing PCR product of Holstein cows FSHR gene. Lane M: marker, lanes 1–14 show result of electrophoresis of Holstein cow FSHR gene with length of 360 bp. Results and DiscussionFigures 1 and 3 are electrophoresis images of PCR FSHR and LHCGR genes in Holstein cows respectively. Figures 2 and 4 are electrophoresis images of PCR-RFLP FSHR and LHCGR genes respectively. Table 1 depicts the polymorphism of the above genes and their weights, and Table 2 shows the hormone concentration measure during the ovulation of Holstein cows. The single nucleotide (G/C) polymorphism presents in the upstream regulatory region of the FSHR gene that alters the expression of FSHR in the granulosa cells. Further, this phenomenon depending on the participating nucleotide affects the fertility of the cell either negatively or positively (User Manual NanoDrop 2000/2000c, Spectrophotometer V1.0., 2019). Regarding the LHCGR gene, it is considered to be a marker for superovulation. In previous studies, three genotypes were observed by both genes (CC, CG, GG for FSHR gene and CC, CT, TT for LHCGR) (Houde et al., 1994). According to Table 1, PCR-RFLP with HhaI enzyme of the LHCGR gene, 100% of animals were carriers of the same alleles (CC genotype). However, by previous works, it is known that there are three genotypes by LHCGR locus. Regarding FSHR PCR-RFLP by AluI enzyme, three genotypes (CC, CG, and GG) were observed. There are significant differences in the number of genotypes of the FSHR gene: 9.03% of animals were carriers of both alleles (CG genotype,) and 67.74% of cows were homozygous for the C allele (CC genotype) and 23.22% of cows were homozygous for the G allele (GG genotype). As a result, the frequency of allele C is greater than the G allele, which was observed in previous studies as well. Yang et al. (2010, 2011) suggested that the cows carrying the CC genotype were observed to have more amount of ova than others and to produce an increased yield of transferable embryos (Albarrán-Portillo and Pollott, 2013). In addition, GG-genotyped cows were associated with more unfertilized oocytes (Pryce et al., 2004). The total sexual cycle in animals is on average 21 days, and on the 17th day, along with the passage of ovulation, the amount of LH and FSH in the blood increases, and these hormones reach a high level (Yang et al., 2011; Hourvitz and Adashi, 2003). The presence of FSH and LH stimulates the secretion of sex hormones estrogen and progesterone. The concentration of the last two hormones is directly proportional to the concentration of stimulating hormones (LH and FSH). Estrogen in the blood of uterine livestock at a normal level creates favorable conditions for the proper course of the ovulation process, fertilization, and transition to the luteal phase (Gomes and Erb, 1965; Lonergan et al., 2015). During ovulation, the amount of LH and FSH should be within the physiological range of 1.0–2.5 ng/ml. If the amount will be less, then due to the lack of sex hormones in the body of the uterine livestock, problems with infertility in the reproductive system arise. For example: if the volume of one hormone progesterone is insufficient, the uterine layer becomes unacceptable for the embryo site. Such a mucous layer of the uterus becomes unacceptable for embryo resorption and leads to embryonic death (Lonergan et al., 2015; Ziba et al., 2017). Thus, the concentration of hormones in the blood during ovulation was used as an indicator of the productivity level of cows with different genotypes. The farm provided the health information of cows and it was mentioned that 16 cows had reproductive problems. However, further details on problems and the nature of those problems were not determined. For this reason, 16 cows with reproductive system problems along with 50 cows with the GG genotype were excluded from the second hormone concentration analysis. The hormone concentration of 89 cows was measured and the average was recorded as a second analysis. In the first analysis, the hormone concentrations of all 155 cows were measured and the average value was taken (Table 3). Results were as expected, the average concentration of hormones (LH and FSH) of cows with CC genotype by FSHR locus was within the physiological range and higher than the average hormone concentration of a group of cows with all genotypes (CC, CG, and GG) (Yang et al., 2010; Cory et al., 2012).
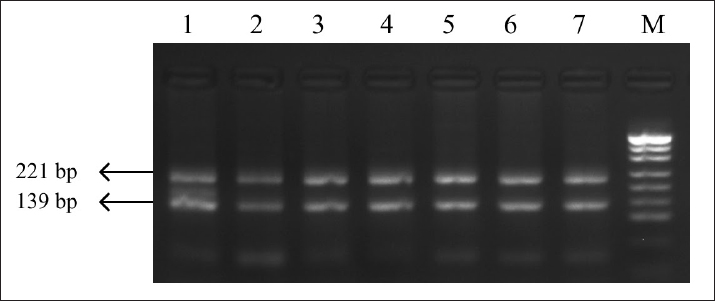
Fig. 2. PCR-RFLP FSHR gene. Electrophoresis results from PCR-RFLP using AluI enzyme in holstein cows FSHR gene. Lane M: marker, lanes 1–7 samples with lengths 139 and 221 bp.
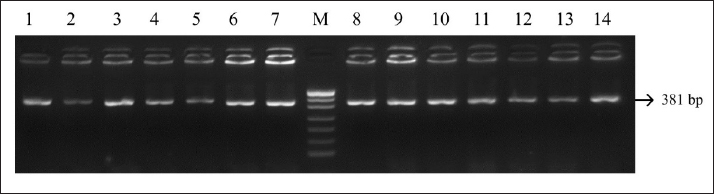
Fig. 3. PCR LHCGR gene. Electrophoresis showing PCR product of Holstein cows LHCGR gene. Lane M: marker, lanes 1–14 samples with length 381 bp.
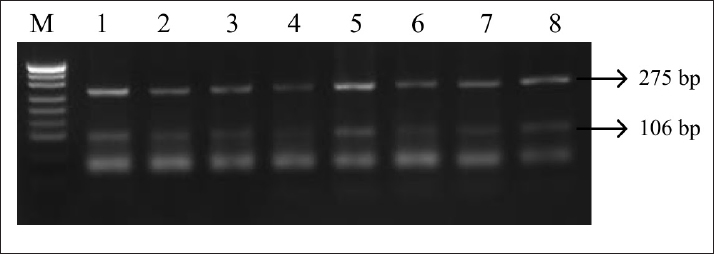
Fig. 4. PCR-RFLP LHCGR gene. Electrophoresis results from PCR-RFLP using HhaI enzyme in holstein cows LHCGR gene. Lane M: marker, lanes 1–7 samples with lengths 106 and 275 bp. Table 1. Genotypes by locus and their weight in Holstein cows.
Table 2. Genotype and allele frequencies of LHCGR and FSHR genes in Holstein cows.
Table 3. The average concentration (in ng/ml) of the hormones LH and FSH in the blood serum of Holstein cows in the first and second analysis.
The authors of the article (Widmer et al., 2021) investigate twin and multiple births in cattle using functional candidate genes LHCGR and FSHR. There was reported for the first time a major quantitative trait locus (QTL) for multiple birth in Holstein cattle and provided evidence for a linked variant in the non-coding region of a functional candidate gene. This discovery, which is a first step towards the understanding of the genetic architecture of this polygenic trait, opens the path for future selection against this undesirable trait, and thus contributes to increased animal health and welfare. The authors (Missio et al., 2022) evaluate the effects of beta-hydroxybutyrate (BHBA) on follicle growth and on ovulatory mechanisms in cattle. The results of this study indicate that the increase in intrafollicular concentrations of BHBA affects follicular growth, but it does not compromise the ovulatory cascade and cellular homeostasis in bovine granulosa cells. The study (Noguchi et al., 2022) predicted the molecules modulating embryo-uterine communication by comparing two sets of differentially expressed genes (DEGs): DEGs in uterine epithelial cells collected from the uterus with and without blastocysts and DEGs between blastocysts developed in vivo and in vitro. This study predicted the molecules that potentially mediate communication between the developing embryo and the uterus in vivo and prepare the uterus for pregnancy. According to the study (Kasimanickam and Kasimanickam, 2021), all genes but MUC1 and all proteins but MUC1 expression was greater in filamentous conceptus and corresponding endometrium versus tubular conceptus and matching endometrium in subclinical endometritis (SCE) and embryo quality grade (QG) groups. Disrupted embryo-uterine communication by altered expression of candidate genes in SCE cows, and in cows following the transfer of poor embryo negatively programs the conceptus development and plausibly affects conceptus survival. The authors of the paper (Kasimanickam et al., 2020) studied the endometrial expression of various genes (ISGs, PPARs, RXRs, and MUC1) in repeat breeder cows. Altered expression of these uterine genes and associated potential impairment in embryo elongation and implantation may promote embryonic loss in repeat breeder cows. Besides, interactions between PPARD, PPARG, and MUC1 may be therapeutically exploitable. ConclusionIn this study, it was expected and proved that cows with CC genotype by FSHR locus will have a more amount of FSH and LH hormones during ovulation and accordingly a healthy reproductive system and higher productivity. According to the result of PCR- RFLP, all 155 Holstein cows had CC genotype by LHCGR locus, and 9.03% of them were carriers of CG genotype, 67.74% of cows were homozygous for the C allele (CC genotype) and 23.22% of cows were homozygous for the G allele (GG genotype) by FSHR locus. In cows with the CC genotype at the FSHR locus, the hormone concentration during ovulation was 1.1–2.5 ng/ml, which is within the physiological range for healthy reproduction. Holstein cows with CC genotype by FSHR gene locus had hormone concentration within the physiological range meaning they have a healthy reproductive system. This work studied the fertility of Holstein heifers with different genotypes and in the future, it is suggested to study the relationship between milk production and polymorphisms. The scientific value of the work is that, based on the results of this work, farms can identify cows with good milk productivity and reproductive function and pay more attention to them. AcknowledgementsThe financial support for this research was provided by the Ph.D. grant (Sarybaev Y.) of the Veterinary Faculty, Kazakhstan National Agrarian Research University. ReferencesAbeygunawardana, D.I., Ranasinghe, R.M.S.B.K., De Silva, S.N.T., Gamika, P.A. and Rajapakse, J. 2022. Effect of LHCGR and FSHR gene polymorphisms on fertility traits and milk yield of cross-bred dairy cows in Sri Lanka. Anim. Biotechnol. 17, 1–8. Albarrán-Portillo, B. and Pollott, G.E. 2013. The relationship between fertility and lactation characteristics in Holstein cows on United Kingdom commercial dairy farms. J. Dairy Sci. 96(1), 635–646. Bautista, V.M., Jiménez Chávez, S.P., Meza Franco, C.D., Ramos, T.I. and Toledo, J.R. 2019. FSH in bovine superovulation. Bionatura 4(1), 1–5. Cherniy, N., Gutyj, B. and Hoffmann, G. 2021. Changes in the spectrum of free fatty acids in blood serum of dairy cows during a prolonged summer heat wave. Animals 11(12), 3391. Cory, A.T., Price, C.A., Lefebvre, R. and Palin, M.F. 2012. Identification of single nucleotide polymorphisms in the bovine follicle-stimulating hormone receptor and effects of genotypes on superovulatory response traits. Anim. Gen. 44(2), 197–201. Desjardins, P. and Conklin, D. 2010. Nanodrop microvolume quantitation of nucleic acids. J. Visual. Exp. Thermo Fisher Sci. 45, 1–5. Gritsienko, Y., Gill, M. and Karatieievа, O. 2022. Connection between gene markers with milk production traits of ukrainian dairy cows. Online J. Anim. Feed Res. 12(5), 302–313. Gomes, W.R. and Erb, R.E. 1965. Progesterone in bovine reproduction: a review. J. Dairy. Sci. 2442, 314–330. Guo, C., Yu, H., Feng, G., Liu, X. and Liu, X. 2021. Associations of FSHR and LHCGR gene variants with ovarian reserve and clinical pregnancy rates. Reprod. Biomed. Online 43(3), 561–569. Gutiérrez-Reinoso, M.A., Aguilera, C.J., Navarrete, F., García-Herreros, M. and Rodríguez-Alvarez, L. 2022. Effects of extra-long-acting recombinant bovine FSH (bscrFSH) on cattle superovulation. Animals 12(2), 153. Houde, A., Lambert, A., Silversides, D.W., Lussier, J.G., Saumande, J. 1994. Structure of the bovine follicle-stimulating hormone receptor complementary DNA and expression in bovine tissues. Mol. Reprod. Dev. 39(2), 127–135. Hourvitz, A. and Adashi, E.Y. 2003. Ovulation. In Encyclopedia of hormones. Eds., Henry H.L. and Norman A.W. New York, NY: Academic Press, vol. 3, pp: 98–106. Kasimanickam, R.K. and Kasimanickam, V.R. 2021. Mrna expressions of candidate genes in gestational day 16 conceptus and corresponding endometrium in repeat breeder dairy cows with suboptimal uterine environment following transfer of different quality day 7 embryos. Animals 11(4), 1092. Kasimanickam, R., Kasimanickam, V. and Grende, K. 2020. Endometrial expression of various genes (ISGs, PPARs, RXRs and MUC1) on day 16 post-ovulation in repeat breeder cows, with or without subclinical endometritis. Theriogenology 142, 251–259. Lonergan, P., Forde, N. and Spencer, T. 2015. Role of progesterone in embryo development in cattle. Reprod. Fertil. Dev. 28(2), 66–74. Missio, D., Fritzen, A., Cupper Vieira, C., Dias Gonçalves, P.B. and Ferreira, R. 2022. Increased β-hydroxybutyrate (BHBA) concentration affect follicular growth in cattle. Anim. Reprod. Sci. 243, 107033. Mylostyvyi, R., Sejian, V., Izhboldina, O., Kalinichenko, O., Karlova, L., Lesnovskay, O., Begma, N., Marenkov, O., Lykhach, V., Mid Noguchi, T., Hayashi, T., Inoue, Y., Shirasuna, K. and Iwata, H. 2022. Predicting of molecules mediating an interaction between bovine embryos and uterine epithelial cells. J. Reprod. Dev. 68(5), 318–323. Ponsart, C., Le Bourhis, D., Knijn, H., Fritz, S., Guyader-Joly, C., Otter, T., Lacaze, S., Charreaux, F., Schibler, L., Dupassieux, D. and Mullaart, E. 2014. Reproductive technologies and genomic selection in dairy cattle. Reprod. Fertil. Dev. 26(1), 12. Pryce, J.E., Royal, M.D., Garnsworthy, P.C. and Mao, I.L. 2004. Fertility in the high-producing dairy cow. Livestock Prod. Sci. 86(1–3), 125–135. Schniewind, H.A., Sattler, L.-M., Haudum, C.W., Obermayer-Pietsch, B. and Schomburg, L. 2021. Autoimmunity to the follicle-stimulating hormone receptor (Fshr) and luteinizing hormone receptor (lhr) in polycystic ovarian syndrome. Int. J. Mol. Sci. 22(24), 13667. Sharifiyazdi, H., Mirzaei, A. and Ghanaatian, Z. 2018. Characterization of polymorphism in the FSH receptor gene and its impact on some reproductive indices in dairy cows. Anim. Reprod. Sci. 188, 45–50. Widmer, S., Seefried, F.R., von Rohr, P., Spengeler, M. and Drögemüller, C. 2021. A major QTL at the LHCGR/FSHR locus for multiple birth in Holstein cattle. Genet. Sel. Evol. 53(1), 57. Yang, W.C., Li, S.J., Tang, K.Q., Hua, G.H., Zhang, C.Y., Yu, J.N., Han, L. and Yang, L.G. 2010. Polymorphisms in the 5′ upstream region of the FSH receptor gene, and their association with superovulation traits in Chinese Holstein cows. Anim. Reprod. Sci. 119(3–4), 172–177. Yang, W.C., Tang, K.Q., Li, S.J., Chao, L.M. and Yang, L.G. 2011. Polymorphisms of the bovine luteinizing hormone/choriogonadotropin receptor (LHCGR) gene and its association with superovulation traits. Mol. Biol. Rep. 39(3), 2481–2487. Younis, L.S., Rasheed, S.T., Aboud, Q.M., Hasan, M.S. and Abid, A.A. 2020. Identification the effect of inhibin ßA/activin A genes polymorphism on superovulation (Calving Rate) in Holstein Friesian cows. Syst. Rev. Pharm. 11(2), 471–481. Ziba, A, Seyed, A.F., Masomeh, F and Shodja, J. 2017. Polymorphisms of the bovine Luteinizing Hormone/ Chorio Gonadotropin Receptor (LHCGR) gene and its association with ovarian follicular cysts. J. Vet. Med. 168(7–9), 143–150. | ||
| How to Cite this Article |
| Pubmed Style Sarybayev Y, Ussenbekov Y, Turebekov O, Bekbolat B, Tutkyshbay I. Genotyping of cows by LHCGR, FSHR loci and determination of the level of ovulation depending on the expression of the studied genes. Open Vet J. 2023; 13(3): 352-357. doi:10.5455/OVJ.2023.v13.i3.12 Web Style Sarybayev Y, Ussenbekov Y, Turebekov O, Bekbolat B, Tutkyshbay I. Genotyping of cows by LHCGR, FSHR loci and determination of the level of ovulation depending on the expression of the studied genes. https://www.openveterinaryjournal.com/?mno=136165 [Access: July 07, 2025]. doi:10.5455/OVJ.2023.v13.i3.12 AMA (American Medical Association) Style Sarybayev Y, Ussenbekov Y, Turebekov O, Bekbolat B, Tutkyshbay I. Genotyping of cows by LHCGR, FSHR loci and determination of the level of ovulation depending on the expression of the studied genes. Open Vet J. 2023; 13(3): 352-357. doi:10.5455/OVJ.2023.v13.i3.12 Vancouver/ICMJE Style Sarybayev Y, Ussenbekov Y, Turebekov O, Bekbolat B, Tutkyshbay I. Genotyping of cows by LHCGR, FSHR loci and determination of the level of ovulation depending on the expression of the studied genes. Open Vet J. (2023), [cited July 07, 2025]; 13(3): 352-357. doi:10.5455/OVJ.2023.v13.i3.12 Harvard Style Sarybayev, Y., Ussenbekov, . Y., Turebekov, . O., Bekbolat, . B. & Tutkyshbay, . I. (2023) Genotyping of cows by LHCGR, FSHR loci and determination of the level of ovulation depending on the expression of the studied genes. Open Vet J, 13 (3), 352-357. doi:10.5455/OVJ.2023.v13.i3.12 Turabian Style Sarybayev, Yktiyar, Yessengali Ussenbekov, Orynbassar Turebekov, Bekbolat Bekbolat, and Ibragim Tutkyshbay. 2023. Genotyping of cows by LHCGR, FSHR loci and determination of the level of ovulation depending on the expression of the studied genes. Open Veterinary Journal, 13 (3), 352-357. doi:10.5455/OVJ.2023.v13.i3.12 Chicago Style Sarybayev, Yktiyar, Yessengali Ussenbekov, Orynbassar Turebekov, Bekbolat Bekbolat, and Ibragim Tutkyshbay. "Genotyping of cows by LHCGR, FSHR loci and determination of the level of ovulation depending on the expression of the studied genes." Open Veterinary Journal 13 (2023), 352-357. doi:10.5455/OVJ.2023.v13.i3.12 MLA (The Modern Language Association) Style Sarybayev, Yktiyar, Yessengali Ussenbekov, Orynbassar Turebekov, Bekbolat Bekbolat, and Ibragim Tutkyshbay. "Genotyping of cows by LHCGR, FSHR loci and determination of the level of ovulation depending on the expression of the studied genes." Open Veterinary Journal 13.3 (2023), 352-357. Print. doi:10.5455/OVJ.2023.v13.i3.12 APA (American Psychological Association) Style Sarybayev, Y., Ussenbekov, . Y., Turebekov, . O., Bekbolat, . B. & Tutkyshbay, . I. (2023) Genotyping of cows by LHCGR, FSHR loci and determination of the level of ovulation depending on the expression of the studied genes. Open Veterinary Journal, 13 (3), 352-357. doi:10.5455/OVJ.2023.v13.i3.12 |