| Original Article | ||
Open Vet J. 2022; 12(2): 197-203 Open Veterinary Journal, (2022), Vol. 12(2): 197–203 Original Research LPS-induced NLRP3 gene expression in chickenAdil Sabr Al-Ogaili1, Samer Sadeq Hameed2* and Noor Noori11Department of Medical Laboratory Techniques, Kut-Technical Institute, Middle Technical University, Baghdad, Iraq 2Department of Pathology and Diseases of Poultry, College of Veterinary Medicine, University of Baghdad, Baghdad, Iraq *Corresponding Author: Samer Sadeq Hameed. Department of Pathology and Diseases of Poultry, College of Veterinary Medicine, University of Baghdad, Baghdad, Iraq. Email: samer.hameed [at] covm.uobaghdad.edu.iq Submitted: 29/10/2021 Accepted: 23/02/2022 Published: 27/03/2022 © 2022 Open Veterinary Journal
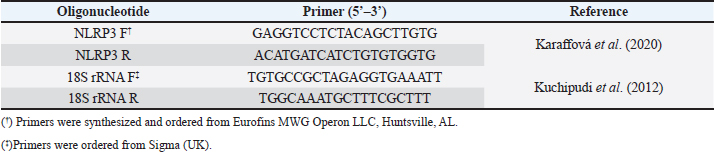
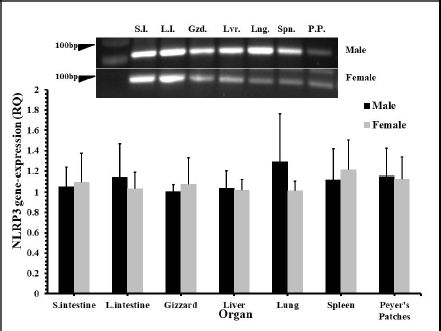
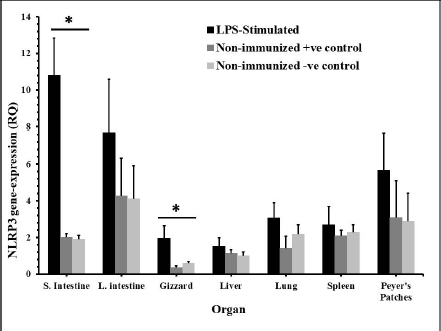
AbstractBackground: NOD-, LRR-, and pyrin domain-containing protein 3 (NLRP3) is a cytosolic sensor that detects many microbial pathogen-associated molecular patterns and damage-associated molecular patterns. It works as a pro-inflammatory cytokine that is encoded in the NLRP3 gene. This protein is profoundly expressed in macrophage and other innate immune system cells, and participates in the assembly of NLRP3 inflammasome. NLRP3 inflammasome activates caspase-1, which in turn renders the inactive precursors of the pro-inflammatory cytokines, the interleukin-1β (IL-1β) and the IL-18, into active forms. This cytokine may trigger many inflammatory responses and cell signaling pathways as well. Little is known about this cytokine in chickens, especially its role in vaccines or induced immune responses. Aim: In this article, we sought to determine the presence of this gene mRNA in selected organs of male and female commercial, brown leghorn egg-type chickens. In addition, we sought to determine this gene’s potential expression in these organs upon stimulation. Methods: Using real-time polymerase chain reaction, first we tested the presence of the NLRP3 gene in the chickens. Second, the levels and the time course of NLRP3 gene expression have been tested after stimulation with bacterial lipopolysaccharide (LPS) for 12, 24, and 48 hours postinoculation (pi). One-hundred twenty, day-old males and females egg-type brown leghorn chickens were used for the study. Results: Our results showed that the gene mRNA is actually present in chickens solely. Also, there were no significant differences in the density of the expression and the distribution of the expression of the NLRP3 between male and female chickens and among different organs. Upon stimulation with LPS administration, however, there were marked elevations in the gene expression rates in small intestine, large intestine, gizzard, liver, lung, spleen, and Peyer’s patches 12 hours pi. This elevation continued to elevate 24 hours pi. However, the significance of the expression was only recorded in the small intestine, large intestine, and with less significance, in Peyer’s patches and spleen. This elevation in expression subsided and almost returned to normal within these organs 48 hours pi. Conclusion: The results suggest that there were no significant differences in the NLRP3 gene expression between male and female. Upon stimulation, the course of the gene expression showed a time-dependent response. First, the dominance in the NLRP3 gene mRNA expression was in the small intestine and gizzard. Similarly, but less profoundly, the large intestine, Peyer’s patches, and spleen expressed NLRP3 mRNA 12 and 24 hours pi with LPS. Secondary immune organs, lungs, small intestine, and large intestine expressed the NLRP3 mRNA significantly 24 hours pi. All the levels had diminished and almost returned to normal 48 hours pi. Keywords: Chickens, NLRP3, Gene expression, Pro-inflammatory cytokine. IntroductionInnate immunity is the first line of defense against invading pathogens. Like other body systems, the innate immune system endures its action through its organs, cells, cytokines, and pathways. Signaling pathways of the immune system are often via specific cytokines herein (Nicholson, 2016; Kelley et al., 2019). In an innate immune response, there is marked integration between inflammatory response and innate immune response (Muñoz-Carrillo et al., 2018). The regulation/collaboration of both responses is through the signaling cytokines (Liu and Cao, 2016). Within and on innate immune cells, there are receptors encoded in the germline called pattern recognition receptors (PRRs). These PRRs are expressed as sequelae of certain stimuli, such as specific receptors on pathogens called pathogen-associated molecular patterns (PAMPs). These PRRs ligands are conserved, unique, and essential structures or molecules on the pathogens. One of the well-known PAMPs is bacterial outer membrane lipopolysaccharide (LPS) (Bowen et al., 2009). In addition, the expression of the PRRs may be stimulated by damage-associated molecular patterns (DAMPs). DAMPs are inflammatory mediators that are produced as a result of stress or inflammatory necrosis (Neerukonda and Katneni, 2020). In an inflammatory response, a group of intracellular mediators serve as cell signaling pathways. Among these is a multimeric protein complex called inflammasome. Inflammasome activates caspase-1, which is a major inflammatory pathway. Therefore, a PRR is actually a platform that acts to activate caspase-1 either through engagement with PAMPs or DAMPs. One of the intracellular signaling molecules for PRRs is a group called nucleotide-binding domain and leucine-rich repeat containing (NLR) and NOD-like receptor superfamily. The NOD-like receptor superfamily has a group of proteins called NLRPs (NLRP1 through NLRP14) (Roh and Sohn, 2018; Kanneganti, 2020). Many members of this family are highly conserved (plants to humans) to act as members in the first line of defense. Among these, NLRP3 is essential in activating and in the proteolysis of pro-inflammatory cytokines, such as caspase-1, which is required in interleukin-1β (IL-1β), and IL-18 proteolysis (Vrentas et al., 2018; Kelley et al., 2019; Paik et al., 2021). In this study, we aimed to record the presence of NLRP3 gene in the mRNA of commercial, brown leghorn egg-type chickens. In addition, we sought to determine if there is a difference in this gene expression regarding the gender of the bird. Finally, we sought to determine if there is a significant elevation in gene expression through various organs after the stimulation with bacterial LPS. Our results showed that upon stimulation with LPS administration, NLRP3 was highly expressed within 12 hours postinoculation (pi) in the small intestine and gizzard and started to subside 24 hours pi. The NLRP3 gene expression almost returned to normal within 48 hours pi in specific organs. The exception, however, was the small intestine, the large intestine, lung, spleen, and Peyer’s patches where the values were significantly higher than the other organs. In addition, we recorded that there were no significant differences in the NLRP3 gene expression regarding the birds’ gender. Materials and MethodsChickensOne-hundred twenty, day-old males and females egg-type brown leghorn chickens were used for this study. Thirty, one-day old chicks were first sexed into males and females (n=15/gender). These birds were euthanized and small intestine, large intestine, gizzard, heart, lung, spleen, and Peyer’s patches were collected and stored under deep freezing (-20°C) for further analyses. The remaining 90 chicks (male and female) were raised in a commingle floor pen (3 × 4 m on wood-shaving litter. All these birds had water and feed ad libitum. On day 7, the birds in the commingle floor pen were divided equally into three groups in three separated floor pens (1.2 × 1.5 m (n=30). The birds were weighed individually. Accordingly, the first group received 10 µg/ml/kg bodyweight (BW) of LPS via oral gavage; the second group received 1 ml of 0.09% sterile normal saline solution via oral gavage (representing the positive control group); and the third group received no treatment (representing the negative control group). Sample collectionAll birds were euthanized and dissected aseptically to collect samples. The first sample collection was before the LPS treatment (time 0). Samples included lungs, liver, small intestine, large intestine, gizzard, spleen and Peyer’s patches. The next sample collection was at 12 hours after the treatment with LPS. 10 birds per group were euthanized and the same organs were collected as with the first collection attempt. All samples were placed in sterile nylon bags and stored under deep freezing (-20°C). 24 hours after LPS treatment, the same protocol was applied. 10 birds per group were euthanized and the visceral organs were collected (see above). This procedure was reapplied 48 hours after LPS treatment for the remaining 10 birds. Bacterial LPSEscherichia coli (026:B6 strain)-derived LPS (Sigma-Aldrich, USA) was used as the challenging bacterial endotoxin (Bowen et al., 2009). 10 mg LPS stock was diluted with deionized distilled H2O (ddH2O) and the final dose was calculated as 10 µg/ml/kg BW. According to the BW of the birds, the LPS calculated dose was administered via oral gavage for each bird in the test group individually. The practice of bird gavaging was carried out using 18G animal gavage ball-end bent needle (cat# CAD7906-12EA, Sigma-Aldrich, USA). RNA isolation, quantitative real-time polymerase chain reaction (RT-PCR), and primers’ designTotal RNA was extracted using Trizol reagent (Life Technologies, Carlsbad, CA) according to the manufacturer’s instructions. The concentration and the quality of the extracted RNA were evaluated by using Take3 micro-volume plate and the Synergy HT multi-mode microplate reader (BioTek, Winooski, VT). RNA was assessed by using (A260/280) ratio, and those RNAs which were with ratio absorbance >1.8 were judged as high quality and high integrity RNAs. The RNA was loaded onto agarose gel electrophoresis for visual assessment. The reverse transcription of the RNA (1 μg) was done with qScript® cDNA Synthesis SuperMix (Quanta Biosciences, Qiagen, Beverly Inc.). The resulting cDNA was amplified by using qPCR (Applied Biosystems 7500 Real-Time System) with SYBR green master mix (Life Technologies, Carlsbad, CA) according to the manufacturer’s instructions. Relative quantification (RQ) of the mRNA expression of the NLRP3 gene was determined using the 2-ΔΔCT calculation (Pfaffl, 2004; Schmittgen and Livak, 2008). All readings were normalized to 18S ribosomal RNA expression as the housekeeping gene (Kuchipudi et al., 2012). Primers designed for mRNA of the NLRP3 and 18S rRNA genes were as described by Kuchipudi et al. (2012) and Karaffová et al. (2020) (Table 1). Primers and conventional PCR procedureFragments of mRNA of the NLRP3 gene and 18S ribosomal RNA (rRNA) were amplified using PCR with specific primers (Table 1). For both genes, PCR buffers and thermal cycling conditions are summarized in Tables 2 and 3. Amplicons were separated and were visualized by using SYBER Safe gel stain and loaded on a 1% agarose gel electrophoresis (Thermo-Fisher Scientific). Images were captured using the FluorChem M MultiFluor System (ProteinSimple, San Jose, CA). Statistical analysesData were analyzed using one-way analysis of variance (JMP software, SAS Inst. 2010). Significant differences were determined using Student’s t-test and Tukey’s HSD test (the experimental unit, n=10). Significances were represented as p ≤ 0.05. All data are shown as means ± SEM. Ethical approvalThe study and all tests, protocols, and procedures were approved by the scientific and animal care committee in the Department of Pathology and Poultry Disease, College of Veterinary Medicine, University of Baghdad. ResultsRelative NLRP3 gene expression in RT-PCR in male and female chicksThe first step of this experiment was designed to determine the possibility of the NLRP3 gene expression in chickens, and whether the gene was differentially expressed by gender-related manner. Our RT-PCR results showed that the NLRP3 gene was actually present in chickens and there were no sex-related differences at the mRNA level (time 0, Fig. 1). For the second hypothesis, however, there was an inequality in the distribution of the mRNA gene level among organs. Relative NLRP3 gene expression in RT-PCR among organs after stimulation with LPSAfter stimulation with bacterial LPS, the studied organs showed marked elevation in the NLRP3 gene level in comparison to the nonimmunized control groups when tested with RT-PCR. However, the significance levels were only in the small intestine and gizzard 12 hours pi (Fig. 2). These levels, however, started to deteriorate to lower RQ levels 24 hours pi (Fig. 3). The small intestine showed significantly higher levels in the NLRP3 gene expression therein. At 48 hours pi, small intestine, large intestine, lung, spleen and Peyer’s patches showed significantly higher levels of the NLRP3 gene expression. Conversely, all remaining organs were drastically reverted to a normal NLRP3 gene level 48 hours pi (Fig. 4). Table 1. Primers’ design for NLRP3 and 18S ribosomal RNA.
Table 2. Quantities for master mixes, supermix, cDNAs, F primers, and R primers for both NLRP3 and 18S rRNA genes.
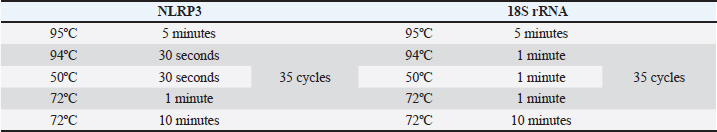
Table 3. Thermal cycles’ conditions for cDNA amplification. For both genes, the nucleic acids were first denatured at 95°C for 5 minutes. After that, 35 thermal cycles followed. Lastly, the final extension at 72°C was for 10 minutes.
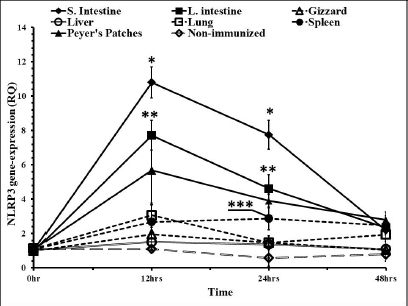
In time course measuring of the levels of the gene expressions, liver and gizzard were with no significant differences when compared to nonimmunized control groups. Peyer’s patches and spleen were with almost the same RQ values along the time course. Both were significantly higher in the RQ level at 12 and 24 hours pi (Fig. 5). The highest level of the NLRP3 gene expression was in the small intestine and large intestine at 12, 24, and 48 hours pi, respectively. Lungs showed higher levels in that gene only 12 hours pi. This level started to diminish 24 and 48 hours pi.
Fig. 1. The distribution of NLRP3 gene mRNA in some organs of male and female chickens. One-day-old male and female eggtype brown leghorn chickens were euthanized and small intestine, large intestine, gizzard, liver, and lung tissues were collected and homogenized to extract mRNA. Using RT-PCR, the expression of mRNA of the NLRP3 gene in these organs was determined.
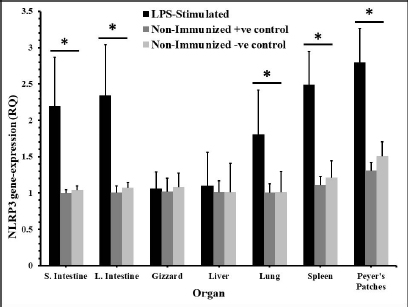
Fig. 2. The distribution of NLRP3 gene in some organs of male and female chickens after stimulation with LPS (12 hours pi). 7-day-old male and female egg-type brown leghorn chickens (n=10/group) were immunized with LPS (10 μg/kg BW). 12 hours later, the birds were euthanized and visceral organs’ tissues were collected (i.e., small intestine, large intestine, gizzard, liver, and lung). The tissues were subjected to the RNA extraction protocol. Using RT-PCR, the expression of the NLRP3 gene in these organs was determined. Asterisks represent significant differences (p ≤ 0.05).
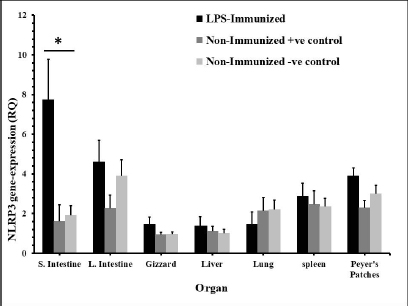
Fig. 3. The distribution of NLRP3 gene in some organs of male and female chickens after stimulation with LPS (24 hours). 7-dayold male and female egg-type brown leghorn chickens were immunized with LPS (10 μg/kg BW). The birds were euthanized 24 hours later and visceral organs’ tissues (i.e., small intestine, large intestine, gizzard, liver, and lung) were collected and homogenized to collect RNA. Using RT-PCR, the expression of the NLRP3 gene in these organs was determined. Asterisks represent significant differences (p ≤ 0.05).
Fig. 4. The distribution of the NLRP3 gene in some organs of male and female chickens after stimulation with LPS (48 hours). 7-day-old male and female egg-type brown leghorn chickens were immunized with LPS (10 μg/kg BW). The birds were euthanized 48 hours later and small intestine, large intestine, gizzard, liver, and lung tissues were collected and homogenized to collect RNA. Using RT-PCR, the expression of the NLRP3 gene in these organs was determined. Asterisks represent significant differences (p ≤ 0.05). DiscussionDistinct stimuli could activate innate immune responses. These stimuli examples are DAMPs and PAMPs. Both groups are ligands for specific receptors on innate immune cells called PRRs. The engagement of these receptors with cognate ligands leads to the stimulation and release of pro-inflammatory cytokines (Kepp et al., 2011). One of the well-studied effects of LPS (as a PAMP) on innate immunity is the consequent release of the pro-inflammatory cytokine (the NLRP3 inflammasome in particular). NLRP3 inflammasome is required for caspase-1 activation. Caspase-1 itself is essential in proteolytic maturation and in the profound release of pro-inflammatory IL-18 and IL-1β (Cornut et al., 2020). We sought to investigate the NLRP3 gene expression using molecular techniques such as Western blotting or histochemistry, but we could not pursue our goal due to the scarcity in the commercial NLRP3 anti-chicken monoclonal antibody. Instead, we investigated this gene at the mRNA level.
Fig. 5. Time course of the NLRP3 gene expression in egg-type brown leghorn chickens. 7-day-old male and female egg-type brown leghorn chickens were immunized with LPS (10 μg/kg BW). The birds were euthanized at 0, 12, 24, and 48 hours and small intestine, large intestine, gizzard, liver, and lung tissues were collected and homogenized to extract the RNA. Using RT-PCR, the expression of the NLRP3 gene in these organs was determined. Our results showed that there were no significant differences between males and females at the NLRP3 gene expression level (Fig. 1). At this young age, however, gender may not be very critical interfering factor in an innate immune response due to lack of sex hormones and other physiological factors among genders (Herrmann et al., 2017; Song et al., 2021). Within 12 hours pi with bacterial LPS, the small intestine showed a profoundly higher level in the NLRP3 mRNA. Gizzard, on the other hand, showed statistically significant levels too when compared to the control groups. Although the large intestine was only twofold less in the NLRP3 level in comparison to small intestine, the differences were insignificant to the control groups. This could be attributed to a nonspecific potential concurrent active inflammatory process at this time. In general, intestines were expressing high levels of NLRP3 due to the presence of high density innate immune cells (Shira and Friedman, 2018) (Fig. 2). Twenty-four hours pi with LPS, the NLRP3 expression level was almost constant in small and large intestines. However, the level in the large intestine was insignificant. Other organs seemed to be with normal RQ levels (Fig. 3). In the last investigation, i.e., 48 hours pi with LPS, small and large intestines, lung, spleen, and Peyer’s patches remained with the highest and significant levels of the NLRP3 mRNA (Fig. 4). The course of the immune response (Fig. 5) showed dominance in the NLRP3 mRNA expression in the small intestine. Similarly, but less profoundly, the large intestine expressed NLRP3 mRNA 12 and 24 hours pi with LPS. Peyer’s patches and spleen showed lower gene expression at 12 and 24 hours; however, the levels were significant in comparison to other organs. Conflict of interestThe authors declare that there is no conflict of interest. Author contributionsThe authors contributed equally to this manuscript. ReferencesBowen, O.T., Dienglewicz, R.L., Wideman, R.F. and Erf, G.F. 2009. Altered monocyte and macrophage numbers in blood and organs of chickens injected i.v. with lipopolysaccharide. Vet. Immunol. Immunopathol. 131, 200–210. Cornut, M., Bourdonnay, E. and Henry, T. 2020. Transcriptional regulation of inflammasomes. Intl. J. Mol. Sci. 21, 8087; doi:10.3390/ijms21218087 Herrmann, M.S., Gallardo, R.A., Bunn, D.A., Bunn, D.A., Kelly, T.R. and Dekkers, J.C.M. 2017. Does gender impact the immune response of chicks? Animal Industry Report: AS 663, ASL R3175. Ames, IA: Iowa State University; doi:10.31274/ans_air-180814-343 Kanneganti, T. 2020. Intracellular innate receptors: life inside the cell. Immunol. Rev. 297, 5–12. Karaffová, V., Revajová, V., Koščová, J., Gancarčíková, S., Nemcová, R., Ševčíková, Z., Herich, R. and Levkut Sr, M. 2020. Local intestinal immune response including NLRP3 inflammasome in broiler chicken infected with Campylobacter jejuni after administration of Lactobacillus reuteri B1/1. Food Agric. Immunol. 31(1), 954–966. Kelley, N., Jeltema, D., Duan, Y. and He, Y. 2019. The NLRP3 inflammasome: an overview of mechanisms of activation and regulation. Int. J. Mol. Sci. 20, 3328; doi:10.3390/ijms20133328. Kepp, O., Galluzzi, L. and Kroemer, G. 2011. Mitochondrial control of the NLRP3 inflammasome. Nature Immunol. 12(3), 199–200. Kuchipudi, S.V., Tellabati, M., Nelli, R.K., White, G.A., Perez, B.B., Sebastian, S., Slomka, M.J., Brookes, S.M., Brown, I.H., Dunham, S.P. and Chang, K. 2012. 18S rRNA is a reliable normalization gene for real time PCR based on influenza virus infected cells. Virol. J. 9, 230. Available via http://www.virologyj.com/content/9/1/230 Liu, J. and Cao, X. 2016. Cellular and molecular regulation of innate inflammatory response. Cell Mol. Immunol. 13, 711–721. Muñoz-Carrillo, J.L., Contreras-Cordero, J.F., Gutiérrez-Coronado, O., Villalobos-Gutiérrez, P.T., Ramos-Gracia, L.G. and Hernández-Reyes, V.E. 2018. Cytokine profiling plays a crucial role in activating the immune system to clear infectious pathogens. In Immune response activation and immunomodulation. Eds., Tyagi, R. and Bisen, P.S., pp: 1–30; InTechOpen, London, UK. doi: 10.5772/intechopen.8084 Neerukonda, S.N. and Katneni, U. 2020. Avian pattern recognition receptor sensing and signaling. Vet. Sci. 7, 14; doi:10.3390/vetsci7010014 Nicholson, L.B. 2016. The immune system. Essay Biochem. 60, 275–301; doi:10.1042/EBC20160017 Paik, S., Kim, J.K., Silwal, P., Sasakawa, C. and Jo, E. 2021. An update on the regulatory mechanisms of NLRP3 inflammasome activation. Cell Mol. Immunol. 18, 1141–1160. Pfaffl, M.W. 2004. Quantification strategies in real-time PCR. In A-Z of quantitative PCR. Eds., Bustin, S.A., La Jolla, CA: International University Line (IUL), pp: 87–112. Roh, J.S. and Sohn, D.H. 2018. Damage-associated molecular patterns in inflammatory diseases. Immune NetW. 18(4), e27; Schmittgen, T. and Livak, K. 2008. Analyzing real-time PCR data by the comparative CT method. Nat. Protoc. 3, 1101–1108. Shira, E.B. and Friedman, A. 2018. Innate immune functions of avian intestinal epithelial cells: response to bacterial stimuli and localization of responding cells in the developing avian digestive tract. PLoS ONE 13(7), e0200393; doi: 10.1371/journal.pone.0200393 Song, B., Tang, D., Yan, S., Fan, H., Li, G., Shahid, M.S., Mahmood, T. and Guo, Y. 2021. Effects of age on immune function in broiler chickens. J. Ani. Sci. Biotech. 12, 42; doi:10.1186/s40104-021-00559-1 Vrentas, C.E., Schaut, R.G., Boggiatto, P.M., Olsen, S.C., Sutterwala, F.S. and Moayeri, M. 2018. Inflammasomes in livestock and wildlife: insights into the intersection of pathogens and natural host species. Vet. Immunol. Immunopathol. 201, 49–56. | ||
| How to Cite this Article |
| Pubmed Style Al-Ogaili AS, Hameed SS, Noori N, . LPS-induced NLRP3 gene-expression in chicken. Open Vet J. 2022; 12(2): 197-203. doi:10.5455/OVJ.2022.v12.i2.7 Web Style Al-Ogaili AS, Hameed SS, Noori N, . LPS-induced NLRP3 gene-expression in chicken. https://www.openveterinaryjournal.com/?mno=136604 [Access: July 27, 2024]. doi:10.5455/OVJ.2022.v12.i2.7 AMA (American Medical Association) Style Al-Ogaili AS, Hameed SS, Noori N, . LPS-induced NLRP3 gene-expression in chicken. Open Vet J. 2022; 12(2): 197-203. doi:10.5455/OVJ.2022.v12.i2.7 Vancouver/ICMJE Style Al-Ogaili AS, Hameed SS, Noori N, . LPS-induced NLRP3 gene-expression in chicken. Open Vet J. (2022), [cited July 27, 2024]; 12(2): 197-203. doi:10.5455/OVJ.2022.v12.i2.7 Harvard Style Al-Ogaili, A. S., Hameed, S. S., Noori, N. & (2022) LPS-induced NLRP3 gene-expression in chicken. Open Vet J, 12 (2), 197-203. doi:10.5455/OVJ.2022.v12.i2.7 Turabian Style Al-Ogaili, Adil Sabr, Samer Sadeq Hameed, Noor Noori, and . 2022. LPS-induced NLRP3 gene-expression in chicken. Open Veterinary Journal, 12 (2), 197-203. doi:10.5455/OVJ.2022.v12.i2.7 Chicago Style Al-Ogaili, Adil Sabr, Samer Sadeq Hameed, Noor Noori, and . "LPS-induced NLRP3 gene-expression in chicken." Open Veterinary Journal 12 (2022), 197-203. doi:10.5455/OVJ.2022.v12.i2.7 MLA (The Modern Language Association) Style Al-Ogaili, Adil Sabr, Samer Sadeq Hameed, Noor Noori, and . "LPS-induced NLRP3 gene-expression in chicken." Open Veterinary Journal 12.2 (2022), 197-203. Print. doi:10.5455/OVJ.2022.v12.i2.7 APA (American Psychological Association) Style Al-Ogaili, A. S., Hameed, S. S., Noori, N. & (2022) LPS-induced NLRP3 gene-expression in chicken. Open Veterinary Journal, 12 (2), 197-203. doi:10.5455/OVJ.2022.v12.i2.7 |