| Research Article | ||
Open Vet J. 2023; 13(5): 588-598 Open Veterinary Journal, (2023), Vol. 13(5): 588–598 Original Research First study on molecular epidemiology of caseous lymphadenitis in slaughtered sheep and goats in Duhok Province, IraqRamadhan Ado Khanamir1, Nawzat Abozaid Issa1* and Rezheen Fatah Abdulrahman21Surgery and Internal Medicine Department, College of Veterinary Medicine, University of Duhok, Duhok, Iraq 2Pathology and Microbiology Department, College of Veterinary Medicine, University of Duhok, Duhok, Iraq *Corresponding Author: Nawzat Abozaid Issa. Surgery and Internal Medicine Department, College of Veterinary Medicine, University of Duhok, Duhok, Iraq. Email: nawzat.issa [at] uod.ac Submitted: 22/02/2022 Accepted: 11/04/2023 Published: 12/05/2023 © 2023 Open Veterinary Journal
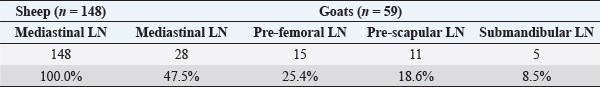
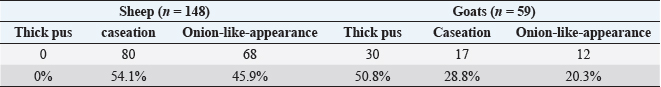
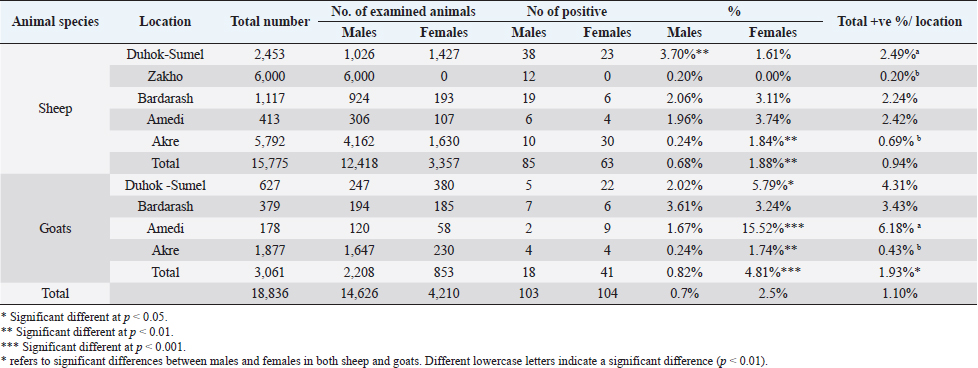
AbstractBackground: Caseous lymphadenitis (CLA) is a chronic suppurative bacterial infection caused by Corynebacterium pseudotuberculosis (C. pseudotuberculosis) affecting superficial and internal lymph nodes and internal organs of small ruminants. Aims: Through the use of molecular methods, this study aimed to estimate the prevalence of CLA and its contributing factors as well as the degree of genetic diversity and epidemiological relationships among C. pseudotuberculosis isolates from slaughtered sheep and goats in various districts of Duhok Province, Iraq. Methods: A total of 18,836 carcasses (15,775 sheep and 3,061 goats) were inspected by veterinarians at slaughterhouses [Duhok-Sumel (2,453 sheep + 627 goats), Zakho (6,000 sheep), Bardarash (1,117 sheep + 379 goats), Amedi (413 sheep + 178 goats) and Akre (5,792 sheep + 1,877 goats)] in Duhok Province for detection the prevalence rate of CLA using molecular techniques. Results: The prevalence of the disease was 0.94% and 1.93% in sheep and goats, respectively. Sheep in Duhok-Sumel and goats in Amedi were at a higher risk of infection than the animals in another location, with a prevalence rate of 4.31% and 6.18%, respectively. Sheep and goats of older age were more susceptible. Females were more susceptible than males in all districts except Duhok-Sumel where the reverse was true. ERIC-PCR analysis grouped the bacterial isolates into 11 different genotypes. The maximum likelihood phylogenetic tree of partial sequences of the 16S rRNA gene sequences of C. pseudotuberculosis revealed no divergent sequences discovered in this study. Conclusion: A strict control program needs to be applied to reduce the entrance of pathogen from neighboring countries. Keywords: Corynebacterium pseudotuberculosis, Sheep and goats, ERIC-PCR, Duhok, Iraq. IntroductionCaseous lymphadenitis (CLA) is a chronic debilitating bacterial infection caused by Corynebacterium pseudotuberculosis manifests as abscesses in internal organs, particularly the lung, as well as lymph nodes throughout the body (Algammal, 2016; Yitagesu et al., 2020). CLA in sheep and goats is a cosmopolitan bacterial infection that lacks effective control measures and has a poor response to treatment. It can persist in the environment, and it is difficult to detect subclinically infected animals (Abebe and Sisay Tessema, 2015). Once the disease is introduced into a flock of sheep or goats, there are no effective control measures in place (Abebe and Sisay Tessema, 2015; Karthik and Prabhu, 2021). The infection results in significant financial losses for sheep and goats industries through reducing milk production, reproductive problems for the affected animals, decreasing wool and meat production, condemnation of portions to the whole carcasses of the affected animals at abattoirs (Ruiz et al ., 2020; Bettini et al., 2022). Because of the lack of a reportable condition, the owners’ ignorance of the disease’s economic significance, and the fact that they seldom seek veterinary care for the disease’s superficial form and abscess development, it is unknown in many countries how common CLA is in sheep and goats (Costa et al., 2020). CLA in small ruminants is diagnosed using a variety of techniques and methods, including clinical signs and characteristics of the lesions, isolation, and identification of the causative agent from pus discharge (Nassar et al., 2016), serological methods (Yitagesu et al., 2020), and PCR (Issa et al., 2021; Terab et al., 2021) which is the most reliable, accurate, and rapid method, and it shows high resolution and repeatability in the genetic discrimination of bacterial strains (Taha, 2022). Accordingly, this study aimed to estimate the prevalence rate of CLA among slaughtered sheep and goats and to determine the role of different associated risk factors such as the animals’ species, age, and sex, as well as the geographical distribution of the disease in different locations in Duhok Province, Iraq. In the context of CLA, nothing, as yet, has been reported in Duhok Province on the prevalence rate of the disease among slaughtered sheep and goats using molecular techniques. In addition, it was unclear what risk factors were linked to the disease’s prevalence and to what extent they may contribute to the disease’s transmission in sheep and goats. Materials and MethodsStudy areasThe research was carried out from June 2021 to October 2022 to estimate the prevalence rate of CLA through bacteriological and molecular techniques in sheep and goats slaughtered at different abattoirs in five districts (Zakho, Duhok-Sumel, Bardarash, Amadi, and Akre) of Duhok Province of Iraq. Duhok is a governorate that lies in the northwest of Iraq and Western part of Kurdistan Region about 470 km north of Baghdad. Duhok has a strategic location since it is considered being a point of joint among three countries (Syria- Turkey- Iraq). Sampling and samples collectionDuring the study period, a total of 18,836 carcasses (15,775 sheep and 3,061 goats) were inspected by veterinarians at slaughterhouses in the study area for detection of CLA with strict adherence to the standard necropsy procedure applied at the local abattoirs. In the inspected animals, mediastinal-, pre-scapular, pre-femoral-, and submandibular lymph nodes were carefully screened for grossly visible abnormalities and texture. In total, 560 samples (enlarged lymph nodes) from sheep (n=399) and goats (n=161) were aseptically removed, collected, and delivered in clean, sterile specimen containers kept in a cool box to the Duhok Research Center at the College of Veterinary Medicine, University of Duhok, Iraq, for gross examination and laboratory investigation. During sample collection, the age and sex of slaughtered animals were recorded by using a preformed questionnaire. Besides, the location and size of lymph nodes were also considered. Isolation and molecular identification of C. pseudotuberculosisTo isolate the bacteria, the pus was first aseptically removed and collected after the surface of infected lymph nodes was cleansed with 70% ethanol, and then it was excised with a sterile disposable surgical blade and cultured onto 5% sheep blood agar. The plates were incubated for 24–48 hours at 37°C. Gram stain, colony shape, and hemolytic activity of the growing bacteria were among the usual microbiological techniques used to initially identify the growth. To gain pure colonies, suspect colonies of C. pseudotuberculosis were chosen, streaked onto a blood agar plate, and then transferred into brain-heart infusion broth containing 50% (v/v) glycerol. These colonies were then refrigerated at −20°C for future investigation. Molecular detection of C. pseudotuberculosis by PCRPCR was used to confirm the identity of the isolated C. pseudotuberculosis. According to Issa et al. (2021), DNA was extracted using the thermal lysis technique. Briefly, isolates were plated out onto 5% sheep blood agar and incubated at 37°C for 48 hours using −20°C glycerol stocks. Five colonies were suspended and thoroughly mixed in 500 µl of distilled water in an Eppendorf tube. In the heating block, thermal lysis was carried out for 15 minutes. The suspension was immediately chilled on ice for 5 minutes, and then centrifuged at 13,000 rpm for 5 minutes. The prepared DNA was kept at −20°C for PCR after being tested for purity using the NanoDrop 2000C spectrophotometer by Thermo Scientific UK. According to Çetinkaya et al. (2002) and Pacheco et al. (2007) the PCR conditions and primers targeting pld genes and 16S rRNA of C. pseudotuberculosis were designed. To amplify the pld gene, a total reaction volume of 25 µl was used, with each reaction containing 12.5 µl of master mix (Ruby Taq Master®, Jena Bioscience, Thuringia, Germany), 2.5 µl of DNA template (Macrogen, South Korea), 1 µl of each forward and reverse primer at a concentration of 10 pmol l−1 (PLD-F: 5´ ATAAGCGTAAGCAGGGAGCA ´3; PLD-R2: 5´ ATCAGCGGTGATTGTCTTCCAGG ´3) and 8 µl of dH2O. Thermo Cycler Gene Amp PCR System 9700 (Applied Biosystems) was used to conduct the reactions. For PCR product, 35 cycles of the following parameters were used: initial denaturation at 94°C for 3 minutes, denaturation at 94°C for 1 minute, annealing at 56°C for 1 minute, extension at 72°C for 90 seconds, and final extension at 72°C for 7 minutes. A 1.5% (w/v) agarose gel containing Prime Safe Dye (GeNet Bio, Korea) alongside DNA ladder (50 bp) (GeneDireX, Inc. Taiwan) was used for electrophoresis to separate the amplified products and then visualized by UV light and amplicons of 203 bp were taken as positive for pld genes of C. pseudotuberculosis. For sequencing, 20 representative positive PCR samples (16 isolates from sheep and 4 isolates from goats) were tested against 16S rRNA which was amplified using a total reaction volume of 50 µl, each reaction was consisted of 5 µl of template DNA, 2 µl of each forward and reverse primer at a concentration of 10 pmol µl−1 (16S-F: 5´ ACCGCACTTTAGTGTGTGTG ´3; 16S-R: 5´ TCTCTACGCCGATCTTGTAT -´3), 25 µl of ready-to-use master mixes (Ruby Taq Master®, Jena Bioscience, Thuringia, Germany), and 16 µl dH2O. The following amplification parameters of 35 cycles were used in a Gene Amp PCR System 9700 Thermo Cycler (Applied Biosystems) to perform the amplification: initial denaturation at 94°C for 3 minutes, denaturation at 94°C for 1 minute, annealing at 56°C for 1 minute, extension at 72°C for 2 minutes, and a final extension step at 72°C for 7 minutes. 1% (w/v) agarose gel containing Prime Safe Dye (GeNet Bio, Korea) alongside DNA ladder (50 bp) (GeneDireX, Inc. Taiwan) was used for electrophoresis to separate the amplified products. The fragment sizes were visualized under UV light and amplicons of 815 bp were taken as positive for 16S rRNA of C. pseudotuberculosis. The PCR products of 16S rRNA were stored refrigerated at 4°C until they were collected; for purification and sequencing, 40 µl of amplified material were sent to a commercial company (Macrogen Inc., South Korea). To create a phylogenetic tree, partial sequences of the 16S rRNA gene of C. pseudotuberculosis isolates from different study locations were aligned using a multiple sequence alignment in the MEGA 11 version. The resulting sequences were compared using the muscle technique to sequence accession numbers obtained from NCBI GenBank. The phylogenetic tree was constructed using the maximum likelihood technique based on the Tamura-Nei Model, and node reliability was calculated using 1,000 bootstrap repetitions. ERIC-PCR fingerprintingTo identify related strains and distinguish between distinct strains, selected isolates of C. pseudotuberculosis were subjected to ERIC-PCR using the primer sequences ERIC1: 5´-ATGTAAGCTCCTGGGGATTCAC 3´ and ERIC2: 5´-AAGTAAGTGACTGGGGTGAGCG-3´ as described by Versalovic et al. (1991). The ERIC-PCR was carried out in a total volume of 25 µl. Each reaction was made of 12 µl of hot start premix (Genedirex, Taiwan), 1 µl of each primer (10 pmol), 2 µl of sample DNA (30–100 ng/l), and 9 µl nuclease-free water (Qiagen, Germany). According to the PCR program used by Bakhshi et al. (2018), the reaction was carried out using the 9700 GeneAmp PCR system (Applied Biosystems, USA) as follows: initial denaturation for 5 minutes at 94°C followed by 35 cycles of repeated steps of denaturation at 94°C for 1 minute, annealing at 54°C for 1 minute, extension at 72°C for 5 minutes, and final extension at 72°C for 10 minutes. On a 1% agarose gel with Prime Safe Dye (GeNet Bio, Korea) at 70 volts for 10 minutes and later at 80 volts for 1 hour, the PCR products were visualized. The GelJ gel analysis program (v. 2.0) was used to analyze the results of ERIC-PCR. The DICE similarity coefficient and a position tolerance of 1.0 were used in the unweighted pair group method with arithmetic averages (UPGMA) approach to create the dendrogram (Heras et al., 2015). Statistical analysisGraphPad Prism software version 8.0.1 was used to carry out the statistical analysis of the data. The statistically significant differences between the variable factors linked with the prevalence of the disease in the examined sheep and goats were determined using the chi-squared and Fisher’s exact tests. When p < 0.05, differences were significant. The collected data were expressed descriptively using the disease prevalence percentages. Ethical approvalThis study was approved by the Local Ethics Committee sets at the College of Veterinary Medicine, University of Duhok (Permit number: VM2021/1560UD). ResultsIsolation and molecular identification of C. pseudotuberculosis isolates from slaughtered sheep and goatsOn blood agar plates, the collected samples were cultured, followed by macroscopic, microscopic, and biochemical testing approaches for initial identification of C. pseudotuberculosis, and then followed by molecular identification. Macroscopically, during 24 hours, colonies were white, tiny, dry, and non-hemolytic; at 48 hours, they were light white to creamy, crumbly, and encircled by a constrictive zone of beta-hemolysis. Gram-stained C. pseudotuberculosis colonies revealed Gram-positive, pleomorphic, club-like bacilli. The isolates were positive for catalase and urease tests. The isolates were further verified by PCR using the pld gene unique to C. pseudotuberculosis; a DNA fragment of 200 bp (Fig. 1) was amplified successfully in isolated C. pseudotuberculosis. The prevalence rates of C. pseudotuberculosis in sheep and goats were 0.94% and 1.93%, respectively. The prevalence rate in goats was significantly (p < 0.001) higher than that in sheep (Table 1). Gross lesions of CLA in infected sheep and goatsIn sheep, gross lesions of CLA were only observed in mediastinal lymph nodes. Whereas, in goats, CLA lesions were observed in mediastinal-, pre-femoral-, pre-scapular-, and submandibular lymph nodes at rates of 47.5% (28/59), 25.4% (15/59), 18.6% (11/59), and 8.5% (5/59), respectively (Table 2). Lymph nodes with CLA showed differences within sizes, ranging from 1 to 15 cm (Fig. 2), with an average of 4.6 cm. The surface of CLA lymph nodes was smooth, while a cut section revealed a rough surface. CLA lymph nodes also revealed texture differences, including thick pus and distinct caseonecrotic dystrophic to calcified masses. The textures were white to grayish. The central zone of the lymph nodes differed as well; in sheep, 54.1% (80/148) had caseation and 45.9% (68/148) had an onion-like appearance. Whereas in goats, the rates of CLA lymph nodes with thick pus, caseation, and onion-like appearance were 50.8%, 28.8%, and 20.3%, respectively (Table 3). Risk factors associated with the prevalence of CLA in sheep and goatsAmong the examined sheep and goats, the factors linked with the occurrence of the infection were considered and analyzed (Table 4). Concerning age, the CLA was only reported in animals older than 1-year-old in both sheep and goats. In terms of the geographical distribution of the disease, the highest rate of the disease in sheep was reported in Duhok-Sumel, which was significantly (p < 0.01) higher than that in Zakho and Akre. Whereas, in goats, the highest prevalence rate was reported in Amadi at a rate of 6.18% (11/178) and the lowest rate of CLA was reported in Akre at 0.43% (8/1887). In terms of gender, the rates of prevalence were significantly higher (p < 0.01) in females than males in both sheep and goats. However, there were differences in the prevalence of disease between the genders in the study locations; in Duhok-Sumel, in sheep; the prevalence of the disease was significantly higher (p < 0.01) in males than females, and vice versa in goats.
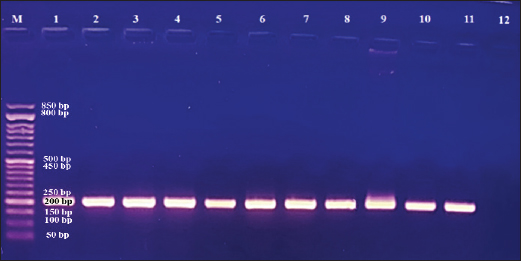
Fig. 1. Amplification of pld gene specific for C. pseudotuberculosis. Lane M: DNA marker, lanes 12: Negative control with water, Lane 1–11: amplified pld (203 bp) gene in isolated C. pseudotuberculosis. Table 1. The isolation rates of C. pseudotuberculosis from inspected slaughtered sheep and goats in abattoirs of Duhok Province, Iraq, based on PCR.
Table 2. Rates of gross lesions of CLA in collected lymph nodes of affected sheep and goats slaughtered at different abattoirs in Duhok Province, Iraq.
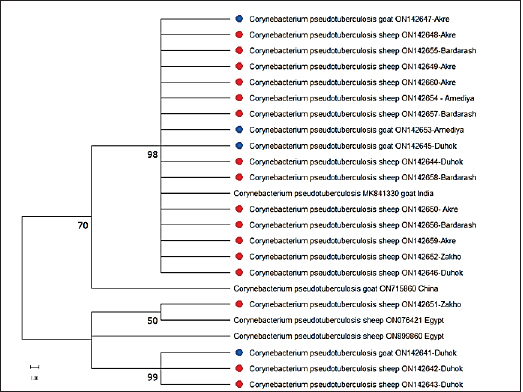
For sequencing, 20 PCR-confirmed isolates (16 sheep isolates and 4 goat isolates) were randomly selected and tested against 16S rRNA, which was successfully amplified by PCR at 815 bp. Based on the nucleotide sequence comparison of partial 16S rRNA sequences, BLAST analysis against globally published sequences of the bacteria revealed that sequence similarity with C. pseudotuberculosis is 100%. The sequences were added to the NCBI GenBank, with accession numbers (ON142641, ON142645, ON142647, and ON142653) for goat isolates, and (ON142642, ON142643, ON142644, ON142646, ON142648, ON142649, ON142650, ON142651, ON142652, ON142654, ON142655, ON142656, ON142657, ON142658, ON142659, and ON142660) for sheep isolates. The C. pseudotuberculosis sequences were used using MEGA-11 to create a maximum likelihood phylogenetic tree. The C. pseudotuberculosis bootstrap consensus tree was derived from 1,000 repetitions with the greatest log probability (−6,309.64). The C. pseudotuberculosis phylogenetic tree demonstrates that the sequences (ON142647, ON142653, and ON142645) from goats and (ON142648, ON142655, ON142649, ON142660, ON142654, ON142657, ON142644, ON142658, ON142650, ON142656, ON142659, ON142652, and ON142646) from sheep were clustered and were in cluster with a sequence previously detected from India (MK841330, goat isolate), and have a clade with a sequence of an isolate from sheep from China (ON715860) and the sequence of (ON142651, sheep isolate) was clustered with a sequence previously detected from Egypt (ON076421). Whereas, the sequences (ON142642, ON142643) from sheep and (ON142641) from goat were cladded with (ON899860, sheep) from Egypt and with that isolated from sheep from Zakho (ON142651, sheep). The data revealed that out of 20 sequenced isolates, 16 isolates were clustered in a clade, the other 4 isolates were also clustered in a clade, and the 2 clades were linked. No divergent sequences were discovered in this study (Fig. 3). Genetic diversity using ERIC-PCR analysisThe ERIC-PCR fingerprinting analysis, based on the variations in the number and size of ERIC sequences in each isolate, revealed that the genetic similarity among the isolates was between 52% and 100%, and the isolates were grouped into 11 different genotypes, from 1 to 11, depending on the 90% cutoff similarity coefficient. The data showed that genotypes 5, 8, and 9 were the most common clones and variants among the isolates, accounting for 12/20 (60%) of the total isolates. Six strains were included in genotype 5; genotype 8 included four stains, and genotype 9 with only two stains, while the remaining genotypes (1, 2, 3, 4, 6, 7, 10, and 11) represented only one C. pseudotuberculosis isolate as shown in (Fig. 4). The identification of 11 different ERIC types in this study it may indicate the diversity of this pathogen in the area. Concerning the animal species and the diversity of the isolated strains, the data found that 9 out of the 20 isolates at a rate of 45% were from sheep with genetic similarity of 56%, where strain 14 was clustered to strains 13, 16, 15, 12 and 11, and strain 9 to strains 8 and 6, Whereas, isolates from goats were diverse and were not clustered.
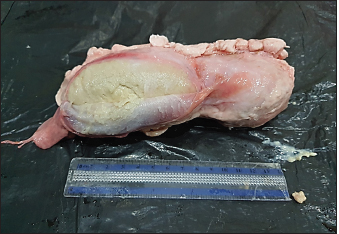
Fig. 2. CLA of mediastinal lymph nodes in infected sheep, size 15 cm. Regarding the geographic distribution of the isolates, the data revealed a relative genetic similarity (grouped under the same genotype or ERIC type), such as (strain No. 14, 13, 16, 15, 12, and 11) from Amedi, Bardarash, and Zakho, (strain No. 9, 8, 7, and 1) from Akre and Duhok, (strain No. 5 with 4) from Duhok. DiscussionThe current study employed molecular methods to estimate the prevalence rate of the disease among slaughtered sheep and goats, and it assessed how risk variables affected the disease’s prevalence. The data found that the rates of CLA in sheep and goats were 0.94% and 1.93%, respectively. These findings are in line with prevalence rates that were reported in slaughtered goats (1.1%) at the local slaughterhouses in Elazig Province, in the east of Turkey (Çetinkaya et al., 2002) and 2.4% in clinically examined goats in Rajasthan, India (Kumar et al., 2012) and in line with the prevalence rate (1.93%) in sheep slaughtered in Sumel district of Duhok Province, Iraq (Issa et al., 2021). Whereas, higher rates of CLA in sheep (26.3%) and goats (45.5%) were reported by Algammal (2016) in Menoufia Governorate, Egypt, and Rizk et al. (2019) who reported the prevalence rate of the disease (13%) in sheep and goat in Beni-Suef Governorate in Egypt. The lower prevalence rates of the infection observed in sheep and goats in the present study are probably due to better management practices and the climatic conditions of the region. The data also revealed that the prevalence of the diseases was significantly higher in goats than in sheep, which is in line with what was reported by Abebe and Sisay Tessema (2015). The higher rate in goats than in sheep is probably because of the variation in management systems and the environmental conditions where sheep and goats were raised. Goats are commonly raised in mountainous regions, whereas sheep are commonly found in plain areas; the mountainous areas of the study areas are colder; the ambient temperature, which may also be attributed to the disease’s endemic nature, significantly influences the survival of the causal agent in the contaminated environment (Augustine and Renshaw, 1986; Sá et al., 2018). Higher environmental temperatures may alter animal immunity and make animals more susceptible to infectious diseases (Al-Gaabary et al., 2009). In addition, goats may scrape their bodies against hard things often, which leads to a high incidence of superficial parotid, mandibular, cervical, and pre-femoral lymph node infections since they drain the shoulder area (Fikre and Abraha, 2014). Table 3. Texture of the affected lymphnodes with CLA in sheep and goats.
Table 4. The prevalence rates of CLA and the risk factors associated with the prevalence of the disease in slaughtered sheep and goats.
Fig. 3. Phylogenetic tree of C. pseudotuberculosis isolates based on partial sequences of 16S rRNA. The blue circles the goat’s strains and the red circles the sheep strains of this work. The numbers near the nodes indicate bootstrap values. In sheep, gross lesions of CLA were only observed in mediastinal lymph nodes. These findings agreed with those of other researchers who reported that the lungs and mediastinal lymph nodes were the two most often infected visceral organs by C. pseudotuberculosis (Oreiby, 2015; Ruiz et al., 2020). Whereas, in goats, CLA lesions were commonly observed in mediastinal- followed by, pre-femoral-, pre-scapular-, and submandibular lymph nodes. The higher rates of mediastinal lymph nodes are probably because of the age of the slaughtered animals; developing internal lesions of CLA increased with age (Oreiby, 2015). The high frequency of external lesions indicates a high risk of transmission that also supports our data of higher prevalence rates in goats than sheep in this study. The reason for increases in superficial lymph is not well understood, but they may be associated with the management system of the goats in mountainous areas, which increases skin abrasions that might facilitate the organism’s entry and could be common in the area. It has been reported that rupture of the granuloma increases the risk of contagion by eight times in those susceptible, since the released caseous content keeps the agent in the environment (Andrade et al., 2012). CLA lesions also revealed texture differences, including the presence of thick pus and distinct caseonecrotic dystrophic to calcified masses. The central zone of the lymph nodes differed as well; in sheep, the rates of lymph nodes with caseation was 50% (68/136) and onion-like appearance was 50% (68/136) which were almost twice that reported in goats with caseation (28.8%), and onion-like-appearance (20.3%). The variation in the rates between the sheep and goats is probably because of the location of the lesions, where caseation “onion ring” which is caused by a process of repetitive necrosis of the lesion capsule by the bacterial growth, followed by capsule reformation as a defense mechanism to limit the infection, is commonly found in mediastinal lymph nodes (Al-Gaabary et al., 2010; Ruiz et al., 2020). Factors linked with the occurrence of the disease in the investigated sheep and goats were considered and assessed. In terms of age, CLA was only found in both sheep and goats of older than 1 year. These findings are consistent with what has been published by Kaba et al. (2011) and Didkowska et al. (2020). The higher rates in older sheep and goats compared to younger animals are most likely owing to the older animals’ increased risk of bacterial infection, which increases CLA infection rates (Ali et al., 2016). Regarding the geographical distribution of the disease, variations were reported in the prevalence rates in sheep, the highest rate of the disease in sheep was reported in Sumel which is probably due to the intensive sheep farming system were practiced in the district. CLA has been observed to be more common in places where intensive husbandry is practiced (Abebe and Sisay Tessema, 2015). Whereas, in goats, the highest prevalence rate was reported in Amadi, and the lowest rate of CLA was reported in Akre. The higher rate in Amedi is probably because of the mountainous nature of the area and its climatic conditions compared to Akre and the other study locations.
Fig. 4. Dendrogram created from ERIC-PCR banding pattern of 20 C. pseudotuberculosis isolates from pus samples taken from slaughtered sheep and goats that have been diagnosed with positive CLA. The similarity analysis was performed using dice coefficient and UPGMA method in GelJ software (v.2.0). Concerning sex, in general, females had a greater prevalence rate than males in both sheep and goats. However, differences in the prevalence of disease between study locations were found in terms of gender. The higher rates in females are probably due to the fact that females of sheep and goats in the study area were usually kept for s longer period than males for breeding purposes which exposes females to predisposing factors like pregnancy, shearing, and interaction with sick animals, as previously reported by Yitagesu et al. (2020) and supported by Issa et al. (2021). Whereas, our data found that the prevalence rate in sheep in Sumel was significantly higher (p < 0.01) in rams than ewes which is probably due to the intensive husbandry where rams physically interact more than ewes during the mating season, the risk of pressing on and rupturing abscesses increases. Furthermore, the quantity of acute-phase proteins, primarily haptoglobin, is significantly higher in males than in females in response to C. pseudotuberculosis; it has been suggested that the development of CLA may be influenced by increased haptoglobin levels in sheep in response to the bacterium (Eckersall et al., 2007; Jeber et al., 2016). However, CLA, on the other hand, is a non-sex-linked illness, implying that its occurrence is based on immunity, which may be influenced by gestation, breastfeeding, immunosuppressive disease, diet, and other management variables that affect both sexes (Oreiby, 2015). We generated a phylogenetic tree using maximum likelihood to verify the phylogenetic relationship of C. pseudotuberculosis isolates in this study and other strains of C. pseudotuberculosis isolated in previous studies by researchers. The phylogenetic tree of C. pseudotuberculosis showed that the sequences from goats and sheep were clustered and were in cluster with a sequence previously detected from India, China, and Egypt. This is most likely because they came from the same source and might have been transmitted by diseased animals; the boundaries of the study area are not well controlled and the animals’ transportation is done by smuggling because of the huge difference in animal prices between these areas. These findings were further confirmed by the ERIC-PCR fingerprinting analysis, as mentioned below. The ERIC-PCR fingerprinting analysis was used to assess the genetic diversity and similarity of 20 strains of C. pseudotuberculosis isolated from slaughtered sheep and goats with CLA in the five districts of Duhok Province. The data revealed a high genetic diversity among the isolated C. pseudotuberculosis strains (11 genotypes) which probably show many clones of C. pseudotuberculosis circulate in our region, possibly from different geographic regions, as Duhok Province receives animals for slaughter from various locations (both native and imported from nearby counties). The spread of a large number of bacterial genotypes into an area resulted from the entrance of animals from various locations (Guimarães et al., 2011; Sellyei et al., 2017). Strain variations among microbial communities occur as a result of the movement of bacteria between different hosts or ecosystems, certain DNA sequences may be added or deleted, causing mutations (Jerome et al., 2011). The similarity between the isolates from the different locations of the study area is probably because the infected animals transferred between the locations for pasturing particularly during the drought seasons, these findings are in line with that reported earlier by Taha (2022). Different genes, such as 16s rRNA (Çetinkaya et al., 2002) and rpoB genes (Khamis et al., 2004; Abdulrahman et al., 2020; Abdulrahman, 2021), have been used to generate a phylogenetic tree in C. pseudotuberculosis isolated from CLA cases in sheep and goats. According to the phylogenetic analysis of 16s rRNA gene sequences, out of 20 isolates of C. pseudotuberculosis, 16 isolates were genetically identical clones, which is not in line with what was found by ERIC fingerprinting analysis, where all strains were grouped into 11 genotypes (11 ERIC types), resulting in a significant level of genetic variation. The distinction between both techniques (the ERIC-PCR technique and 16S rRNA gene sequencing) is probably the size and area of the chromosomes sequenced for the genetic diversity assessment. Where 16S rRNA gene sequencing depends on a specific area of the chromosome, the possibility of detecting genetic changes through 16S rRNA gene sequence analysis is limited (Ogier et al., 2019). In contrast, because the ERIC primers include many annealing sites on the bacterial genome, it can be used to analyze the whole chromosome for any DNA sequence variation (Wilson and Sharp, 2006), which may offer the best chance to evaluate genomic diversity using ERIC-PCR. Thus, when compared to other molecular typing tools (ribotyping), the finding suggests that ERIC PCR is a useful genotyping technique for the study of genetic diversity in bacterial species. This study’s findings support the assumption that ERIC PCR possesses a strong capacity for discrimination and is an effective molecular typing method for detecting genetic diversity, specifically for identifying the source of infection (Taha, 2022). ConclusionIt is concluded that the prevalence of the diseases in sheep and goats in Duhok Province is low, but the risk of increasing its rate is high; more biosecurity measures on imported animals must be applied to minimize or maintain the disease’s prevalence rate. In Duhok Governorate, numerous C. pseudotuberculosis clones are circulating. Multivariable factors were found to play roles in the disease’s prevalence rate in sheep and goats. AcknowledgmentsThe authors are grateful for the support provided by Duhok University’s College of Veterinary Medicine in completing this study project. Conflicts of interestThe authors declare that there is no conflict of interest. ReferencesAbdulrahman, R.F. 2021. Virulence potential, antimicrobial susceptibility and phylogenetic analysis of Corynebacterium pseudotuberculosis isolated from caseous lymphadenitis in sheep and goats in Duhok City, Iraq. Adv. Anim. Vet. Sci. 9(6), 919–925. Abdulrahman, R.F., Abdullah, M.A., Kareem, K.H., Najeeb, Z.D. and Hameed, H.M. 2020. Histopathological and molecular studies on caseous lymphadenitis in sheep and goats in Duhok City, Iraq. Explor. Anim. Med. Res. 10, 134–140. Abebe, D. and Sisay Tessema, T. 2015. Determination of Corynebacterium pseudotuberculosis prevalence and antimicrobial susceptibility pattern of isolates from lymph nodes of sheep and goats at an organic export abattoir, Modjo, Ethiopia. Lett. Appl. Microbiol. 61(5), 469–476. Al-Gaabary, M.H., Osman, S.A., Ahmed, M.S. and Oreiby, A.F. 2010. Abattoir survey on caseous lymphadenitis in sheep and goats in Tanta, Egypt. Small Rumin. Res. 94(1–3), 117–124. Al-Gaabary, M.H., Osman, S.A. and Oreiby, A.F. 2009. Caseous lymphadenitis in sheep and goats: clinical, epidemiological and preventive studies. Small Rumin. Res. 87(1–3), 116–121. Algammal, A. 2016. Molecular characterization and antibiotic susceptibility of Corynebacterium pseudotuberculosis isolated from sheep and goats suffering from caseous lymphadenitis. Zagazig Vet. J. 44(1), 1-8. Ali, A., Mahmoud, A., Khadr, A., Elshemey, T. and Abdelrahman, A. 2016. Corynebacterium pseudotuberculosis: disease prevalence, lesion distribution, and diagnostic comparison through microbiological culture and molecular diagnosis. Alex. J. Vet. Sci. 51(2), 189. Andrade, J.S.L., Azevedo, S.S., Teles, J.A.A., Higino, S.S.S. and Azevedo, E.O. 2012. Ocorrência e fatores de risco associados à infecção por Corynebacterium pseudotuberculosis em caprinos e ovinos do semiárido paraibano. Pesq. Vet. Bras. 32(2), 116–120. Augustine, J.L. and Renshaw, H.W. 1986. Survival of Corynebacterium pseudotuberculosis in axenic purulent exudate on common barnyard fomites. Am. J. Vet. Res. 47(4), 713–715. Bakhshi, B., Afshari, N. and Fallah, F. 2018. Enterobacterial repetitive intergenic consensus (ERIC)-PCR analysis as a reliable evidence for suspected Shigella spp. outbreaks. Braz. J. Microbiol. 49(3), 529–533. Bettini, A., Mancin, M., Mazzucato, M., Schanung, A., Colorio, S. and Tavella, A. 2022. A seroepidemiological survey of Corynebacterium pseudotuberculosis infection in South Tyrol, Italy. Pathogens. 11, 1314. Çetinkaya, B., Karahan, M., Atil, E., Kalin, R., De Baere, T. and Vaneechoutte, M. 2002. Identification of Corynebacterium pseudotuberculosis isolates from sheep and goats by PCR. Vet. Microbiol. 88, 75–83. Costa, L., Huerta, B., Galán-Relaño, Á., Gómez-Gascón, L., Almeida, A., Viegas, I. and Maldonado, A. 2020. Utility assessment of an enzyme-linked immunosorbent assay for detection of subclinical cases of caseous lymphadenitis in small ruminant flocks. Vet. Med. Sci. 6(4), 796-803. Didkowska, A., Zmuda, P., Kwiecień, E., Rzewuska, M., Klich, D., Krajewska-Wȩdzina, M., Witkowski, L., Zychska, M., Kaczmarkowska, A., Orłowska, B. and Anusz, K. 2020. Microbiological assessment of sheep lymph nodes with lymphadenitis found during post-mortem examination of slaughtered sheep: implications for veterinary-sanitary meat control. Acta Vet. Scand. 62(1), 2–7. Eckersall, P.D., Lawson, F.P., Bence, L., Waterston, M.M., Lang, T.L., Donachie, W. and Fontaine, M.C. 2007. Acute phase protein response in an experimental model of ovine caseous lymphadenitis. BMC Vet. Res. 3, 1–6. Fikre, Z. and Abraha, G.K. 2014. Caseous lymphadenitis in goats from Borena Range Land South Ethiopia slaughtered at Luna Export Abattoir. Vet. Med. Anim. Health. 6, 168–173. Guimarães, A. de S., Dorneles, E.M.S., Andrade, G.I., Lage, A.P., Miyoshi, A., Azevedo, V., Gouveia, A.M.G. and Heinemann, M.B. 2011. Molecular characterization of Corynebacterium pseudotuberculosis isolates using ERIC-PCR. Vet. Microbiol. 153(3–4), 299–306. Heras, J., Domínguez, C., Mata, E., Pascual, V., Lozano, C., Torres, C. and Zarazaga, M. 2015. GelJ—a tool for analyzing DNA fingerprint gel images. BMC Bioinform. [Preprint] 16(1), 1–8. Issa, N.A., Abdulrahman, R.F., Taha, Z.M., Hussain, M.M., Kareem, K.H., Hamadamin, H.I., Najeeb, Z.D., Ahmed, B.M. and Hameed, H.M. 2021. Prevalence and molecular investigation of caseous lymphadenitis among the slaughtered sheep at Duhok Abattoirs; experimental infection with Corynebacterium pseudotuberculosis in rabbits. Iraqi J. Vet. Sci. 35, 263–270. Jeber, Z.K.H., MohdJin, Z., Jesse, F.F., Saharee, A.A., Sabri, J., Yusoff, R. and Wahid, H. 2016. Influence of Corynebacterium pseudotuberculosis infection on level of acute phase proteins in goats. BMC Vet. Res. 12(1), 2–5. Jerome, J.P., Bell, J.A., Plovanich-Jones, A.E., Barrick, J.E., Brown, C.T. and Mansfield, L.S. 2011. Standing genetic variation in contingency loci drives the rapid adaptation of Campylobacter jejuni to a novel host. PLoS One 6(1), 1–11. Kaba, J., Nowicki, M., Frymus, T., Nowicka, D., Witkowski, L., Szaluś-Jordanow, O., Czopowicz, M. and Thrusfield, M. 2011. Evaluation of the risk factors influencing the spread of caseous lymphadenitis in goat herds. Pol. J. Vet. Sci. 14(2), 231–237. Karthik, K. and Prabhu, M. 2021. Bacterial diseases of goat and its preventive measures. In: Goat science—environment, health and economy, pp: 1–8; doi:10.5772/intechopen.97434 Khamis, A., Raoult, D. and La Scola, B., 2004. rpoB gene sequencing for identification of Corynebacterium species. J. Clin. Microbiol. 42, 3925–3931. Kumar, J., Singh, F., Tripathi, B.N., Kumar, R., Dixit, S.K. and Sonawane, G.G. 2012. Epidemiological, bacteriological and molecular studies on caseous lymphadenitis in Sirohi goats of Rajasthan, India. Trop. Anim. Health Prod. 44(7), 1319–1322. Nassar, A.F. de C., Daniel, G.T., Ruiz, R., Miyashiro, S., Scannapieco, E.M., Souza Neto, J. and Gregory, L. 2016. Diagnostic comparison of Corynebacterium pseudotuberculosis through microbiological culture and PCR in sheep samples. Arq. Inst. Biol. 82(0), 1–6. Ogier, J.C., Pagès, S., Galan, M., Barret, M. and Gaudriault, S. 2019. RpoB, a promising marker for analyzing the diversity of bacterial communities by amplicon sequencing. BMC Microbiol. 19(1), 1–16. Oreiby, A.F. 2015. Diagnosis of caseous lymphadenitis in sheep and goat. Small Rumin. Res.123(1), 160–166. Pacheco, L.G.C., Pena, R.R., Castro, T.L.P., Dorella, F.A., Bahia, R.C., Carminati, R., Frota, M.N.L., Oliveira, S.C., Meyer, R., Alves, F.S.F., Miyoshi, A. and Azevedo, V. 2007. Multiplex PCR assay for identification of Corynebacterium pseudotuberculosis from pure cultures and for rapid detection of this pathogen in clinical samples. J. Med. Microbiol. 56, 480–486. Rizk, A., Abd El-Tawab, A., AFIFI, S. and Mohamed, S. 2019. Corynebacterium pseudotuberculosis infection in small ruminant and molecular study of virulence and resistance genes in Beni-Suef Governorate. Benha Med. J. 37(1), 122–127. Ruiz, H., Ferrer, L.M., Ramos, J.J., Baselga, C., Alzuguren, O., Tejedor, M.T., de Miguel, R. and Lacasta, D. 2020. The relevance of caseous lymphadenitis as a cause of culling in adult sheep. Animals 10(11), 1–13. Sá, M.C.A., Samily, A.S.O., Edmilson, M.D., Gisele, V.G., João, J.S.G., Josir, L.A.V. and Mateus, M.C. 2018. Resistance of Corynebacterium pseudotuberculosis in the Brazilian Semiarid Environment. Pesq. Vet. Bras. 38(6), 1091–1096. Sellyei, B., Bányai, K., Bartha, D., Hajtós, I., Fodor, L. and Makrai, L. 2017. Multilocus sequencing of Corynebacterium pseudotuberculosis biotype ovis strains. Biomed. Res. Int. 2017, 1–7. Taha, Z.M. 2022. Genotyping and genetic diversity of Corynebacterium pseudotuberculosis strains isolated from caseous lymphadenitis in sheep and goat. Explor. Anim. Med. Res. 12, 85–90. Terab, A.M.A., Abdel Wahab, G.E.D., Ishag, H.Z.A., Khalil, N.A.H., El Tigani-Asil, E.T.A., Hashem, F.M., Khalafalla, A.I., Shah, A.A.M. and Al Muhairi, S.S.M. 2021. Pathology, bacteriology and molecular studies on caseous lymphadenitis in Camelus dromedarius in the Emirate of Abu Dhabi, UAE, 2015-2020. PLoS One 16(6), 1–14. Versalovic, J., Koeuth, T. and Lupski, R. 1991. Distribution of repetitive DNA sequences in eubacteria and application to finerpriting of bacterial enomes. Nucleic Acids Res. 19(24), 6823–6831. Wilson, L.A. and Sharp, P.M. 2006. Enterobacterial repetitive intergenic consensus (ERIC) sequences in Escherichia coli: evolution and implications for ERIC-PCR. Mol. Biol. Evol. 23(6), 1156–1168. Yitagesu, E., Alemnew, E., Olani, A., Asfaw, T. and Demis, C. 2020. Survival analysis of clinical cases of caseous lymphadenitis of goats in North Shoa, Ethiopia. Vet. Med. Int. 2020, 1–8. | ||
| How to Cite this Article |
| Pubmed Style Khanamir RA, Issa NA, Abdulrahman RF. First study on molecular epidemiology of caseous lymphadenitis in slaughtered sheep and goats in Duhok Province, Iraq. Open Vet J. 2023; 13(5): 588-598. doi:10.5455/OVJ.2023.v13.i5.11 Web Style Khanamir RA, Issa NA, Abdulrahman RF. First study on molecular epidemiology of caseous lymphadenitis in slaughtered sheep and goats in Duhok Province, Iraq. https://www.openveterinaryjournal.com/?mno=143797 [Access: July 02, 2025]. doi:10.5455/OVJ.2023.v13.i5.11 AMA (American Medical Association) Style Khanamir RA, Issa NA, Abdulrahman RF. First study on molecular epidemiology of caseous lymphadenitis in slaughtered sheep and goats in Duhok Province, Iraq. Open Vet J. 2023; 13(5): 588-598. doi:10.5455/OVJ.2023.v13.i5.11 Vancouver/ICMJE Style Khanamir RA, Issa NA, Abdulrahman RF. First study on molecular epidemiology of caseous lymphadenitis in slaughtered sheep and goats in Duhok Province, Iraq. Open Vet J. (2023), [cited July 02, 2025]; 13(5): 588-598. doi:10.5455/OVJ.2023.v13.i5.11 Harvard Style Khanamir, R. A., Issa, . N. A. & Abdulrahman, . R. F. (2023) First study on molecular epidemiology of caseous lymphadenitis in slaughtered sheep and goats in Duhok Province, Iraq. Open Vet J, 13 (5), 588-598. doi:10.5455/OVJ.2023.v13.i5.11 Turabian Style Khanamir, Ramadhan Ado, Nawzat Aboziad Issa, and Rezheen Fatah Abdulrahman. 2023. First study on molecular epidemiology of caseous lymphadenitis in slaughtered sheep and goats in Duhok Province, Iraq. Open Veterinary Journal, 13 (5), 588-598. doi:10.5455/OVJ.2023.v13.i5.11 Chicago Style Khanamir, Ramadhan Ado, Nawzat Aboziad Issa, and Rezheen Fatah Abdulrahman. "First study on molecular epidemiology of caseous lymphadenitis in slaughtered sheep and goats in Duhok Province, Iraq." Open Veterinary Journal 13 (2023), 588-598. doi:10.5455/OVJ.2023.v13.i5.11 MLA (The Modern Language Association) Style Khanamir, Ramadhan Ado, Nawzat Aboziad Issa, and Rezheen Fatah Abdulrahman. "First study on molecular epidemiology of caseous lymphadenitis in slaughtered sheep and goats in Duhok Province, Iraq." Open Veterinary Journal 13.5 (2023), 588-598. Print. doi:10.5455/OVJ.2023.v13.i5.11 APA (American Psychological Association) Style Khanamir, R. A., Issa, . N. A. & Abdulrahman, . R. F. (2023) First study on molecular epidemiology of caseous lymphadenitis in slaughtered sheep and goats in Duhok Province, Iraq. Open Veterinary Journal, 13 (5), 588-598. doi:10.5455/OVJ.2023.v13.i5.11 |