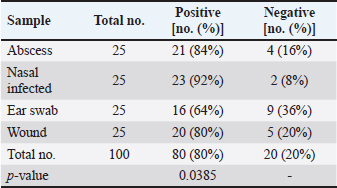
| Research Article | ||
Open Vet J. 2023; 13(8): 1021-1026 Open Veterinary Journal, (2023), Vol. 13(8): 1021-1026 Original Research Role of nuc gene in Staphylococcus aureus to phagocytic activity in different cattle infectionsHanaa Khaleel Ibraheim1, Rana A. Fayez1, Alyaa S. Jasim1 and Hasanain A. J. Gharban2*1Microbiology Department, College of Veterinary Medicine, University of Basrah, Basrah, Iraq 2Department of Internal and Preventive Veterinary Medicine, College of Veterinary Medicine, University of Wasit, Wasit, Iraq *Corresponding Author: Hasanain A. J. Gharban. Department of Internal and Preventive Veterinary Medicine, College of Veterinary Medicine, University of Wasit, Wasit, Iraq. Email: hghirban [at] uowasit.edu.iq Submitted: 22/05/2023 Accepted: 30/07/2023 Published: XX/08/2023 © 2023 Open Veterinary Journal
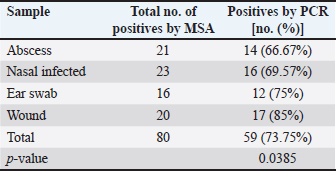
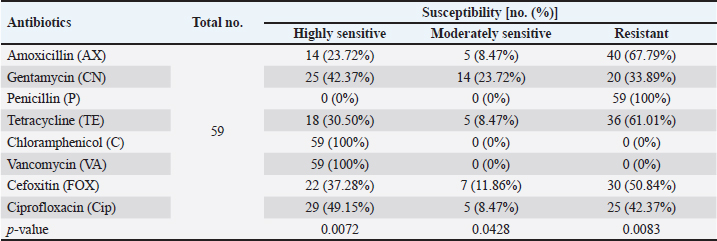
AbstractBackground: Staphylococcus aureus is a typical pathogenic agent that causes several morbidities and mortalities which variable largely following the severity of bacteria and activity of host immunity. Aim: Isolation of S. aureus from different cattle infections, molecular detection of nuc gene in positive S. aureus isolates, and identification of the effectiveness of the phagocytic activity. Methods: Totally, 100 cattle with various infections (25 wounds, 25 abscesses, 25 nasal discharges, and 25 ear swaps) were selected and subjected to collection of swabs under controlled conditions. All collected samples were cultured on mannitol salt agar (MSA) and assessed by biochemical tests. Targeting the nuc gene, all study MSA positive isolates were examined molecularly by conventional polymerase chain reaction (PCR), and then subjected to antibiotic susceptibility test. Jugular venous blood was collected from all infected animals in addition to 20 healthy cattle that were selected as a control group to estimate the phagocytic activity of S. aureus isolates. Results: The findings of MSA culture revealed a total of 80 positive samples of S. aureus as 23, 21, 20, and 16 positive isolates for nasal discharge, abscess, wound, and ear swab, respectively; based on its morphology, cultural trait, and biochemical test. Subsequently, PCR assaying of MSA-positive isolates demonstrated an overall 59 positive samples as 14, 16, 12, and 17 positive isolates for nasal discharge, abscess, wound, and ear swabs, respectively. Antibiotic susceptibility testing of S. aureus-positive PCR isolates reported a significantly high sensitivity to chloramphenicol and vancomycin, and a high resistance to penicillin. Finally, there was a considerable decline in phagocytic activity in particular 2 weeks post-infection as a result of bacterial invasion. Conclusion: This study shows a high prevalence of S. aureus in cattle infections, and the protocol includes a regular screening of cattle infection and suitable therapy based on antibacterial susceptibility test is of great importance in the long-term control of the pathogen. However, additional molecular studies targeting other genes of S. aureus and the role of immune markers in different infections should be aimed. Keywords: Antibiotic susceptibility, Bovine bacterial infection, Molecular test, Polymerase chain reaction, Iraq. IntroductionStaphylococcus is a member of the Staphylococcaceae family, which forms irregular clusters resembling bunches of grapes during culture and is positive for Gram stain (Markey et al., 2013). In general, Staphylococcus was described as a facultative, non-spore-forming, not-motile, aerobic, and oxidase-negative but catalase-positive bacterium (Quinn et al., 2015). Coagulation of plasma areas surrounding bacterial isolates could play a protective role in the prevention of phagocytosis by the host immune system (van Dalen et al., 2020). Although there were at least 43 species in the Staphylococcus genus, S. aureus appeared to be the most important one in both humans and different animal species because it represents a typical pathogenic agent that mostly can secrete coagulase enzyme and ferment lactic acid (Seo and Bohach, 2013; Shoen et al., 2019; Pickering et al., 2021). The main gene builds the framework of the S. aureus genome and is highly conserved. The accessory region that accounts for 25% of S. aureus genome was having mobile genetic elements as the transposons, chromosome-cassette tapes, pathogenicity islands, genome tropical islands, and horizontally acquired prophages (Rocha et al., 2019; Wildeman et al., 2020). In the field, different S. aureus strains have the ability to produce a wide range of exo-enzymes, with the existence of several virulence factors (Generalov, 2017; Cheung et al., 2021). The nuc gene encodes thermonuclear and is widely used as a specific target for the detection of S. aureus by polymerase chain reaction (PCR) (Wongboot et al., 2013). Numerous PCR-based studies were carried out targeting the nuc gene alone or in combination with the mecA gene for rapid identification or recognition of methicillin-susceptible S. aureus and methicillin-resistant S. aureus (González-Domínguez et al., 2020). Also, the nuc gene has been extensively utilized as an individual marker for detecting S. aureus contamination (Mahros et al., 2021). Phagocytosis has found to play a significant role in innate and adaptive immune responses to infection through recognizing and binding to receptors on the cell surface (Naik and Harrison, 2013). In addition, it is essential to start the adaptive immune response since phagocytic targets may attract lymphoid cells by encouraging the production of pro-inflammatory cytokines (Caillon et al., 2019). Hence, this study aimed to isolate S. aureus from different cattle infections, molecular detection of nuc gene in positive S. aureus isolates, and identification of the effectiveness of phagocytic activity. Material and MethodsCollection of samplesA total of 100 infected cattle reported at the veterinary clinics in Al-Basrah province (Iraq), were selected for the current study and categorized as follows: 25 cattle having wounds, 25 cattle having abscesses, 25 cattle having nasal discharges, and 25 cattle having ear infections. All study animals were subjected to direct swab sampling from the lesions using sterile absorbent swabs soaked in nourishing broth. Then, the swabs were transported to the laboratory of the College of Veterinary Medicine (University of Basrah, Basra, Iraq) using a cooled plastic box to be cultured on mannitol salt agar (MSA) as soon as possible. IsolationAll collected samples were cultured on MSA as described by Nocera et al. (2021). Then, the isolates were used to prepare the slide smears that were stained with the Gram stain and examined under light microscope at × 100 (Sizar and Unakal, 2021). Biochemical testingTube coagulase test Approximately 0.1 ml of bacterial culture was mixed with 0.3 ml of rabbit plasma, and the mixture was incubated in tubes of the brain-heart infusion broth at 37°C for 4 hours. The presence of clotting confirmed the successful outcome comparable to control (Chauhan et al., 2020). Catalase test It was performed by transferring few amount of pure growth from MSA on a clean slide, and then, adding a drop of H2O2. The presence of gas bubbles indicates favorable outcome (Becker et al., 2015). Oxidase test This test assays to indicate the presence of cytochrome oxidase in organism, and oxidizing reagent “tetra-methyl-p-phenylenediamine dihydrochloride” from colorless to dark blue (Fern et al., 2021). Molecular examinationAll positive MSA samples were used for the extraction of DNAs according to the manufacturer’s instructions for the Genomic DNA Mini Kit (Geneaid, Korea). Nanodrop System (Thermo-Scientific, UK) was employed to evaluate the concentration and purity of extracted DNAs at an optical density of 260/280 nm. For preparation of MasterMix tubes, one set of primers was designed targeting the nuc gene [F: (5'-GCT TGC TAT GAT TGT GGT AGC C-3') and R: (5'-TCT CTA GCA AGT CCC TTT TCC A-3')] and prepared at 20 µl final volume following the manufacturer’s instructions of AccuPower PCR PreMic Kit (Bioneer, Korea). Thermocycler reaction was carried out under the following conditions: 1 cycle for initial denaturation (5 minutes at 95°C), 30 cycles for denaturation (40 seconds at 95°C), annealing (35 seconds at 58°C), and extension (35 seconds at 72°C), and 1 cycle for final extension (5 minutes at 72°C). The PCR products were separated on 2% agarose gel stained with ethidium bromide at 100 Volt, 80 mAm, for 1 hour, and visualized under the light UV transilluminator to detect the positive samples at 423 bp. Free nuclease water was used as a negative control; while DNAs of previously confirmed S. aureus isolates were used as a positive control during gel-electrophoresis. Antimicrobial susceptibilityColonies of S. aureus-positive isolates by both MSA and PCR assay were selected to preparing of bacterial suspension that was inoculated in nutrient broth, overnight. The suspension was then mixed by a vortex so that the turbidities could be compared and adjusted fit 0.5 McFarland standards by adding additional colonies. The bacterial suspension containing 1 × 108–2 × 108 CFU/ml is equivalent to 0.5 McFarland standards. Ready to use antimicrobial susceptibility test kit (bioMérieux, India) was applied to Mueller-Hinton agar using the disk diffusion technique (Yang et al., 2019). Phagocytic activityInitially, a total of 20 healthy cattle were selected as a negative control group. Then, all positively S. aureus infected cattle in addition to cattle of negative control groups were subjected to sampling 2 ml of jugular venous blood under aseptic conditions into a heparinized tube. As described by Seki et al. (1989), S. aureus colonies were grown on heart infusion agar at 37ºC overnight, and then harvested, extensively washed with saline, heated in boiling water for 10 minutes, and adjusted to 109 CFU/ml in saline. One drop of heparinized blood was laid on the bacterial thin layer in a plastic dish, and the phagocytic activity was represented as blank spaces of bacterial thin-layer usually containing phagocytic leukocytes (plague). The number of plaques was counted using the light microscope at 100 × to detect phagocytic activity. Statistical analysisThe t-test and one-way analysis of variance in the GraphPad Prism version 6.0.1 Software (GraphPad Inc, USA), were used to detect significant differences between values at p < 0.05 (*), p < 0.01 (**), p < 0.001 (***) (Gharban, 2022). Ethical approvalThe study was approved by the College of Veterinary Medicine and College of Medicine at the University of Basrah (Basra, Iraq) and the College of Veterinary Medicine at the University of Wasit (Wasit, Iraq) following international ethical rules. ResultsIdentification of S. aureus isolatesThe findings of MSA culture revealed a total of 80% positive samples to S. aureus as 92%, 84%, 80%, and 64% positive isolates for nasal discharge, abscess, wound, and ear swabs, respectively (Table 1). The positive S. aureus isolates were distinguished in this study following the morphology and cultural traits of grown colonies in addition to the results of biochemical tests that generated positive coagulase and catalase, but negative to oxidase. Molecular examinationPCR assaying of 80 MSA-positive isolates demonstrated an overall 59 positive samples as 66.67%, 69.57%, 75%, and 85% positive isolates for nasal discharge, abscess, wound, and ear swab, respectively (Table 2, Fig. 1). Antimicrobial susceptibility testingSignificantly, positive S. aureus isolates by both MSA and PCR assays (total no.: 59 samples) showed a variation (p < 0.05) in their susceptibility to eight antibiotics used in the current study (Table 3). However, the study isolates have appeared highly sensitive to chloramphenicol (100%) and vancomycin (100%); and highly resistant to penicillin (100%) when compared to other selected antibiotics (Table 3). Table 1. Total results of traditional diagnosis of 100 swab samples obtained from different infections on MSA.
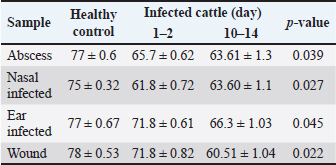
Phagocytic activityA significant decrease (p < 0.05) in phagocytic activity was showed in values (mean ± standard errors) of both periods of post-infections but more significantly in 10–14 days post-infection when compared to controls (Table 4). DiscussionIn this study, isolates of S. aureus grown on MSA appeared as large, golden, and yellow colonies surrounded by wide yellow zones, which is in agreement with that detected by several studies (Bhunia and Bhunia, 2018; Aubaid et al., 2020; Al-Shuwaili and Saab, 2022). Using the Gram-stain, S. aureus isolates examined under the microscope were seen as positive cocci arranged in clusters resembling grapes or in other strange patterns throughout the colonies, which is similar to other observations (Kengne et al., 2019; Andhare and Shinde, 2020; Taylor and Unakal, 2022). Biochemical testing of positive isolates demonstrated the production of gas bubbles throughout the catalase test indicating the successful outcome. These findings were similar to other researchers (Lucia et al., 2017; Shen et al., 2019; Senthil and Aravind, 2023). In the present study, 73.75% of positive MSA isolates were positive for PCR assay. These findings were compatible with the findings of other studies (Sizar and Unakal, 2021; Qian et al., 2022; Cao et al., 2023). The high specificity of the nuc gene in the detection of S. aureus means that molecular assaying targeting this gene can be used instead of the traditional diagnosis of S. aureus. In Iraq, several authors demonstrated the efficacy of molecular assay in the confirmation of S. aureus isolates by targeting the nuc gene (Hasan and Hoshyar, 2019; Sayhood et al., 2022; Sheet, 2022). PCR is a crucial method for identifying bacteria isolates and detecting specific genes as it is simple, rapid, valuable, and more accurate than traditional diagnostic methods. This suggests that the application of PCR in combination with the additional techniques can increase the sensitivity of diagnosis (Sheet, 2022).
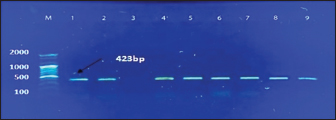
Fig. 1. Electrophoresis of 2% agarose gel of PCR products targeting the nuc gene. Lane (M): Marker ladder (100–2,000 bp). Lane (3): Negative control. Lane (5): Positive control. Lanes (1, 2, 4, 6, 7, 8, and 9): Some positive PCR products at 423 bp. Table 2. Total positive results of S. aureus isolates obtained from different swab samples using the MSA and PCR.
Table 3. Result of antibiotic susceptibility for S. aureus positive isolates by MSA and PCR (total no. 59).
Table 4. Phagocytic activity in cattle infected in wounds, abscesses, nasal, and ear infected.
In this study, all S. aureus isolates showed a high sensitivity to chloramphenicol and vancomycin as reported by other studies (Deyno et al., 2017; Shahid et al., 2021; Rasmi et al., 2022). In Iraq, almost S. aureus isolates were concluded to be vancomycin-sensitive (Abdrabaa and Flayyih, 2019; Jafar and Abed, 2021; Anwaar et al., 2023). In general, several authors, studying bacterial sensitivity to different antibiotics, discovered that some S. aureus strains were susceptible to antibiotics; indicating the effect of bacterial status, inoculum size, antibiotic concentrations, and host interactions (Ali et al., 2010; King et al., 2017). However, the presence of different resistance mechanisms might be attributed to variations in plasmodial or chromosomal genetic elements. Numerous factors including excessive use of antibiotics and poor environmental animal hygiene conditions could account for the spread of resistant bacteria (Li et al., 2017). King et al. (2017) mentioned that the low sensitivity of S. aureus isolates to penicillin might belong to mutation or production of penicillinase enzyme (a type of β-lactamase) that hydrolyses the beta-lactam ring of penicillin. In this study, the phagocytic activity of infected animals was significantly reduced when compared to healthy animals indicating the effect of infection on the cell functions including phagocytosis. Pavlou et al. (2017) showed that macrophage phenotype and function are related to their states, whereas Fountain et al. (2021) recorded that phagocytosis could be designed following bacterial infection. Vollrath et al. (2021) concluded that the phagocytic activity and capacity show significantly different alterations depending on the pathogen strain, and thus potentially at certain and possibly more relevant infections caused after infection. ConclusionThis study shows a high prevalence of S. aureus in cattle infections, and the protocol includes a regular screening of cattle infection and suitable therapy based on an antibacterial susceptibility test is of great importance in long-term control of pathogen and to ensure the suitable therapy. However, additional molecular studies targeting other genes of S. aureus and the role of immune markers in different infections should be aimed. AcknowledgmentsWe really appreciate the support provided by the College of Veterinary Medicine and University of Basrah in completing our study. Conflict of interestThe authors declare that there is no conflict of interest FundingThere is no external fund (private funding). Data availabilityAll data supporting the findings of this study are available within the manuscript. Author’s contributionHKI: Collection of samples, culture, and traditional diagnosis. RAF: Biochemical testing. ASJ: Phagocytic activity. HAJG: Statistical analysis and copy editing of the manuscript. All authors contributed to molecular examination, as well as to reading and approving the final copy of manuscript. ReferencesAbdrabaa, M.K. and Flayyih, M.T. 2019. Autolysis activity of vancomycin resistance Staphylococcus epidermidis. Iraqi. J. Biotechnol. 18(2), 1–10. Ali, S.Q., Zehra, A., Naqvi, B.S., Shah, S. and Bushra, R. 2010. Resistance pattern of ciprofloxacin against different pathogens. Oman Med. J. 25(4), 294–298. Al-Shuwaili, M. and Saab, K.Z. 2022. Evaluation of the bacterial load in the raw dairy products in Baghdad, Iraq. Instit. Razi. Arch. 77(6), 2319–2328. Andhare, A.A. and Shinde, R.S. 2020. In vitro studies on antimicrobial sensitivity of Staphylococcus aureus from Prosopis juliflora, Cassia occidentalis and Tephrosia purpurea. Anti-Infect. Agents 18(4), 384–390. Anwaar, F., Ijaz, M., Rasheed, H., Shah, S.F.A., Haider, S.A.R. and Sabir, M.J. 2023. Evidence and molecular characterization of multidrug resistant Staphylococcus aureus isolated from equines in Pakistan. J. Equine. Vet. Sci. 126, 104498. Aubaid, A.H., Mahdi, Z.H., Abd-Alraoof, T.S. and Jabbar, N.M. 2020. Detection of mecA, vanA and vanB genes of Staphylococcus aureus isolated from patients in Al Muthanna province hospitals. Indian. J. Forensic. Med. Toxicol. 14(2), 1002–1008. Becker, K., Skov, R.L. and von Eiff, C. 2015. Staphylococcus, Micrococcus, and other catalase-positive cocci. Manual of clinical microbiology. Washington, DC: ASM Press, pp: 354–382. Bhunia, A.K. and Bhunia, A.K. 2018. Staphylococcus aureus. Foodborne microbial pathogens: mechanisms and pathogenesis. New York, NY: Springer, pp: 181–192. Caillon, A., Paradis, P. and Schiffrin, E.L. 2019. Role of immune cells in hypertension. Br. J. Pharmacol. 176(12), 1818–1828. Cao, X., Chang, Y., Tao, C., Chen, S., Lin, Q., Ling, C. and Zhang, H. 2023. Cas12a/Guide RNA-based platforms for rapidly and accurately identifying Staphylococcus aureus and methicillin-resistant S. aureus. Microbiol. Spect. 11(2), e04870-22. Chauhan, A., Jindal, T., Chauhan, A. and Jindal, T. 2020. Biochemical and molecular methods for bacterial identification. Microbiological methods for environment, food and pharmaceutical analysis. Cham, Switzerland: Springer, pp 425–468. Cheung, G.Y., Bae, J.S. and Otto, M. 2021. Pathogenicity and virulence of Staphylococcus aureus. Virulence 12(1), 547–569. Deyno, S., Fekadu, S. and Astatkie, A. 2017. Resistance of Staphylococcus aureus to antimicrobial agents in Ethiopia: a meta-analysis. Antimicrob. Resist. Infect. Control. 6(1), 1–15. Fern, Q.M., Cordeiro, L.V. and de rade Júnior, F.P. 2021. Main laboratory methods used for the isolation and identification of Staphylococcus spp. Rev. Colomb. Cien. Químico-Farmacéut. 50(1), 5–28. Fountain, A., Inpanathan, S., Alves, P., Verdawala, M.B. and Botelho, R.J. 2021. Phagosome maturation in macrophages: eat, digest, adapt, and repeat. Adv. Biol. Regul. 82, 100832. Generalov, I.I. 2017. Medical microbiology, virology and immunology. Vitebsk, Belarus: Ministry of Health of the Republic of Belarus Higher Educational Establishment, pp: 587–588. Gharban, H.A. 2022. Clinical and serological diagnosis of bovine hypodermosis in Wasit Province. Rev. Electron. Vet. 23(3), 457–466. González-Domínguez, M.S., Carvajal, H.D., Calle-Echeverri, D.A. and Chinchilla-Cárdenas, D. 2020. Molecular detection and characterization of the mecA and nuc genes from Staphylococcus species (S. aureus, S. pseudintermedius, and S. schleiferi) isolated from dogs suffering superficial pyoderma and their antimicrobial resistance profiles. Front. Vet. Sci. 7, 376-381. Hasan, N.S. and Hoshyar, D.F. 2019. Detection of enterotoxigenic Staphylococcus aureus strains in raw milk of cows reared in Erbil Province, Iraq. Zanco. J. Pure. Appl. Sci. 31(4), 50–60. Jafar A.S. and Abed, A.R. 2021. The Differences in antibiotic-resistance among several Staphylococcus aureus strains in Iraq. Med. Legal. Upd. 21(3), 1–10. Kengne, M., Fotsing, O., Ndomgue, T. and Nwobegahay, J.M. 2019. Antibiotic susceptibility patterns of Staphylococcus aureus strains isolated at the Yaounde Central Hospital, Cameroon: a retro prospective study. Pan. Afr. Med. J. 32(1), 1–8. King, D.T., Sobhanifar, S. and Strynadka, N.C. 2017. The mechanisms of resistance to β-lactam antibiotics. Handbook. Antimicrob. Resist. 23, 177–201. Li, J., Xie, S., Ahmed, S., Wang, F., Gu, Y., Zhang, C. and Cheng, G. 2017. Antimicrobial activity and resistance: influencing factors. Front. Pharmacol. 8, 364. Lucia, M., Rahayu, S., Haerah, D. and Wahyuni, D. 2017. Detection of Staphylococcus aureus and Streptococcus agalactiae: subclinical mastitis causes in dairy cow and dairy buffalo (Bubalus bubalis). Am. J. Biomed. Res. 5(1), 8–13. Mahros, M.A., Abd-Elghany, S.M. and Sallam, K.I. 2021. Multidrug-, methicillin-, and vancomycin-resistant Staphylococcus aureus isolated from ready-to-eat meat sandwiches: an ongoing food and public health concern. Int. J. Food Microbiol. 346, 109165. Markey, B., Leonard, F., Archambault, M., Cullinane, A. and Maguire, D. 2013. Introduction to the pathogenic fungi. In Clinical veterinary microbiology, 2nd ed. London, UK: Mosby, pp: 457–470. Naik, U. and Harrison, R.E. 2013. Phagocytosis. In Colloquium series on building blocks of the cell: cell structure and function. San Rafael, CA: Morgan & Claypool Life Sciences, pp: 1–105. Nocera, F.P., Ferrara, G., Scandura, E., Ambrosio, M., Fiorito, F. and De Martino, L. 2021. A Preliminary study on antimicrobial susceptibility of Staphylococcus spp. and Enterococcus spp. grown on mannitol salt agar in european wild boar (Sus scrofa) hunted in Campania Region-Italy. Animal 12(1), 85–99. Pavlou, S., Wang, L., Xu, H. and Chen, M. 2017. Higher phagocytic activity of thioglycollate-elicited peritoneal macrophages is related to metabolic status of the cells. J. Inflamm. 14, 1–6. Pickering, A.C., Yebra, G., Gong, X., Goncheva, M.I., Wee, B. A., MacFadyen, A.C. and Fitzgerald, J.R. 2021. Evolutionary and functional analysis of coagulase positivity among the Staphylococci. Msphere 6(4), e00381-21. Qian, J., Huang, D., Ni, D., Zhao, J., Shi, Z., Fang, M. and Xu, Z. 2022. A portable CRISPR Cas12a based lateral flow platform for sensitive detection of Staphylococcus aureus with double insurance. Food Control 132, 108485. Quinn, P.J., Markey, B.K., Leonard, F.C., FitzPatrick, E.S. and Fanning, S. 2015. Concise review of veterinary microbiology, 2nd ed. Chichester, UK: Wiley, pp: 65. Rasmi, A.H., Ahmed, E.F., Darwish, A.M.A. and Gad, G.F. M. 2022. Virulence genes distributed among Staphylococcus aureus causing wound infections and their correlation to antibiotic resistance. BMC. Infect. Dis. 22(1), 652–659. Rocha, L.S., Silva, D.M., Silva, M.P., Vidigal, P.M.P., Silva, J.C.F., Guerra, S.T. and Ribon, A.D.O. 2019. Comparative genomics of Staphylococcus aureus associated with subclinical and clinical bovine mastitis. PLoS. One. 14(8), e0220804. Sayhood, M.H., Mohammed, A.L., Abdulhameed, M.F. and Jori, M. M. 2022. Classical and molecular identification of Staphylococcus aureus isolated from infestation cattle wounds with myiasis in Basrah governorate, Iraq. Iraqi J. Vet. Sci. 36(3), 641–646. Seki, K., Murai, M., Sakurada, J., Shirahige, A., Kobayashi, N., Hwang, S. M. and Masuda, S. 1989. A simple method for observation of phagocytosis on bacterial thin-layer. Microbiol. Immunol. 33(1), 81–85. Senthil, G. and Aravind, J. 2023. Nanobubbles: a promising efficient tool for therapeutic delivery of antibacterial agents for the Staphylococcus aureus infections. Appl. Nanosci. 2023, 1–14. Seo, K.S. and Bohach, G.A. 2013. Staphylococcus aureus. In Microbiology: fundamentals and frontiers, 4th ed. Eds., Doyle, M.P. and Buchanan, R.L. Washington, DC: Food ASM Press, pp: 547–593. Shahid, A.H., Nazir, K.N.H., El Zowalaty, M.E., Kabir, A., Sarker, S.A., Siddique, M.P. and Ashour, H.M. 2021. Molecular detection of vancomycin and methicillin resistance in Staphylococcus aureus isolated from food processing environments. One Health 13, 100276. Sheet, O.H. 2022. Molecular detection of mecA gene in methicillin-resistant Staphylococcus aureus isolated from dairy mastitis in Nineveh governorate, Iraq. Iraqi J. Vet. Sci. 36(4), 939–943. Shen, J., Zhang, H., Xu, Z., Zhang, Z., Cheng, C., Ni, G. and Chu, P.K. 2019. Preferential production of reactive species and bactericidal efficacy of gas-liquid plasma discharge. Chem. Eng. J. 362, 402–412. Shoen, H.R., Rose, S.J., Ramsey, S.A., de Morais, H. and Bermudez, L.E. 2019. Analysis of Staphylococcus infections in a veterinary teaching hospital from 2012 to 2015. Com. Immunol. Microbiol. Infect. Dise. 66, 101332. Sizar, O. and Unakal, C.G. 2021. Gram positive bacteria. Treasure Island, FL: StatPearls Publishing, pp: 227. Taylor, T.A. and Unakal, C.G. 2022. Staphylococcus aureus. Treasure Island, FL: StatPearls Publishing, pp: 42. van Dalen, R., Peschel, A. and van Sorge, N.M. 2020. Wall teichoic acid in Staphylococcus aureus host interaction. Trends Microbial. 28(12), 985–998. Vollrath, J.T., Klingebiel, F., Bläsius, F.M., Greven, J., Bolierakis, E., Janicova, A. and Relja, B. 2021. Alterations of phagocytic activity and capacity in granulocytes and monocytes depend on the pathogen strain in porcine polytrauma. Frontiers. Med. 8, 645589. Wildeman, P., Tevell, S., Eriksson, C., Lagos, A.C., Söderquist, B. and Stenmark, B. 2020. Genomic characterization and outcome of prosthetic joint infections caused by Staphylococcus aureus. Sci. Rep. 10(1), 5938. Wongboot, W., Chomvarin, C., Engchanil, C. and Chaimanee, P. 2013. Multiplex PCR for detection of superantigenic toxin genes in methicillin-sensitive and methicillin-resistant Staphylococcus aureus isolated from patients and carriers of a hospital in northeast Thailand. Southeast. Asian. J. Trop. Med. Public. Health. 44, 660–671. Yang, X., Wang, D., Zhou, Q., Nie, F., Du, H., Pang, X. and Xu, Y. 2019. Antimicrobial susceptibility testing of Enterobacteriaceae: determination of disk content and Kirby-Bauer breakpoint for ceftazidime/avibactam. BMC. Microbial. 19(1), 1–7. | ||
| How to Cite this Article |
| Pubmed Style Ibraheim HK, Fayez RA, Jasim AS, Gharban HAJ. Role of nuc gene in Staphylococcus aureus to phagocytic activity in different cattle infections. Open Vet J. 2023; 13(8): 1021-1026. doi:10.5455/OVJ.2023.v13.i8.8 Web Style Ibraheim HK, Fayez RA, Jasim AS, Gharban HAJ. Role of nuc gene in Staphylococcus aureus to phagocytic activity in different cattle infections. https://www.openveterinaryjournal.com/?mno=154206 [Access: July 03, 2025]. doi:10.5455/OVJ.2023.v13.i8.8 AMA (American Medical Association) Style Ibraheim HK, Fayez RA, Jasim AS, Gharban HAJ. Role of nuc gene in Staphylococcus aureus to phagocytic activity in different cattle infections. Open Vet J. 2023; 13(8): 1021-1026. doi:10.5455/OVJ.2023.v13.i8.8 Vancouver/ICMJE Style Ibraheim HK, Fayez RA, Jasim AS, Gharban HAJ. Role of nuc gene in Staphylococcus aureus to phagocytic activity in different cattle infections. Open Vet J. (2023), [cited July 03, 2025]; 13(8): 1021-1026. doi:10.5455/OVJ.2023.v13.i8.8 Harvard Style Ibraheim, H. K., Fayez, . R. A., Jasim, . A. S. & Gharban, . H. A. J. (2023) Role of nuc gene in Staphylococcus aureus to phagocytic activity in different cattle infections. Open Vet J, 13 (8), 1021-1026. doi:10.5455/OVJ.2023.v13.i8.8 Turabian Style Ibraheim, Hanaa Khaleel, Rana A. Fayez, Alyaa S. Jasim, and Hasanain A. J. Gharban. 2023. Role of nuc gene in Staphylococcus aureus to phagocytic activity in different cattle infections. Open Veterinary Journal, 13 (8), 1021-1026. doi:10.5455/OVJ.2023.v13.i8.8 Chicago Style Ibraheim, Hanaa Khaleel, Rana A. Fayez, Alyaa S. Jasim, and Hasanain A. J. Gharban. "Role of nuc gene in Staphylococcus aureus to phagocytic activity in different cattle infections." Open Veterinary Journal 13 (2023), 1021-1026. doi:10.5455/OVJ.2023.v13.i8.8 MLA (The Modern Language Association) Style Ibraheim, Hanaa Khaleel, Rana A. Fayez, Alyaa S. Jasim, and Hasanain A. J. Gharban. "Role of nuc gene in Staphylococcus aureus to phagocytic activity in different cattle infections." Open Veterinary Journal 13.8 (2023), 1021-1026. Print. doi:10.5455/OVJ.2023.v13.i8.8 APA (American Psychological Association) Style Ibraheim, H. K., Fayez, . R. A., Jasim, . A. S. & Gharban, . H. A. J. (2023) Role of nuc gene in Staphylococcus aureus to phagocytic activity in different cattle infections. Open Veterinary Journal, 13 (8), 1021-1026. doi:10.5455/OVJ.2023.v13.i8.8 |