| Original Article | ||
Open Vet J. 2022; 12(2): 221-230 Open Veterinary Journal, (2022), Vol. 12(2): 221–230 Original Research Generation of an inactivated vaccine for avian pathogenic Escherichia coli using microarrays: A more rational approach to inactivated vaccine designXiangmei Zhou1,2†, Philip Richards1,3†, Daniel Windhorst4, Ariel Imre1,5, Agnes Bukovinski1, Jessica Ruggeri6, Altayeb Elazomi7, and Paul Barrow1,8*1School of Veterinary Medicine and Science, University of Nottingham, Sutton Bonington Campus, Loughborough, UK 2Department of Veterinary Pathology, College of Veterinary Medicine, China Agricultural University, Beijing, China 3School of Biosciences, University of Nottingham, Sutton Bonington Campus, Loughborough, UK 4Cargill Animal Nutrition, Rotterdam, Netherlands 5CEVA Phylaxia, Budapest, Hungary 6Veterinary Services, ATS Brescia, Brescia, Italy 7Medical Laboratories Department, Medical Technology Faculty, University of Zawia, Zawiya, Libya 8School of Veterinary Medicine, University of Surrey, Guildford, UK †Both authors contributed equally to this work *Corresponding Author: Paul Barrow. School of Veterinary Medicine, University of Surrey, Guildford, UK. Email: nakaichi [at] yamaguchi-u.ac.jp Submitted: 30/11/2021 Accepted: 20/03/2022 Published: 04/04/2022 © 2022 Open Veterinary Journal
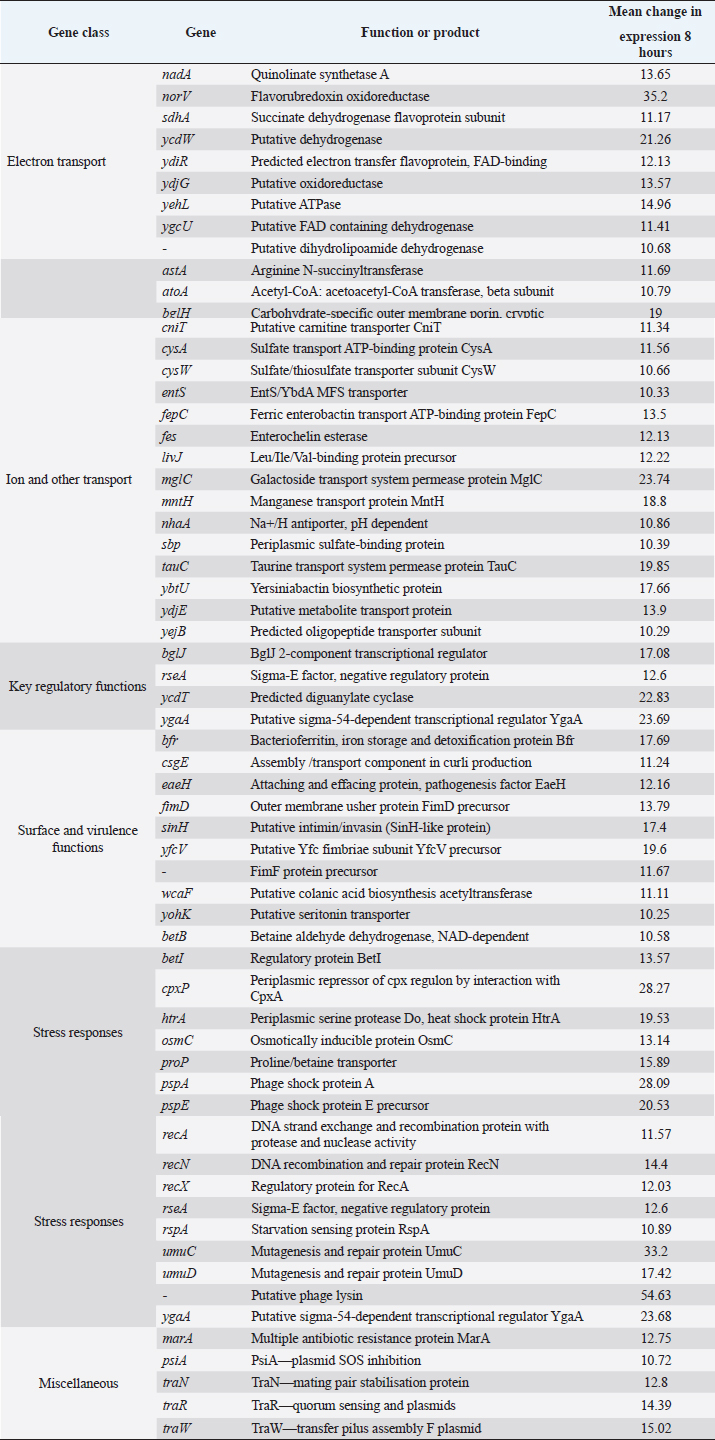
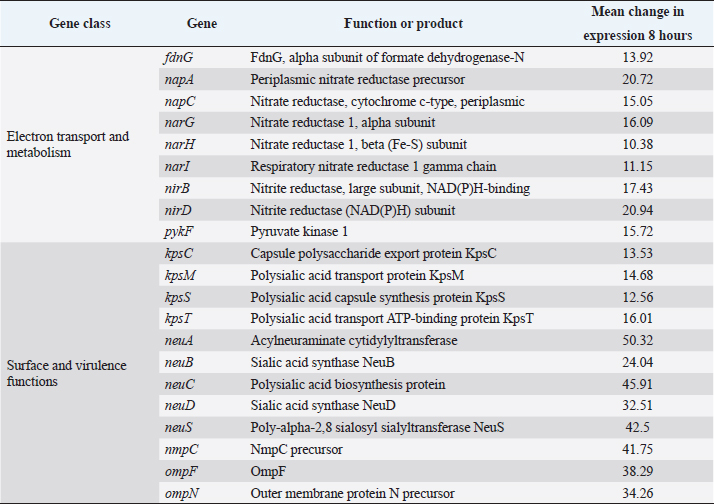
AbstractBackground: Escherichia coli remains a major pathogen of poultry. Most vaccines are inactivated and produced empirically. Although inactivated Salmonella vaccines have been produced by culture under conditions of Fe deprivation, no vaccines have been produced which are likely to express all the proteins expressed during infection of antigen-presenting cells. Aim: The aim was to produce a more protective inactivated vaccine by culturing the avian E. coli in a synthetic medium that resembled the environment of the phagolysosome. Methods: Global gene expression in a pathogenic avian O78:K80 strain of E. coli, harvested from infected avian macrophage-like HD11 cells, was compared by microarray with bacteria cultured in a tissue culture medium. A liquid synthetic medium was produced based on the environmental conditions identified to which the bacteria were exposed intracellularly. A bacterin was produced from this strain and its protective ability was assessed in chickens. Results: The changes in E. coli gene expression observed included the use of different electron acceptors and carbon sources such as ethanolamine, β-glucosides, galactonate, dicarboxylic acids, and amino acids, up-regulation of genes associated with Fe and Mn uptake, and up-regulation of type-1 and curli fimbriae, other adhesion genes and down-regulation of sialic acid synthesis genes. The bacterin produced in the synthetic medium was statistically more protective than a bacterin prepared from bacteria cultured in the nutrient broth when tested in vaccinated chickens challenged with a different virulent E. coli O78:K80 strain. Conclusion: The approach of using gene expression to produce synthetic media for the generation of more effective bacterins could be used for a number of intracellular bacteria pathogens including Enteroinvasive E. coli, Salmonella, and the Pasteurella/Riemerella/Mannheimia group of organisms. Keywords: Microarray, Vaccine, Chicken, Gene expression, Synthetic medium. IntroductionAvian pathogenic Escherichia coli (APEC) is a major poultry pathogen causing septicemia and polyserositis in the immediate post-hatching period (Nolan et al., 2013) or following infectious bronchitis (Dho-Moulin and Fairbrother, 1999) or turkey rhinotracheitis metapneumovirus infection (Picault et al., 1987). As a taxon, APEC comprises related clusters of strains expressing a range of virulence determinants associated with individual syndromes (Gyles and Fairbrother, 2010). No single combination of virulence determinants characterizes virulence. Identified determinants include individual fimbriae and other adhesins and invasins, serum resistance factors, and toxins (Nolan et al., 2013). The availability of several annotated genome sequences (Johnson et al., 2007; Dziva and Stevens, 2008; Dziva et al., 2013; Mangiamele et al., 2013), has facilitated the identification of new candidate virulence genes. Some serotypes are associated with both avian and human infection (Johnson et al., 2007). Although septicemia is characteristic of many poultry infections, the highest numbers of bacteria are isolated from the spleen, in addition to other organs, most probably located within macrophages (Smith et al., 1985; Barrow et al., 1998). The microbial behavior in this site and within antigen-presenting cells (APCs) is also important in initiating the immune response. Control of infection by chemotherapy inevitably selects for antibiotic resistance, especially in countries where regulation is less stringent (Wang et al., 2010). Inactivated vaccines (bacterins) are generally regarded as being less effective than live, attenuated vaccines because the latter stimulates both humoral and cellular immunity. However, issues including the use of genetically manipulated vaccines suggest that inactivated vaccines will be used for the foreseeable future. Attempts have been made to develop more rational inactivated bacterial vaccines (Woodward et al., 2002) by culturing the bacteria under conditions of iron restriction. However, studies with Salmonella typhimurium infecting mouse macrophages (Eriksson et al., 2003) and with avian serovars in chicken macrophages (Imre et al., 2013) indicate that Fe restriction is not the only feature characterizing the intracellular environment. We hypothesized that an analysis of the conditions inside macrophages using data generated by bacterial transcriptional analysis could lead to the development of a medium reproducing those conditions more accurately. A bacterin produced in this way should more closely resemble bacteria within macrophages and other APCs antigenically than bacteria cultured in nutrient broth (NB) and should be more protective. The Objective was therefore to infect the avian macrophage-like cell line HD11 with an APEC strain, carry out transcriptional analysis on the harvested bacteria by microarray and identify from the pattern of gene expression the environmental conditions to which the bacteria are subjected intracellularly. These conditions would be reproduced in a synthetic medium to culture the bacteria which would then be evaluated as a bacterin in chickens challenged with a different APEC strain. Material and MethodsBacterial strains and cultureEscherichia coli O78:K80 strains F31 and F135 were isolated from cases of avian colibacillosis. Both are virulent for chickens when inoculated parenterally (Barrow, unpublished results; Smith et al., 1985). NB (Oxoid, CM67) cultures were incubated at 37°C for 24 hours. These contained between 1 and 3 × 109 cfu/ml. Bacterial enumeration was made using the method of Miles et al. (1938) culturing on MacConkey agar plates (Oxoid CM0007). Cell culture and in vitro infection modelAvian macrophage-like HD11 cells (Beug et al., 1979) were grown in RPMI-1640 medium supplemented with 20 mM L-glutamine (Gibco), 2.5% fetal calf serum (Gibco), 2.5% chicken serum (Sigma) and 10% tryptose phosphate broth (Sigma). For each bacterial infection, a total of 3 × 107 HD11 cells were seeded in each of three tissue culture flasks (175 cm2, Nunc). Overnight NB cultures of bacterial strain F31 were diluted 20-fold in the cell culture medium described above and grown statically for 2 hours (5% CO2), then added onto the HD11 cells at a Multiplicity of Infection of ≈ 100. After co-incubation for 1 hour under the above conditions, the medium was replaced with a medium supplemented with 100 μg/ml gentamicin (Gm, Gibco). After the first 1 hour incubation, the medium was replaced with one containing 15 μg/ml Gm. Sampling points were at 0, 4, 8, 12, 24 and 48 hours post-infection. Cells were lysed (Barrow and Lovell, 1989) for bacterial enumeration. RNA extraction and processingThe RNA extraction protocol for F31 bacteria extracted from the infected cells and broth culture was that of Eriksson et al. (2003) and used by Imre et al. (2013). At 4 and 8 hours after infection, cells were lysed with 0.1% SDS, 1% phenol, and 19% ethanol in water for 30 minutes on ice. Bacteria were collected by centrifugation (5,000 g, 10 minutes, 4°C), treated with protective lysozyme and Proteinase K, and total RNA was prepared using the RNeasy Mini Kit (Qiagen). Bacterial RNA was also harvested from bacteria cultured in RPMI for 2 hours, which had been inoculated with a 20-fold dilution from the overnight NB culture, to produce bacterial numbers similar to those applied to the cell monolayers. RNA from bacteria grown in a cell culture medium was isolated using the same RNA purification kit. The quality of bacterial RNA and host RNA contamination was checked by a 2100 Bioanalyzer (Agilent). The RNA was amplified using the MessageAmp™ II-Bacteria Kit (Ambion), resulting in aminoallyl-UTP labeled amplified RNA (aRNA). For labeling, 6 µg aliquots of the aRNA samples were coupled with the fluorescent dyes Cy3 and Cy5 (Amersham). Cy3 was coupled to in vitro control RNA, while Cy5 was used to label bacterial RNA extracted from macrophages. Microarray design and data analysisThe sequence used for the array design was the APEC O1:K1:H7 reference strain (NC_008563) (Johnson et al., 2007). All predicted ORFs were designated for probe design. The web-based Agilent eArray system (Agilent Technologies, https://earray.chem.agilent.com/earray/) was used with the following settings during the microarray probe design: Tm (70°C) matching methodology, 60-mer probe length, 3 probes/target. The protocol, experimental setup, RNA extraction, amplification, labelling, and hybridization are described in detail at http://www.ebi.ac.uk/arrayexpress/. Data analysis was done using GeneSpring GX 10.0 (Agilent). The ultimate purpose of this analysis was to develop a synthetic medium reproducing these conditions as near as possible. For this reason, a full analysis of gene expression was not carried out by COGs gene classes. They were grouped according to genes likely to affect intracellular survival, response to stress, and virulence gene expression as indicated below. Synthetic Macrophage Medium (SMM-1)The composition of the SMM-1 was: 100 mM Tris-Cl, pH 5.0, 2 mM D-glucose, 2 mM D(+)-galactose, 20 mM glycerol, 20 mM glycerol-3-phosphate, 20 mg/; L-valine, 50 mg/l L-leucine, 50 mg/l L-isoleucine, 65 mg/l L-cysteine 2HCl, 5 mg/l L-tryptophan, 1 mM MgCl2, 200 uM 2,2-dipyridyl, 500 uM Cacl2, 3 mM ZnSO4, 50 mM K2SO4, 100 mM NaCl, 7.5 mM (NH4)2SO4 in 100 ml ultrapure water. The pH was adjusted to 5 with hydrochloric acid and the final medium filter sterilized. Bacterin productionA 5 ml aliquot of an overnight NB culture of E. coli F31 was added to 95 ml RPMI1640 medium (Invitrogen Ltd., Paisley, UK) pre-warmed to 37°C. This was incubated statically for 2 hours at 37°C in 5% CO2. Bacterial cells were recovered and washed twice in fresh SMM-1 medium or NB before being resuspended in fresh SMM-1 or NB respectively. These cultures were incubated statically at 37°C under aerobic conditions for 4 hours and the viable counts were estimated on nutrient agar (NA). A 0.2 ml aliquot of 40% formalin was added to 10 ml of the SMM-1 and NB cultures and allowed to stand for approximately 12 hours at ambient temperature followed by a 24 hours period at 4°C. Both formalinised preparations were centrifuged at 1,500 g for 30 minutes and both pellets were resuspended separately in 10 ml PBS. 100 µl aliquots were removed and cultured in NB (37°C, overnight) to determine sterility. These formalin-treated cell suspensions were adjusted to a density equivalent to ~1 × 109 CFU/ml before use as vaccine. Vaccination studiesThree groups (A, B, and C) of 25 1-day-old Hy-Line layers were housed in separate rooms on solid floors with shavings. At 4 and 18 days of age groups, A and B were inoculated with the bacterins without adjuvant. Birds were vaccinated simultaneously orally (100 µl with a blunt-ended needle) and intra-muscularly (100, 50 µl into each breast muscle), in the case of group A, with the SMM-1 bacterin, group B with the NB bacterin and group C with PBS. At 29 days of age, all birds were challenged intravenously with 1 × 105 cfu of a 24 hours NB culture of an O78:K80 E. coli strain F135. At 30, 31, 33 days of age, 5 birds, and at 36 days, 10 birds were killed from each group and the numbers of the inoculated challenge strain in homogenized samples of liver, spleen, and blood were counted on MacConkey agar. The protective effect was measured as reductions in the severity of frequency of morbidity if these occurred or by reductions in bacterial numbers in the blood, liver, and spleen. Ethical approvalAnimal experiments were carried out under a UK government Home Office Project and Personal Licence held by Prof. Barrow and reviewed internally by the University of Nottingham Animal Welfare and Ethical Review Body prior to initiation of the work. ResultsThe viable numbers of the F31 strain in the cultured HD11 cells fell from Log10 3.8 at 4 hours to 3.7 at 12 hours. RNA was therefore recovered 8 hours post-infection. Transcriptional profile of bacteria harvested from HD11 cellsThe patterns of gene expression of strain F31 in the intra-macrophage environment 8 hours after infection and grouped by activity, are shown in Tables 1 and 2. Genes showing statistically significant increases in expression of more than 10-fold are shown in Table 1. A more limited selection of genes of microbiological interest that were similarly significantly down-regulated is shown in Table 2. The ultimate purpose of this analysis was to develop a synthetic medium reproducing these conditions as near as possible. For this reason, a full analysis of gene expression was not carried out by COGs gene classes. They were grouped according to genes likely to affect intracellular survival, response to stress and virulence gene expression as indicated below (i – vi). (i) Electron transport. Upregulation of putative oxido-reductases and dehydrogenases coincided with downregulation of nitrate and nitrite reductases including nirBD (nitrite reductase), narGHI (nitrate reductase), and napAC (nitrate reductase with additional functions). Pyruvate kinase I (pykF) and formate dehydrogenase sub-unit I (fdnG) were also down-regulated. This suggested that the macrophage phagolysosome environment had a relatively high redox value as indicated by Turner et al. (2003). (ii) Carbon sources. A number of different loci associated with carbon sources utilized by the bacteria showed changes in expression although there was no clear picture. There was evidence that dicarboxylates were utilised as carbon sources suggested by up-regulation of the dcuB transporter gene and a diacid regulator cdaR. The gntR gene which regulates the gnt1 operon associated with gluconate utilization was up-regulated. The involvement of gluconate was emphasised by the up-regulation of idnK, a gluconate kinase. Expression of glcC was up-regulated. This is an activator of the glc operon which is associated with the transformation of glycolate to glyoxalate, and therefore may also be associated with osmotic stress rather than a prime carbon source (Núñez et al., 2001). Its regulation requires Integration Host Factor which affects virulence and responses to stressful conditions. Glyoxalate is also an important intermediate in the glyoxalate bypass involved in acetate or fatty acids as major energy sources (Pellicer et al., 1999). Other carbohydrates such as ethanolamine, mannose, and xylan may also have been important although the evidence through up-regulation of single genes eutA, gmd, and yieL, respectively was not strong as indicated by low changes in gene expression. Table 1. Changes in gene expression in E. coli F31 cultured in HD11 cells measured by microarray which were up-regulated by greater than 10-fold after 8 hours culture in HD11 cells.
(iii) Transport. Many of the transport genes up-regulated in macrophages were associated with the uptake of carbohydrates and other potential carbon sources. These included galactoside transport permease (mglC), a C4-dicarboxylate binding protein (dcuB, mentioned above), a putative hexuronate transporter, and transport proteins for oligopeptides (yejB), and long-chain fatty acids (fadL). Up-regulation of genes encoding ion transport proteins for sulfate (cysA, cysW, sbp, tauC), manganese (mntH), and ferric iron (fepC, entS, fes, ybtU), and iron storage (bfr) indicated restriction for these ions. (iv) Stress. In comparison with growth in NB, the bacteria were subjected to higher osmotic stress as indicated by a number of genes related to the biosynthesis of betaine (betAB), its regulator (betL), proP which transports both proline and betaine and osmC. The heat stress-related proteins htrA and cpxP were up-regulated. Stresses to DNA are indicated by up-regulation of recA and its regulator recX, recN, and umuC, and umuD. This is probably also linked to increased prophage activity with a number of genes up-regulated including pspA and pspE, a prophage integrase, intD and a putative phage lysin. (v) Surface and virulence proteins. The major virulence genes up-regulated were associated with fimbriae elaboration (csgE, fimD, fimF, and yfcV) and adhesins/invasins (eaeH and sinH). One of the colanic acid biosynthesis genes wcaF was up-regulated. These changes were accompanied by down-regulation of genes associated with sialic acid capsule formation. Genes encoding surface porins OmpF, NmpC, and the OmpN precursor were also down-regulated. Confirmation of levels of gene expression by qRT-PCR was not carried out. Although there are some limitations in the range of signal intensity obtainable with microarrays, most publications show very good correlation between levels of expression measured by microarray and by qRT-PCR or other quantitative methods, including E. coli in vitro (Richmond et al., 1999), Enterohaemorrhagic E. coli in contact with rabbit cells (Dahan et al., 2004), S. typhimurium in murine macrophages (Eriksson et al., 2003), in human and canine epithelial cells (Hautefort et al., 2008), the chicken intestine (Dhawi et al., 2011; Harvey e t a l., 2011) and with four different Salmonella enterica serovars in HD11 cells (Imre et al., 2013). Synthetic medium and vaccine developmentThe microarray results showed changes in gene expression indicative of availability of gluconate, succinate, and cysteine with Fe and Mn restriction and osmotic and heat stress, although the preferred carbon source was unclear. This information combined with published literature, indicating low phagosome pH, (Rathman et al., 1996), was used to develop a synthetic medium (SMM-1), the full composition which is described in Materials and Methods. This was used to generate a bacterin of E. coli F31 for vaccination studies. Vaccination experimentThe protective effects of the F31 bacterin produced by culture in medium SMM-1 against intravenous challenge of an O78:K80 strain F135 compared to those induced by a bacterin prepared using the same strain cultured in NB are shown in Table 3. Table 2. Changes in gene expression in E. coli F31 cultured in HD11 cells measured by microarray which were down-regulated by greater than 10-fold after 8 hours culture in HD11 cells.
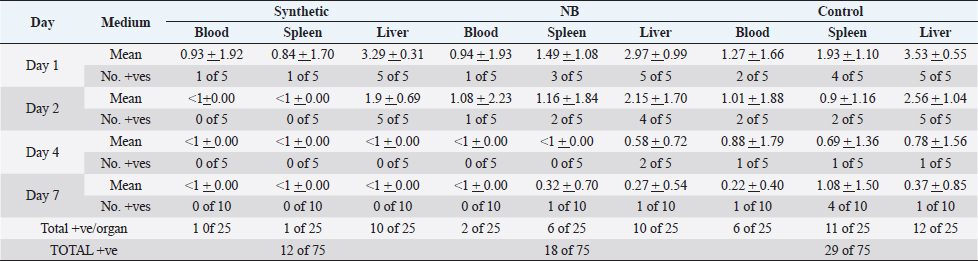
None of the birds showed any signs of illness. Tissue samples from a proportion of the birds from both the unvaccinated birds and those which had been vaccinated with the bacterin prepared from NB remained infected while the birds vaccinated with the bacterin cultured in the synthetic SMM-1 medium had cleared the challenge bacteria from their tissues almost by day 4 post-challenge. A comparison of the number of samples from which the challenge strain was isolated revealed a statistically significant difference between control unvaccinated and the NB vaccine group (χ2 =3.75, p =0.05), and between the control and synthetic medium group (χ2 =8.45, p =<0.01) but no statistically significant difference between the nutrient both and synthetic medium groups (χ2 =1.01, p =0.4). The differences were more marked for the blood and spleen samples than for the bacterial counts in the liver. Compared with the control group the level of significance for the synthetic medium group was higher than for the NB bacterin group. DiscussionVaccination is more desirable than chemotherapy to control APEC. Early protection mediated by high titre circulating specific antibodies may be important as indicated by the protection against parenteral avian Salmonella infections using the commercial Salenvac vaccine which stimulates high titer antibodies (Woodward et al., 2002). That high titer antibody correlates to some extent with protection is indicated by the fact that passively transferred antibody may protect against challenge with the homologous strain (Arp, 1980; Bolin and Jensen, 1987; Kariyawasam e t al., 2004). The ultimate purpose of this analysis was to develop a synthetic medium reproducing these conditions as near as possible. For this reason, a full analysis of gene expression was not carried out by COGs gene classes. They were grouped according to genes likely to affect intracellular survival, response to stress, and virulence gene expression. Our microarray studies here were designed to identify genes likely to affect intra-cellular survival, response to stress, and virulence gene expression. Although detailed analyses of gene expression were not carried out it was clear that the experiments showed that E. coli O78:K80 when established within avian macrophages, show patterns of physiology that are different from growth in liquid RPMI medium. These changes include a trend toward respiration with oxygen as a terminal electron acceptor indicating that oxygen was available as the preferred electron acceptor. There were indications from individual genes (gntR, gmd, glcC, yieL, and dcuB) that different carbohydrates (gluconate, mannose, glycolate, xylan, and dicarboxylate acids, respectively) may have been used but the picture was not clear from the small number of genes involved in each case. Table 3. The effect of vaccination with bacterins produced by culture in NB or SMM-1 on the numbers of E. coli F135 in the tissues of chickens challenged intravenously. Log10 (± SD) of E. coli F135 in the blood, liver or spleen together with the number of chickens in each group from which the challenge strain was isolated.
Osmotic stress was indicated by the expression of osmY and osmC (Yim and Villarejo, 1992) and the proU operon encoding a transport system for glycine, betaine, and proline, two osmo-protectant compounds (Haardt et al., 1995). In the absence of solid information on the importance of amino acids to E. coli within the macrophage, information was taken from the literature and from Imre et al. (2013) to develop SMM-1. Manganese was omitted and iron availability was restricted by the addition of 2, -2-dipyridyl. Osmolarity was adjusted with K+ and Na+ to a water activity less than that in RPMI. The SMM-1 medium clearly did not reflect all of the conditions within the macrophage. This was an approximation since the major carbon sources were unclear and we modeled this to include a reduction in glucose availability. Further studies could evaluate this in more detail and include incubation at a temperature closer to that of birds (41.5°C) which could also lead to even better immunogenicity and protection. The bacterin produced by culture in the synthetic medium produced a level of protection above the control which was more statistically significant than the bacterin produced by culture in NB. We propose that such an approach may be used for several bacterial pathogens for which inactivated vaccines are still produced regularly and this approach merits further investigation. FundingThe corresponding author would like to acknowledge the financial support of the Lower Saxony Federal Government (SLP137) and of a Biotechnology and Biological Sciences Research Council (BBSRC), UK China Partnership Award (CPA 1497) to Barrow. Conflict of interestThe authors have no conflict of interest associated with this work. Authors contributionAll authors consented to participation in the work and its publication. Barrow and Windhorst conceived the study and Barrow and Elazomi wrote and edited the manuscript. The work was carried out by Zhou, Imre, Bukovinski and Ruggeri (microarray work), Richards (vaccine development) and Barrow (animal work). ReferencesArp, L.H. 1980. Consequences of active or passive immunization of turkeys against Escherichia coli O78. Avian Dis. 24, 808–815. Barrow, P.A. and Lovell, M.A. 1989. Invasion of Vero cells by the Salmonella genus. J. Med. Microbiol. 28, 59–67. Barrow, P.A., Lovell, M.A. and Berchieri, A. Jnr. 1998. Use of lytic bacteriophage for control of experimental Escherichia coli septicaemia and meningitis in chickens and calves. Clin. Diagnost. Lab. Immunol. 5, 294–298. Beug, H., von Kirchbach, A., Doderlein, G., Conscience, J.F. and Graf, T. 1979. Chicken hematopoietic cells transformed by seven strains of defective avian leukemia viruses display three distinct phenotypes of differentiation. Cell 18, 375–390. Bolin, C.A. and Jensen, A.E. 1987. Passive immunization with antibodies against iron-regulated outer membrane proteins protects turkeys from Escherichia coli septicemia. Infect. Immun. 55, 1239–1242. Dahan, S., Knutton, S., Shaw, R.K., Crepin, V.F., Dougan, G. and Frankel, G. 2004. Transcriptome of Enterohaemorrhagic Escherichia coli O157 adhering to eukaryotic plasma membranes. Infect. Immun. 72, 5452–5459. Dhawi, A.A., Elazomi, A., Jones, M., Lovell, M.A., Li, H., Emes, R.D. and Barrow, P.A. 2011. Adaptation to the chicken intestine in Salmonella Enteritidis PT4 studied by transcriptional analysis. Vet. Microbiol. 153, 198–204. Dho-Moulin, M. and Fairbrother, J.M. 1999. Avian pathogenic Escherichia coli (APEC). Vet. Res. 30, 299–316. Dziva, F., Hauser, H., Connor, T.R., van Diemen, P.M., Prescott, G., Langridge, G.C., Eckert, S., Chaudhuri, R.R., Ewers, C., Mellata, M., Mukhopadbyay, S., Curtiss II, R., Dougan, G., Wieler, L.W., Thomson, N.R., Pickard, D.J. and Stevens, M.P. 2013. Sequencing and functional annotation of avian pathogenic Escherichia coli serogroup O78 strains reveal the evolution of E. coli lineages pathogenic for poultry via distinct mechanisms. Infect. Immun. 81, 838–849. Dziva, F. and Stevens, M.P. 2008. Colibacillosis in poultry: unravelling the molecular basis of virulence of avian pathogenic Escherichia coli in their natural hosts. Avian Pathol. 37, 355–366. Eriksson, S., Lucchini, S., Thompson, A., Rhen, M. and Hinton, J.C. 2003. Unravelling the biology of macrophage infection by gene expression profiling of intracellular Salmonella enterica. Mol. Microbiol. 47, 103–118. Gyles, C.L. and Fairbrother, J.M. 2010 Escherichia coli. In: Pathogenesis of bacterial infections in animals. Eds., Gyles, C.L., Prescott, J.F., Songer, J.G. and Thoen, C.O. Ames, IO: Wiley-Blackwell, pp: 267–308. Haardt, M., Kempf, B., Faatz, E. and Bremer, E. 1995. The osmoprotectant proline betaine is a major substrate for the binding-protein-dependent transport system ProU of Escherichia coli K-12. Mol. Gen. Genetics 246, 783–786. Harvey, P.C., Watson, M., Hulme, S., Jones, M.A., Lovell, M.A., Berchieri, A. Jr., Young, J., Bumstead, N. and Barrow, P.A. 2011. Salmonella enterica serovar typhimurium colonizing the lumen of the chicken intestine grows slowly and upregulates a unique set of virulence and metabolism genes. Infect. Immun. 79, 4105–4121. Hautefort, I., Thompson, A., Eriksson-Ygberg, S., Parker, M. L., Lucchini, S., Danino, V., Bongaerts, R.J., Ahmad, N., Rhen, M. and Hinton, J.C. 2008. During infection of epithelial cells Salmonella enterica serovar typhimurium undergoes a time-dependent transcriptional adaptation that results in simultaneous expression of three type 3 secretion systems. Cell. Microbiol. 10, 958–984. Imre, A., Bukovinski, A., Lovell, M.A., Li, H., Zhou, X. and Barrow, P.A. 2013. Gene expression analysis of Salmonella enterica SPI in macrophages indicates differences between serovars that induce systemic disease from those normally causing enteritis. Vet. Microbiol. 167, 675–679. Johnson, T.J., Kariyawasam, S., Wannemuehler, Y., Mangiamele, P., Johnson, S.J., Doetkott, C., Skyberg, J.A., Lynne, A.M., Johnson, J.R. and Nolan, L.K. 2007. The genome sequence of avian pathogenic Escherichia coli strain O1:K1:H7 shares strong similarities with human extraintestinal pathogenic E. coli genomes. J. Bacteriol. 189, 3228–3236. Kariyawasam, S., Wilkie, B.N. and Gyles, C.L. 2004. Resistance of broiler chickens to Escherichia coli respiratory tract infection induced by passively transferred egg-yolk antibodies. Vet. Microbiol. 98, 273–284. Mangiamele, P., Nicholson, B., Wannemuehler, Y., Seemann, T., Logue, C.M., Li, G., Tivendale, K.A. and Nolan, L.K. 2013. Complete genome sequence of the avian pathogenic Escherichia coli strain APEC O78. Genome Announc. 1(2), e0002613; doi: 10.1128/genomeA.00026-13 Miles, A.A., Misra, S.S. and Irwin, J.O. 1938. The estimation of the bactericidal power of the blood. J. Hygiene (Lond) 38, 732–749. Nolan, L.K., Barnes, H.J., Vaillancourt, J.P., Abdul-Aziz, T. and Logue, C.M. 2013. Colibacillosis. In: Diseases of poultry. Ed. Swayne, D.E. 13th ed. Ames, IO: Wiley-Blackwell. pp: 751–805. Núñez, M.F., Pellicer, M.T., Badía, J., Aguilar, J. and Baldomà, L. 2001. The gene yghK linked to the glc operon of Escherichia coli encodes a permease for glycolate that is structurally and functionally similar to L-lactate permease. Microbiology 147, 1069–1077. Pellicer, M.T., Fernandez, C., Badía, J., Aguilar, J., Lin, E.C. and Baldom, L. 1999. Cross-induction of glc and ace operons of Escherichia coli attributable to pathway intersection. Characterization of the glc promoter. J. Biol. Chem. 274, 1745–1752. Picault, J.P., Giraud, P., Drouin, P., Guittet, M., Bennejean, G., Lamande, J., Toquin, D. and Gueguen, C. 1987. Isolation of a TRTV-like virus from chickens with swollen-head syndrome. Vet. Rec. 121, 35. Rathman, M., Sjaastad, M.D. and Falkow, S. 1996. Acidification of phagosomes containing Salmonella typhimurium in murine macrophages. Infect. Immun. 64, 2765–2773. Richmond, C.S., Glasner, J.D., Mau, R., Jin, H. and Blattner, F.R. 1999. Genome-wide expression profiling in Escherichia coli K-12. Nucleic Acids Res. 27, 3821–3835. Smith, H.W., Cook, J.K.A. and Parsell. Z.E. 1985. The experimental infection of chickens with mixtures of infectious bronchitis virus and Escherichia coli. J. Gen. Virol. 66, 777–786. Turner, A.K., Barber, L.Z., Wigley, P., Muhammad, S., Jones, M.A., Lovell, M.A., Hulme, S. and Barrow, P.A. 2003. The contribution of proton-translocating proteins to the virulence of Salmonella enterica serovars typhimurium, S. gallinarum and S. Dublin in chickens and mice. Infect. Immun. 71, 3392–3401. Wang, X.M., Liao, X.P., Zhang, W.J., Jiang, H.X., Sun, J., Zhang, M.J. 2010. Prevalence of serogroups, virulence genotypes, antimicrobial resistance, and phylogenetic background of avian pathogenic Escherichia coli in south of China. Foodborne Pathogens Dis. 7, 1099–1106. Woodward, M.J., Gettinby, G., Breslin, M.F., Corkish, J.D. and Houghton, S. 2002. The efficacy of Salenvac, a Salmonella enterica subsp. Enterica serotype Enteritidis iron-restricted bacterin vaccine, in laying chickens. Avian Pathol. 31, 383–392. Yim, H.H. and Villarejo, M. 1992. osmY, a new hyperosmotically inducible gene, encodes a periplasmic protein in Escherichia coli. J. Bacteriol. 174, 3637–3644. | ||
| How to Cite this Article |
| Pubmed Style Zhou X, Richards P, Windhorst D, Imre A, Bukovinski A, Ruggeri J, Elazomi A, Barrow P. Generation of an inactivated vaccine for avian pathogenic Escherichia coli using microarrays; a more rational approach to inactivated vaccine design. Open Vet J. 2022; 12(2): 221-230. doi:10.5455/OVJ.2022.v12.i2.10 Web Style Zhou X, Richards P, Windhorst D, Imre A, Bukovinski A, Ruggeri J, Elazomi A, Barrow P. Generation of an inactivated vaccine for avian pathogenic Escherichia coli using microarrays; a more rational approach to inactivated vaccine design. https://www.openveterinaryjournal.com/?mno=31285 [Access: July 27, 2024]. doi:10.5455/OVJ.2022.v12.i2.10 AMA (American Medical Association) Style Zhou X, Richards P, Windhorst D, Imre A, Bukovinski A, Ruggeri J, Elazomi A, Barrow P. Generation of an inactivated vaccine for avian pathogenic Escherichia coli using microarrays; a more rational approach to inactivated vaccine design. Open Vet J. 2022; 12(2): 221-230. doi:10.5455/OVJ.2022.v12.i2.10 Vancouver/ICMJE Style Zhou X, Richards P, Windhorst D, Imre A, Bukovinski A, Ruggeri J, Elazomi A, Barrow P. Generation of an inactivated vaccine for avian pathogenic Escherichia coli using microarrays; a more rational approach to inactivated vaccine design. Open Vet J. (2022), [cited July 27, 2024]; 12(2): 221-230. doi:10.5455/OVJ.2022.v12.i2.10 Harvard Style Zhou, X., Richards, . P., Windhorst, . D., Imre, . A., Bukovinski, . A., Ruggeri, . J., Elazomi, . A. & Barrow, . P. (2022) Generation of an inactivated vaccine for avian pathogenic Escherichia coli using microarrays; a more rational approach to inactivated vaccine design. Open Vet J, 12 (2), 221-230. doi:10.5455/OVJ.2022.v12.i2.10 Turabian Style Zhou, Xiangmei, Philip Richards, Daniel Windhorst, Ariel Imre, Agnes Bukovinski, Jessica Ruggeri, Altayeb Elazomi, and Paul Barrow. 2022. Generation of an inactivated vaccine for avian pathogenic Escherichia coli using microarrays; a more rational approach to inactivated vaccine design. Open Veterinary Journal, 12 (2), 221-230. doi:10.5455/OVJ.2022.v12.i2.10 Chicago Style Zhou, Xiangmei, Philip Richards, Daniel Windhorst, Ariel Imre, Agnes Bukovinski, Jessica Ruggeri, Altayeb Elazomi, and Paul Barrow. "Generation of an inactivated vaccine for avian pathogenic Escherichia coli using microarrays; a more rational approach to inactivated vaccine design." Open Veterinary Journal 12 (2022), 221-230. doi:10.5455/OVJ.2022.v12.i2.10 MLA (The Modern Language Association) Style Zhou, Xiangmei, Philip Richards, Daniel Windhorst, Ariel Imre, Agnes Bukovinski, Jessica Ruggeri, Altayeb Elazomi, and Paul Barrow. "Generation of an inactivated vaccine for avian pathogenic Escherichia coli using microarrays; a more rational approach to inactivated vaccine design." Open Veterinary Journal 12.2 (2022), 221-230. Print. doi:10.5455/OVJ.2022.v12.i2.10 APA (American Psychological Association) Style Zhou, X., Richards, . P., Windhorst, . D., Imre, . A., Bukovinski, . A., Ruggeri, . J., Elazomi, . A. & Barrow, . P. (2022) Generation of an inactivated vaccine for avian pathogenic Escherichia coli using microarrays; a more rational approach to inactivated vaccine design. Open Veterinary Journal, 12 (2), 221-230. doi:10.5455/OVJ.2022.v12.i2.10 |