| Original Article | ||
Open Vet J. 2022; 12(3): 391-398 Open Veterinary Journal, (2022), Vol. 12(3): 391–398 Original Research Identification, serotyping, and antimicrobial susceptibility of Riemerella anatipestifer isolated from ducks in VietnamThanh-Thin Vo1*, Van-Tuan Dang1, Dinh-Hai Le1 and Trong-Hai Nguyen1,21Bacteriology Department, Institute of Veterinary Research and Development, Nha Trang city, Khanh Hoa province, 57000, Vietnam 2Vaccine and Infectious Disease Organization, University of Saskatchewan, 120 Veterinary Road Saskatoon, SK, S7N 5E3, Canada *Corresponding Author: Thanh-Thin Vo. Bacteriology Department, Institute of Veterinary Research and Development. Nha Trang, Vietnam. Email: vothanhthin [at] gmail.com Submitted: 18/01/2022 Accepted: 27/05/2022 Published: 17/06/2022 © 2022 Open Veterinary Journal
AbstractBackground: Septicemia caused by Riemerella anatipestifer (R. anatipestifer) is a serious problem in the duck industry worldwide, and it is currently one of the major concerns for duck farming in Vietnam.. Aim: This study was conducted to identify the causative agent of septicemia in ducks in Vietnam. The antimicrobial susceptibility and serotypes of R. anatipestifer isolates were also determined to provide valuable information for disease treatment and vaccine development. Methods: Riemerella anatipestifer was isolated using blood agar and chocolate agar media. The commercial API 20NE microtest system and the partial nucleotide sequence analysis of the 16s rRNA were used to identify R. anatipestifer strains. Serotypes were determined by slide agglutination test using standard antisera against R. anatipestifer. The disk diffusion method was utilized to investigate the antimicrobial susceptibility of R. anatipestifer isolated strains. Results: A total of 408 samples were collected from ducks with typical symptoms of septicemia for R. anatipestifer isolation. Sixty-nine R. anatipestifer strains were identified. Serotyping results showed that 30 out of 69 bacterial strains were classified as serotypes 1, 6, 8, 10, and 20, with serotype 10 being the most prevalent. The antimicrobial susceptibility test revealed that 100% of the bacterial isolates were susceptible to Amoxicillin/clavulanic acid and Imipenem. On the contrary, the majority of R. anatipestifer strains were resistant to Nalidixic acid (89.9%), Streptomycin (75.4%), and Norfloxacin (72.5%). Conclusion: This is the first ever report in terms of identification, serotyping, and antimicrobial susceptibility tests of R. anatipestifer causing septicemia in ducks of Vietnam, providing useful scientific information for treatment as well as vaccine development to control the disease. Keywords: Antimicrobial susceptibility, Ducks, R. anatipestifer, Serotyping. IntroductionRiemerella anatipestifer causes septicemia in ducks, turkeys, geese, and other avian species. It is a Gram-negative, immobile, nonspore-forming, and rod-shaped bacterium (Huang et al., 2002). In ducks, the contagious septicemic disease usually occurs in acute or chronic septicemia, showing typical clinical symptoms, such as visual disturbance, nasal discharge, coughing, sinusitis, diarrhea, and neural signs, including torticollis, head tremor, and movement disorder. Without proper treatment, the disease may take several weeks and cause high mortality rates in affected ducks (Ruiz and Sandhu, 2013). The infection has led to significant economic losses for duck farming worldwide (Sun et al., 2012). Several studies on R. anatipestifer causing septicemia in ducks and its antimicrobial susceptibility profiles have been published in recent years. Riemerella anatipestifer strains were reported to be resistant to a wide range of antibiotics, including Aztreonam, Cefepime, Oxacillin, Penicillin G, Ceftazidime, Trimethoprim/sulfamethoxazole (Zhong et al., 2009), Flumequine, Tetracycline, Erythromycin, and Streptomycin (Gyuris et al., 2017). More than 79% of R. anatipestifer strains in Taiwan were resistant to at least 3 commonly used antibiotics (Chang et al., 2019). In Vietnam, duck farming has been becoming increasingly popular, contributing to a large proportion of the country’s food supply. However, R. anatipestifer infection has frequently occurred in many duck farms across the country in recent years, resulting in significant financial losses. Antibiotics are mainly used for the treatment and control of septicemia caused by R. anatipestifer and other infectious diseases. Improper use of antibiotics, however, has resulted in the alarming phenomenon of antibiotic resistance, making treatment ineffective. In the present study, we identified the causative agent of septicemia in ducks farmed in Vietnam and evaluated its antibiotic susceptibility. The results of this study should provide useful information for developing effective treatment and preventative techniques in the duck industry in Vietnam, adding to the global effort to combat the disease. Materials and MethodsSample collection and R. anatipestifer isolationSpecimen samples, including brains, hearts, lungs, blood, and livers, were collected from 408 ducks with specific symptoms of septicemia farmed in 4 different provinces, including Thai Binh, Thanh Hoa in the north, Binh Dinh in the center, and Long An in the south of Vietnam (Fig. 1) from 2018 to 2020 for R. anatipestifer isolation. The bacteria was isolated by streaking on blood agar (Trypticase soy agar with 5% sheep blood) and/or chocolate agar and incubating in aerobic condition at 37°C for 24–48 hours. All suspected colonies of R. anatipestifer were subcultured on brain heart infusion broth and blood agar for Gram staining and biological test using the commercial API 20NE microtest system (BioMerieux, France). Molecular identification of R. anatipestiferRiemerella anatipestifer isolates were verified by polymerase chain reaction (PCR), amplifying the partial region of the igene. Total DNA was extracted from the R. anatipestifer culture, using the Wizard Genomic DNA Purification Kit (Promega, USA). The expected 662 bp DNA fragment of 16s rRNA gene was amplified using primer pair F-5′-CAGCTTAACGTAGAACTGC-3′/R-5′-TCGAGATTTG-CATCACTTCG-3′ (Tsai et al., 2005). PCR reactions were carried out on a thermal cycler (Bio-Rad), under the following conditions: initial denaturation at 94°C for 5 minutes, followed by 35 cycles of denaturation at 94°C for 1 minute, annealing at 60°C for 1 minute, extension at 72°C for 1 minute, and ended with a final extension of 72°C for 10 minutes. The amplified PCR products were electrophoresed on 1.5% (w/v) agarose gels stained with ethidium bromide. The 16s rRNA amplified fragments were subsequently sequenced (Nam Khoa Co. Ltd, Ho Chi Minh City, Vietnam), using an automated DNA sequencer (Applied Biosystems, USA) to reconfirm the identification of R. anatipestifer. Nucleotide sequences were then aligned and compared with the R. anatipestifer 16s rRNA sequences retrieved from GenBank using ClustalW program software within BioEdit Sequence Alignment Editor (version 7.2.5). All the nucleotide sequences were finally submitted to GenBank for R. anatipestifer identification, using the BLAST program on the NCBI website (https://www.ncbi.nlm.nih.gov/).
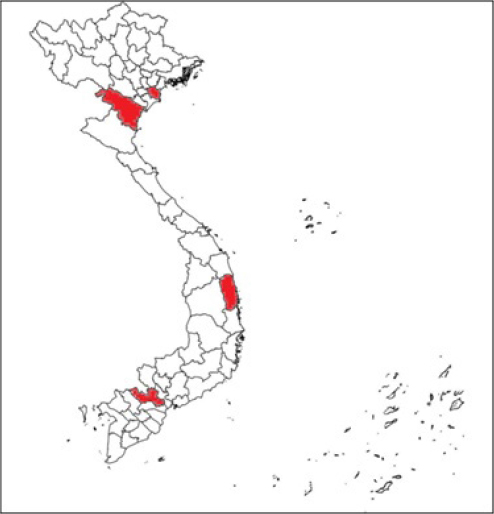
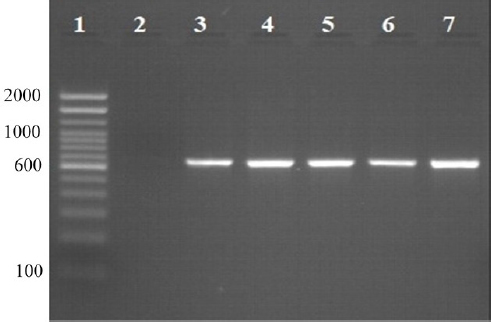
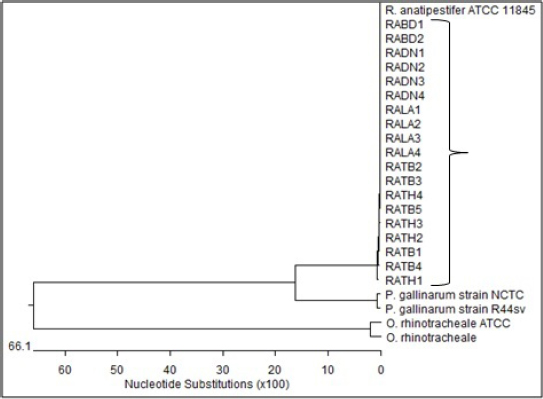
Fig. 1. Locations of the four provinces (shaded) in Vietnam where samples were collected (from the North to the South: Thai Binh, Thanh Hoa, Binh Dinh, and Long An). Serotyping of R. anatipestifer isolatesRiemerella anatipestifer isolates were serotyped by the slide agglutination method, using a series of 21 standard antisera against R. anatipestifer, which was kindly provided by the Animal Health Research Institute, Taiwan. In short, 20 μl of the standard antiserum for R. anatipestifer serotyping was mixed with the same volume of single colony suspension from each bacterial isolate. The reaction is positive if clumping of bacteria was observed within 1 or 2 minutes. Riemerella anatipestifer isolates would be considered nonserotypable, if not agglutinated with any of the 21 tested antisera. Antimicrobial susceptibilityThe disk diffusion on Mueller-Hinton agar method was utilized to test the antibiotic susceptibility of R. anatipestifer isolates (Hudzicki, 2009). Twenty-five antibiotic disks (Oxoid, UK) were used, including Amikacin (30 µg), Amoxicillin/clavulanic acid (30 µg), Cefepime (30 µg), Cefoperazone (30 µg), Ceftazidime (30 µg), Ceftiofur (30 µg), Ceftriaxone (30 µg), Cephalexin (30 µg), Cephalothin (30 µg), Ciprofloxacin (5 µg), Doxycycline (30 µg), Erythromycin (15 µg), Florfenicol (30 µg), Gentamicin (10 µg), Imipenem (10 µg), Kanamycin (30 µg), Nalidixic acid (30 µg), Neomycin (30 µg), Norfloxacin (10 µg), Oxacilin (5 µg), Penicillin G (10 UI), Piperacillin (30 µg), Streptomycin (10 µg), Tetracyline (30 µg), and Trimethoprime/sulfamethoxazol (23.75/1.25 µg). The inhibition zone diameters were interpreted according to the criteria of the Clinical and Laboratory Standards Institute (USA) guidelines (CLSI, 2019). ResultsSample collection and R. anatipestifer isolationRiemerella anatipestifer was isolated from the samples, including brains, hearts, lungs, blood, and livers. Of the total 408 ducks, which were having typical clinical symptoms of septicemia, 69 ducks tested positive with R. anatipestifer in at least 1 kind of specimens. In aerobic conditions, the bacteria grow well on blood agar, forming round, convex shape, slightly wet mucus, transparent white, and nonhemolytic colonies with about 1–2 mm in diameter after 24–48 hours of incubation at 37°C (Fig. 2). On chocolate agar, R. anatipestifer formed white to gray, wet, slightly mucus colonies, and had almost the same size as on blood agar medium (Fig. 3). The morphology of R. anatipestifer cells was observed under an optical microscope at 1,000× magnification. Typically, R. anatipestifer cells were Gram-negative, nonspore-forming, and regular rod-shaped with round ends (Fig. 4). Molecular identification of R. anatipestifer isolatesAll 69 R. anatipestifer isolates were further verified by PCR amplifying 16s rRNA gene. As expected, a 662-base pairs PCR product was successfully amplified from all of 69 tested isolates (Fig. 5). The PCR amplicons were subsequently sequenced from both directions, using the same primers for PCR. All the nucleotide sequences were aligned, using ClustalW program software within BioEdit Sequence Alignment Editor (version 7.2.5). Multiple sequences alignments were generated, using gap open and gap extension penalties of 10 and 1, respectively. Nucleotide sequence identity or similarity was from 99.0% to 100% among the 69 R. anatipestifer isolates, as well as compared with other R. anatipestifer strains, including the reference strains ATCC 11845 published on the GenBank, using BLAST tool on the NCBI (data not shown). Analyzing the phylogenetic tree based on the nucleotide sequences of the 16s rRNA gene of 19 representative R. anatipestifer isolates showed that all isolates were grouped in a single cluster. Riemerella anatipestifer isolates from samples in the three different geological regions were highly identical. All of the 19 representative R. anatipestifer isolates in this study were completely separated from the 2 other close genetic relationship species, Pasteurella gallinarum and Ornithobacterium rhinotracheale. The partial 16s rRNA nucleotide sequences of all 19 R. anatipestifer isolates in this study, which showed in the phylogenetic tree (isolates in the bracket) (Fig. 6), were submitted to GenBank with accession numbers MT012264, MT012265, MT012266, MT012267, MT012268, MT012269, MT012270, MT012271, MT012272, MT012273, MT012274, MT012275, MT012276, MT012277, MT012278, MT012279, MT012280, MT012281, and MT012282.
Fig. 2. Colonies of R. anatipestifer on the blood agar.
Fig. 3. Colonies of R. anatipestifer on the chocolate agar.
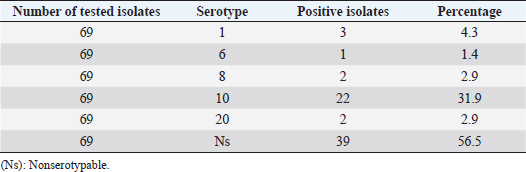
Fig. 4. Gram stain of R. anatipestifer (1,000×). Serotyping of R. anatipestiferA total of 69 R. anatipestifer isolates, which have been verified by PCR and sequencing, were serotyped. The results show that 30 isolates were classified into 5 different serotypes, including serotypes 1, 6, 8, 10, and 20. Among these, serotype 10 comprised the majority of the isolates, with 22 isolates, accounting for 31.9%. Serotype 1 was represented by 3 strains (4.3%), whereas this number was 2 isolates (2.9%) for both serotypes 8 and 20. Finally, serotype 6 was determined in only 1 out of 69 R. anatipestifer isolates, accounting for 1.4%. It is worth noting that 39 R. anatipestifer isolates (56.5%) were unable to determine serotypes. These strains showed negative reaction with all 21 antisera tested (Table 1). In addition, of the total 69 R. anatipestifer isolates tested, serotypes 2, 3, 4, 5, 7, 9, 11, 12, 13, 14, 15, 16, 17, 18, 19, and 21 were not observed (data not shown).
Fig. 5. PCR amplicons (662 bp) of 16s rRNA gene of R. anatipestifer. Lane 1: DNA marker 100 bp; lane 2: negative control; lane 3: positive control; and lane 4–7: R. anatipestifer isolates.
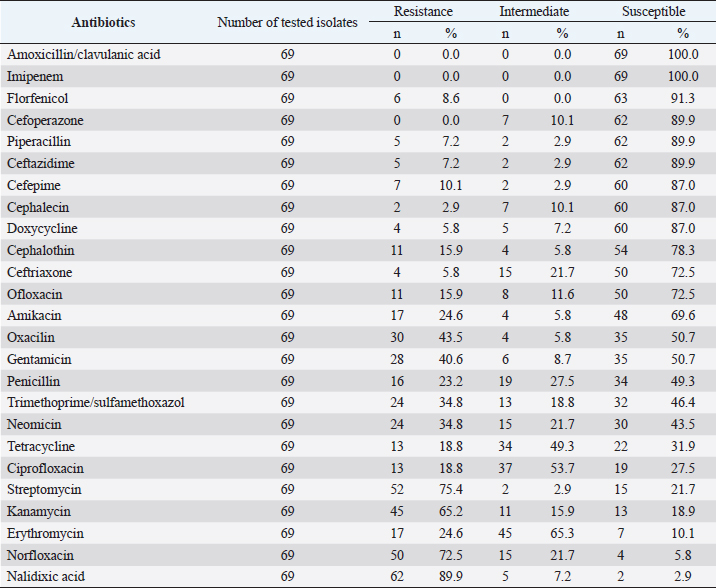
Fig. 6. Phylogenetic relationship between R. anatipestifer isolates, R. anatipestifer reference strain, P. gallinarum, and O. rhinotracheale based on the partial sequences of 16s rRNA gene. Antimicrobial susceptibilityA total of 69 R. anatipestifer isolates were examined for antimicrobial susceptibility to 25 commonly used antibiotics by disk diffusion method. The results indicated that all isolates (100%) were susceptible to Amoxicillin/clavulanic acid and Imipenem, and 63 isolates (91.3%) were susceptible to Florfenicol. Ceftriaxone was found less effective as only 50 out of 69 isolates (72.5%) were susceptible to this antibiotic. On the other hand, the majority of R. anatipestifer isolates were resistant to Nalidixic acid (89.9%), Streptomycin (75.4%), and Norfloxacin (72.5%) (Table 2). DiscussionIn this study, R. anatipestifer was successfully isolated from at least one organ, mostly from the brain, heart, or lungs of 69 out of 408 ducks, which were having typical symptoms of septicemia. The rate of ducks positive with R. anatipestifer was 16.9% (data not shown). There were differences in the rate of R. anatipestifer infection in previous reports. In Denmark, the percentage of ducks infected with R. anatipestifer was 80% (Ryll et al., 2001). Riemerellosis in ducks was reported in India with an infection rate of 40% and 75% mortality (Priya et al.,2008). The rate of R. anatipestifer infection in ducks was 16.7% in Egypt (Heba et al., 2015). A study in China revealed that 26 out of 56 duck farms suffered from R. anatipestifer infection (Wang et al., 2012). Table 1. Serotyping of R. anatipestifer isolates.
Table 2. The susceptibility of R. anatipestifer isolates to commonly used antibiotics.
According to Ruiz and Sandhu (2013), ducks infected with some different bacterial species such as P. multocida, R. anatipestifer, O. rhinotracheale, or Escherichia coli usually show similar clinical signs, so that it is difficult to correctly diagnose Riemerellosis based on those clinical symptoms. Therefore, isolation and genetic-based identification of R. anatipestifer from the suspected ducks are necessary. The 16s rRNA is usually used for the determination of the genetic relationship between bacteria because it is a stable fragment on the chromosome, and highly conserved in all organisms (Tsai et al., 2005). In the present study, sequence analysis of 662 bp fragment of 16s rRNA showed a close similarity between 19 representative R. anatipestifer isolates recovered from ducks farmed in different geographical origins in Vietnam. In comparison with R. anatipestifer strains published in GenBank, the divergences were also extremely low, ranging from 0.0% to 1.0%. This result suggests that the PCR primers used in this study could be applied for screening or diagnostic of R. anatipestifer infection in Vietnam. In this study, 30 out of 69 R. anatipestifer isolates were classified into 5 serotypes, including serotypes 1, 6, 8, 10, and 20. However, up to 39 out of 69 isolates were nonserotypable, accounting for 56.5%. According to Rubbenstroth et al. (2013), 21 serotypes of R. anatipestifer have been identified; however, some of the R. anatipestifer strains have not been serotyped or do not belong to the 21 identified serotypes. Research conducted by Cheng et al. (2003) showed that R. anatipestifer strains isolated from ducks in China mostly belong to serotypes 1, 2, 3, 4, 5, 6, 7, 8, 10, 11, 13, and 14. However, this study also found several strains that belong to 4 different serotypes other than 21 identified serotypes and proposed as serotypes 22, 23, 24, and 25. The serotypes 3, 21, and B, not belonging the 21 reported serotypes, were also found to be the 3 dominant serotypes among R. anatipestifer strains isolated from waterfowl slaughterhouses in Taiwan (Chang et al., 2019). In 2013, serotype B was reported as one of the three most predominant serotypes of R. anatipestifer strains isolated from sick waterfowls from 2008 to 2012 in Taiwan (Chen et al., 2013). The predominance of nonserotypable R. anatipestifer strains was also found in the survey conducted in Southern Taiwan duck farms. Among 11 tested strains, 6 of them were nonserotypable, accounting for 54.54% (Phonvisay et al., 2017). Previous studies reported that several serotypes of R. anatipestifer strains could exist at the same time in a field or even in a farm. Moreover, the serotype of R. anatipestifer strains commonly changes by the times within a farm. Noticeably, there was a high proportion of nonserotypable R. anatipestifer strains, and no cross-protection between the different serotypes (Gyuris et al., 2017; Chang et al., 2019). In Vietnam, no studies on R. anatipestifer and riemerellosis were conducted previously. This is the first research on the serotype of R. anatipestifer strains isolated from ducks with riemerellosis symptoms. Therefore, the result of R. anatipestifer serotyping in this study is critically important, providing the foundation for further studies, especially for vaccine development and disease prevention. In the present study, the susceptibility of R. anatipestifer to 25 most commonly used antibiotics was examined. Our findings are consistent with those of Chang et al. (2019), who observed 98% of the R. anatipestifer isolates to be susceptible to Amoxicillin/clavulanic acid, while 71.4% of the tested strains resistant to Streptomycin. Zhong et al. (2009) reported that the percentage of R. anatipestifer strains susceptible to Imipenem was 96.7%. In another study conducted by Gyuris et al. (2017), 97% of the R. anatipestifer isolates were susceptible to Florfenicol, whereas 68.4% were resistant to Nalidixic acid. However, we identified some discrepancies in the antibiotic susceptibility profiles of R. anatipestifer isolates; for example, a higher percentage of R. anatipestifer strains were resistant to Norfloxacin (70%) compared to the resistance rate (21.1%) in a previous study (Chang et al., 2019). The disparity could be attributed to bacterial strains isolated at different times, or from duck farms with different breeding procedures and antibiotic usage in disease prevention and treatment. Interestingly, there was 100% of 69 R. anatipestifer isolates tested susceptible to Amoxicillin/clavulanic acid and Imipenem. These two antibiotics are members of the β-Lactam group, which inhibits bacterial growth by interfering with the process of bacterial cell wall function. Amoxicillin/clavulanic acid is a combination of Amoxicillin and Clavulanic acid – the β-lactamases inhibitor (Evans et al., 2021). Imipenemis is a sub-β-Lactam antibiotic and belongs to the Carbapenem family. It is particularly stable in the presence of β-lactamases produced by bacteria (Codjoe and Donkor, 2018). This could explain why these two antimicrobial drugs were particularly efficient in suppressing R. anatipestifer strains in this investigation. Antibiotics have been widely used in animal husbandry in Vietnam to treat and prevent infectious diseases. This remains an important disease control tool as no vaccines have been developed to prevent septicemia caused by R. anatipestifer in ducks and other avian species. Antibiotic resistance in bacteria, on the other hand, has become an alarming problem. Therefore, antibiotic susceptibility of bacterial pathogens must be assessed to help veterinarians and farm owners use antibiotics effectively and avoid antibiotic overuse or abuse on their farms. AcknowledgmentThe authors would like to thank the Animal Health Research Institute, Taiwan, for kindly offering standard antiserum for R. anatipestifer serotyping. The authors also would like to thank Nam Khoa Co. Ltd (Ho Chi Minh City, Vietnam) for technical support. This research was funded by the Ministry of Agriculture and Rural Development of Vietnam. Conflict of interestThe authors declare that there is no conflict of interest. Authors’ contributionsThanh-Thin Vo and Trong-Hai Nguyen conceived and designed the experiments. Van-Tuan Dang and Dinh-Hai Le performed the experiments. Thanh-Thin Vo and Dinh-Hai Le analyzed the data. Thanh-Thin Vo and Van-Tuan Dang wrote the manuscript. All authors read and approved the final manuscript. ReferencesChang, F.F., Chen, C.C., Wang, S.H. and Chen C.L. 2019. Epidemiology and antibiogram of Riemerella anatipestifer isolated from waterfowl slaughterhouses in Taiwan. J. Vet. Res. 63(1), 79–86. Chen, Y., Lee, S. and Tsai, H. 2013. Serotyping of Riemerellar anatipestifer isolates from waterfowl in Taiwan between 2008 and 2012. Exp. Rep. Taiwan AHRI. 48, 21–28. Cheng, A., Wang, M., Chen, X., Zhu, D., Huang, C., Liu, F., Zhou, Y., Guo, Y., Liu, Z. and Fang, P. 2003. Epidemiology and new serotypes of Riemerella anatipestifer isolated from ducks in China and studies on their pathogenic characteristics. Chinese J. Vet. Sci. 23(4), 320–323. CLSI. 2019. Performance standards for antimicrobial susceptibility testing, 29th ed. Wayne, PA: Clinical and Laboratory Standards Institute. Codjoe, F.S. and Donkor, E.S. 2018. Carbapenem resistance: a review. Med. Sci. 6(1), 1. Evans, J., Hannoodee, M. and Wittler, M. 2021. Amoxicillin clavulanate. In StatPearls. Treasure Island, FL: StatPearls Publishing. Gyuris, É., Wehmann, E., Czeibert, K. and Magyar, T. 2017. Antimicrobial susceptibility of Riemerella anatipestifer strains isolated from geese and ducks in Hungary. Acta. Vet. Hung. 65(2), 153–165. Heba, D., Samir, A., Mohamed, K,F. and El-Jakee, J. 2015. Identification of duck septicemia in Egypt. Glob. Vet. 15, 397–400. Huang, B., Kwang, J., Loh, H., Frey, J., Tan, H.M. and Chua, K.L. 2002. Development of an ELISA using a recombinant 41 kDa partial protein (P45N′) for the detection of Riemerella anatipestifer infections in ducks. Vet. Microbiol. 88(4), 339–349. Hudzicki, J. 2009. Kirby-Bauer Disk Diffusion Susceptibility Test Protocol. American Society for Microbiology, USA. Phonvisay, M., Liou, J.J., Cheng, L.T., Chen, Y.P., Wu, H.C., Liu, C.H., Lee, J.W. and Chu, C.Y. 2017. Survey of a Riemerella anatipestifer outbreak in southern Taiwan duck farms. Taiwan Vet. J. 43(3), 165–170. Priya, P., Pillai, D.S., Balusamy, C., Rameshkumar, P. and Senthamilselvan, P. 2008. Studies on outbreak of new duck disease in Kerala, India. Int. J. Poult. Sci.7(2), 189–190. Rubbenstroth, D., Ryll, M., Knobloch, J.K.M., Köhler, B. and Rautenschlein, S. 2013. Evaluation of different diagnostic tools for the detection and identification of Riemerella anatipestifer. Avi. Path. 42(1), 17–26. Ruiz, J. and Sandhu, T. 2013. Riemerella anatipestifer infection. In Diseases of poultry, 13th ed. Eds., Swayne, D.E., Glisson, J.R., McDougald, L.R., Nolan, L.K., Suarez, D.,L., and Nair, V., Hoboken, NJ: Wiley Blackwell, pp: 824–828. Ryll, M., Christensen, H., Bisgaard, M., Christensen, J.P., Hinz, K.H. and Köhler, B. 2001. Studies on the prevalence of Riemerella anatipestifer in the upper respiratory tract of clinically healthy ducklings and characterization of untypable strains. J. Vet Med. Ser. B. 48(7), 537–546. Sun, N., Liu, J.H., Yang, F., Lin, D.C., Li, G.H., Chen, Z.L. and Zeng, Z.L. 2012. Molecular characterization of the antimicrobial resistance of Riemerella anatipestifer isolated from ducks. Vet. Microbiol. 158(3), 376–383. Tsai, H.J., Liu, Y.T., Tseng, C.S. and Pan, M.J. 2005. Genetic variation of the ompA and 16s rRNA genes of Riemerella anatipestifer. Avi. Path. 34(1), 55–64. Wang X, Zhu D, Wang M, Chen, A., Jia, R., Zhou, Y., Chen, Z., Luo, Q., Liu, F., Wang, Y. and Chen X.Y. 2012. Complete genome sequence of Riemerella anatipestifer reference strain. J. Bacteriol. 194(12), 3270–3271. Zhong, C.Y., Cheng, A.C., Wang, M.S., Zhu, D.K., Luo, Q.H., De Zhong, C., Li, L. and Duan, Z. 2009. Antibiotic susceptibility of Riemerella anatipestifer field isolates. Avian Dis. 53(4), 601–607. | ||
| How to Cite this Article |
| Pubmed Style Vo T, Dang V, Le D, Nguyen T, . Identification, serotyping and antimicrobial susceptibility of Riemerella anatipestifer isolated from ducks in Vietnam. Open Vet J. 2022; 12(3): 391-398. doi:10.5455/OVJ.2022.v12.i3.13 Web Style Vo T, Dang V, Le D, Nguyen T, . Identification, serotyping and antimicrobial susceptibility of Riemerella anatipestifer isolated from ducks in Vietnam. https://www.openveterinaryjournal.com/?mno=5360 [Access: July 26, 2024]. doi:10.5455/OVJ.2022.v12.i3.13 AMA (American Medical Association) Style Vo T, Dang V, Le D, Nguyen T, . Identification, serotyping and antimicrobial susceptibility of Riemerella anatipestifer isolated from ducks in Vietnam. Open Vet J. 2022; 12(3): 391-398. doi:10.5455/OVJ.2022.v12.i3.13 Vancouver/ICMJE Style Vo T, Dang V, Le D, Nguyen T, . Identification, serotyping and antimicrobial susceptibility of Riemerella anatipestifer isolated from ducks in Vietnam. Open Vet J. (2022), [cited July 26, 2024]; 12(3): 391-398. doi:10.5455/OVJ.2022.v12.i3.13 Harvard Style Vo, T., Dang, V., Le, D., Nguyen, T. & (2022) Identification, serotyping and antimicrobial susceptibility of Riemerella anatipestifer isolated from ducks in Vietnam. Open Vet J, 12 (3), 391-398. doi:10.5455/OVJ.2022.v12.i3.13 Turabian Style Vo, Thanh-Thin, Van-Tuan Dang, Dinh-Hai Le, Trong-Hai Nguyen, and . 2022. Identification, serotyping and antimicrobial susceptibility of Riemerella anatipestifer isolated from ducks in Vietnam. Open Veterinary Journal, 12 (3), 391-398. doi:10.5455/OVJ.2022.v12.i3.13 Chicago Style Vo, Thanh-Thin, Van-Tuan Dang, Dinh-Hai Le, Trong-Hai Nguyen, and . "Identification, serotyping and antimicrobial susceptibility of Riemerella anatipestifer isolated from ducks in Vietnam." Open Veterinary Journal 12 (2022), 391-398. doi:10.5455/OVJ.2022.v12.i3.13 MLA (The Modern Language Association) Style Vo, Thanh-Thin, Van-Tuan Dang, Dinh-Hai Le, Trong-Hai Nguyen, and . "Identification, serotyping and antimicrobial susceptibility of Riemerella anatipestifer isolated from ducks in Vietnam." Open Veterinary Journal 12.3 (2022), 391-398. Print. doi:10.5455/OVJ.2022.v12.i3.13 APA (American Psychological Association) Style Vo, T., Dang, V., Le, D., Nguyen, T. & (2022) Identification, serotyping and antimicrobial susceptibility of Riemerella anatipestifer isolated from ducks in Vietnam. Open Veterinary Journal, 12 (3), 391-398. doi:10.5455/OVJ.2022.v12.i3.13 |