| Original Article | ||
Open Vet. J.. 2022; 12(4): 451-462 Open Veterinary Journal, (2022), Vol. 12(4): 451–462 Original Research Molecular diagnosis of three outbreaks during three successive years (2018, 2019, and 2020) of Lumpy skin disease virus in cattle in Sharkia Governorate, EgyptElshaima Mohamed Fawzi1*, AbdelKarem Mansour Morsi2 and Eman Beshry Abd-Elfatah11Infectious Diseases, Department of Animal Medicine, Faculty of Veterinary Medicine, Zagazig University, Zagazig, Egypt 2Internal Medicine, Department of Animal Medicine, Faculty of Veterinary Medicine, Zagazig University, Zagazig, Egypt Submitted: 14/03/2022 Accepted: 13/06/2022 Published: 12/07/2022 *Corresponding Author: Elshaima Mohamed Fawzi. Infectious Diseases, Department of Animal Medicine, Faculty of Veterinary Medicine, Zagazig University, Zagazig, Egypt. Email: a.fawzy [at] zu.edu.eg; elshaimafawzi [at] yahoo.es © 2022 Open Veterinary Journal
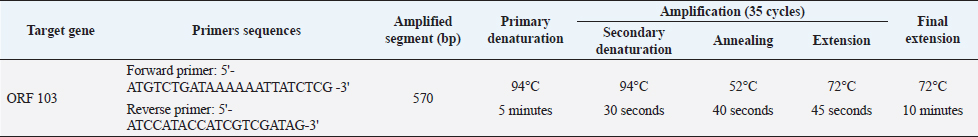
AbstractBackground: Lumpy skin disease (LSD) is endemic in Egypt despite the Egyptian authorities’ annual mass vaccination of cattle with sheeppox vaccine (Veterinary Serum and Vaccine Research Institute, Egypt), and the LSD virus (LSDV) continues to thrive practically every summer. The disease has a huge economic impact on the trade of the animal and its by-product. Aim: This paper study the molecular characterization of LSDV strains that have been circulating in Sharkia Governorate, Egypt, for three successive years (2018, 2019, and 2020). Methods: A total of 61 specimens (26 skin nodules and 35 oculonasal swabs) were collected from a clinic in the hospital of veterinary medicine, Zagazig University, during the summer months (July, August, and September) of three outbreaks in 2018, 2019, and 2020. These were examined by polymerase chain reaction (PCR) based on the open reading frame 103 (ORF103) gene to confirm the suspected cases and determine the degree of homology between the three different outbreaks during three successive years between each other and between the derived sequences of GenBank. Results: Cattle is thought to be infected with LSDV due to the presence of scattered local or diffuse circumscribed skin nodules along with fever and lymph node enlargement and sometimes leg edema. The PCR approach proved rapid, sensitive, and specific for the detection of LSDV and confirmative diagnosis of the disease. Forty-six were detected to be positive by PCR (75.4%). The seven sequenced samples were translated to amino acid and registered in GenBank under accession number MW357655-MW357661. A single nucleotide mutation and amino acid variation were observed at positions 161 C (2020)/T (2018&2019) and consequently, a change in the amino acid at position 54 P (2020)/L (2018&2019) between the outbreak in 2020 and those in 2018 and 2019, respectively. The field LSDV isolates from Egypt cattle were more closely related to other LSDV sequences from Africa (Kenya), Asia, Europe, and the United States. These findings highlight the necessity of ongoing surveillance and characterization of circulating strains and the need to improve procedures for distinguishing vaccine strains from field viruses. Keywords: Lumpy skin disease, Egypt, Cattle, Phylogenetic analysis, PCR. IntroductionLumpy skin disease virus (LSDV) is a double-stranded DNA virus that encodes 30 structural and nonstructural homologs of pox viral proteins. It is closely genetically and antigenically related to Sheeppox virus (SPPV) and Goatpox virus (GTPV) (Tulman et al., 2001, 2002), which belongs to the genus Capripoxvirus (CaPV), subfamily Chordopoxvirinae, and family Poxviridae that can infect a wide variety of animals especially cattle, buffalo and wild ruminant (Sharawi and Abd El-Rahim, 2011). Due to its potential rapid spread and economic impact, the World Organization for Animal Health (OIE) has designated LSD as a notifiable disease (Shen et al., 2011). The transmission of the LSDV mainly occurs mechanically via insect vectors, including mosquites as Aedes species especially female Aedes aegypti (Chihota et al., 2001) and by ticks (Tuppurainen et al., 2011), also Hodhod et al. (2020) detected the virus by polymerase chain reaction (PCR) from collected tick samples. However, there are also minor routes of transmission that need to take into consideration, such as direct contact with an infected animal through the mucosa and indirect contact by sharing the same food and water troughs (Mikhael et al., 2017). Therefore, the restriction of the movement of cattle along with vaccination and the control of insect vector may be considered as the good procedure that limit occurrence of new outbreaks of LSD in near future (Sharawi and Abd El-Rahim, 2011; Aleksandr et al., 2020). LSDV infection is clinically manifested by fever, circumscribed nodules (2–5 cm in diameter) on the skin and mucous membranes with deep eroded and crusted lesions, and enlargement of superficial lymph nodes. In severe clinically diseased animal lesions may extend to the respiratory and gastrointestinal tracts, emaciation, and sometimes death (Carn and Kitching, 1995; Gari et al., 2011; Constable et al., 2017; Abdallah et al., 2018). Animals that recover from this severe disease may experience mastitis, pneumonia, and the formation of necrotic skin plugs leaving deep holes in the hide (Tuppurainen et al., 2017). LSDV is worldwide distribution. In Egypt, it was first recorded between 1988 and 1989 and again in 2006, 2011, and 2014 (Salib and Osman, 2011; Elhaig et al., 2017). In addition, the disease reemerged during the summer of 2016 and 2018 even in animals vaccinated with the Romanian SPPV vaccine (Elhaig et al., 2017; Allam et al., 2020). LSDV continues to be noted in Egypt every summer as recorded by Keshta et al. (2020) and Rouby et al. (2021). LSDV infection leads to financial-economic losses in affected countries due to the restrictions on applied trade of diseased animals and its products (Tuppurainen et al., 2017), temporary or permanent loss in milk production and condition, infertility, abortion, and permanently damaged of hides (Irons et al., 2005; Salib and Osman, 2011). Further economic loss is due to restrictions in animal movement, vaccination costs, and costs of treating secondary bacterial infections (Molla et al., 2017). In different circumstances, the morbidity of LSD ranges from 3% to 85%, whereas the mortality rate fluctuates between 1% and 40% depending on the nature of the disease, either endemic or not (Coetzer and Tustin, 2004; Constable et al., 2017). The wide ranges of morbidity and mortality are due to genetic diversity in livestock breeds leading to a variable degree of susceptibility to disease, virulence of the isolated virus, and a wide range of transmission routes (Tuppurainen et al., 2017). Furthermore, LSDV can survive for a further 120 days in infected tissue and the usage of quarantine regulations is limited (Awadin et al., 2011). Rapid and accurate diagnostic techniques are needed to achieve a putative diagnosis for the effective management or eradication of LSD in endemic and non-endemic countries. The LSD laboratory tests typically include virus isolation (VI), fluorescent antibody test, electron microscopy, PCR, virus neutralization test, and enzyme-linked immunosorbent assay, as described in the OIE Terrestrial Manual (OIE, 2018). Serological tests have shown low specificity due to the cross-reactivity between Parapoxvirus and CaPV (Tian et al., 2010). Fast, sensitive, and specific molecular PCR methods targeting P32, RPO30 and GPCRs, and ORF103 genes have been used for genotyping and phylogenetic analysis of LSDV and other CaPVs (Zhou et al., 2012; Zhu et al., 2013). Vaccination is considered an appropriate means of controlling LSD (Carn and Kitching, 1995; Kitching, 2003; Constable et al., 2017). Various types of CaPVs vaccine strains are currently used in vaccination programs; despite regular LSD vaccination of cattle, vaccination failures and recurrences of multiple outbreaks have been recorded (Gari et al., 2011). Several CaPVs vaccine strains are applied for the prevention and control of LSD, and these vaccines are live-attenuated CaPV vaccine strains, including the Neethling strain of LSDV, Kenyan SPPV and GTPV, Yugoslavian strain of SPPV, Romanian strain of SPPV, and Gorgan strain of GTPV (Carn and Kitching, 1995; Kitching, 2003; Gari et al., 2015). In Egypt, vaccination with an attenuated sheeppox vaccine (Veterinary Serum and Vaccine Research Institute, Egypt) is considered an effective procedure for LSD control by the Egyptian authorities (Elhaig et al., 2017; Tuppurainen et al., 2017). This study was conducted to molecularly characterize LSDV strains that have been circulating in Sharkia Governorate, Egypt, for three successive years (2018, 2019, and 2020). Moreover, this study aimed to perform phylogenetic analysis of the LSDV ORF103 gene, amplified from cattle samples obtained during these outbreaks, and determine the relatedness of sequenced samples during the three outbreaks of LSDV in three different years with each other and within published sequences available in GenBank. Materials and MethodsAnimal selectionThe cattle under investigation were selected based on the presence of clinical signs consistent with LSD that occurred in summer months during three outbreaks in three consecutive years (2018, 2019, and 2020) in Sharkia Governorate, Egypt. All suspected cases were clinically examined according to Constable et al. (2017). The cattle used in this investigation were previously vaccinated with the Romanian strain of SPPV (103.5 TCID 50/dose) in March 2018, 2019, and 2020. SamplesA total of 61 diseased cattle with suspected LSDV were admitted to the Zagazig Veterinary Clinic of Zagazig University during the summer months (July, August, and September) during the LSDV outbreaks in 2018, 2019, and 2020. A total of 61 samples were collected from diseased cattle, 19 in 2018 (11 from scabs and nodules and 8 oculonasal swabs), 22 in 2019 (6 from scabs and nodules and 16 oculonasal swabs), and 20 in 2020 (9 from scabs and nodules and 11 oculonasal swabs), to demonstrate the degree of similarity and variability of LSDV during the three outbreaks and the cause of recurrent occurrence of outbreaks despite regular vaccination. Scab and nodule samples (n=26) were aseptically collected from cattle with LSD-like lesions in sterile tubes containing phosphate-buffered saline (PBS). With a sterile scalpel, the surrounding nodules were removed, and a tiny punch was created deeply into the skin to incorporate all skin layers as described in the OIE Terrestrial Manual (OIE, 2018). Samples were transported to the laboratory of the Virology Department, Animal Health Research Institute, Dokki, El-Giza. DNA extractionGathered samples (skin biopsies, scabs, and oculonasal swabs) were thawed at room temperature. Skin biopsies and scab specimens were cut into small pieces of approximately 400 mg with a sterile scalpel blade, homogenized with 500 µl sterile 1 × PBS solution (pH 7.4) with 10% antibiotic solution (penicillin G sodium 100 IU/ml and streptomycin sulfate 100 mg/ml) using a tissue homogenizer, and finally centrifuged at 344 g at 4°C for 10 minutes (Roche Diagnostics, Germany). Total DNA was extracted from tissue homogenates and 200 μl swab aliquots using the QIAamp DNA Mini Kit (Qiagen, Germany, GmbH) with modifications from the manufacturer’s recommendations. Next, PCR was performed to confirm the presence of LSDV-specific nucleic acids by amplifying the 570 bp region of the ORF103 gene using primer pairs provided by metabion (Germany). PCR conditions are shown in Table 1 as described by Zhu et al. (2013). In an Applied Biosystems 2720 thermal cycler, the reaction was set up on a 25 µl final volume, including 1 µl of each primer of 20 pmol concentration, 12.5 µl of Emerald Amp Max PCR Master Mix (Takara, Japan), 6 µl of DNA template, and 4.5 µl of water. Analysis of the PCR productsPCR products were viewed on 1.5% agarose gel electrophoresis (AppliChem, Germany, GmbH) in 1 × TBE buffer at room temperature using gradients of 5 V/cm. For gel analysis, 15 µl of the products was loaded in each gel slot. GelPilot 100 bp DNA Ladder (Qiagen, Germany, GmbH) was used to confirm LSDV-positive samples, with a fragment size of 570 bp. A gel documentation system (Alpha Innotech, Biometra) was used to photograph the gel, and the data were processed using computer software. Nucleotide sequencing and analysisA molecular weight marker was used to identify amplification products of the expected size. DNA bands of the correct size were excised and purified using gel purification (Qiagen, Germany) according to the manufacturer’s instructions and then forwarded to Inqaba Biotec in South Africa for Sanger sequencing. The quality of the acquired sequences was evaluated, and the ends of the sequences were trimmed using BioEdit software (Ibis Biosciences, Carlsbad, CA). Using the National Center for Biotechnology Information’s web-based Basic Local Alignment Search Tool, the trimmed sequences were compared with other LSDV/ORF103 sequences in GenBank (BLASTn). Table 1. Primers sequences sets, target genes, amplicon sizes, and optimized cycling conditions concerning conventional PCR assay of ORF 103 gene.
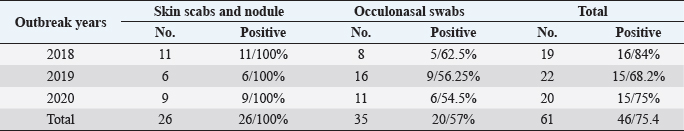
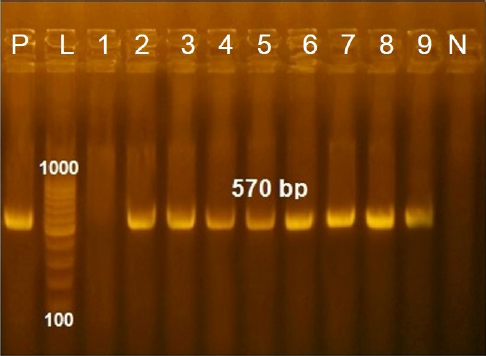
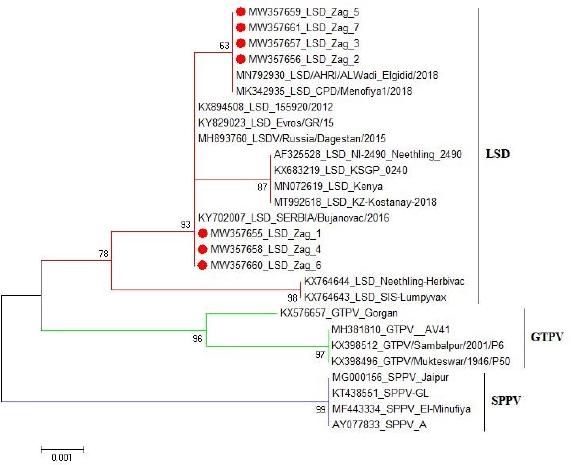
These nucleotide sequences were then translated to amino acid sequences and multiple sequence alignment using MUSCLE from the EMBL-EBI web server to check for LSDV-specific signatures. Phylogenetic analysis was performed using Molecular Evolutionary Genetics Analysis version 6 (Pennsylvania, USA) (Thompson et al., 1994). All sequences were submitted to GenBank and can be found under the accession number MW357655-MW357661. Phylogenetic analysisA total of 27 CaPVs/ORF103 sequences (used to root tree) were selected from GenBank for phylogenetic analysis. After BLAST, LSDV sequences were selected based on the nucleotide similarity and origin of isolates to have representative sequences from East Africa, the rest of Africa, and Eurasia. We also selected sequences from LSDV vaccine strains, GTPV and SPPV. The maximum likelihood method was used to create a phylogenetic tree based on the Tamura 3 parameter model with 1,000 bootstrap replications. The tree was drawn to scale, with branch lengths in the same units as those of the evolutionary distances used to estimate the phylogenetic trees (Tamura et al., 2013), and the heat map was generated using R software (package: plot. matrix). Ethical approvalNo ethical approval was needed for the collection of specimens from clinically infected cattle. It was for the routine diagnosis of the disease under the usual veterinary service work in Egypt and according to the national standards within the country. ResultsThe field observation of the three outbreaks in 3 years (2018, 2019, and 2020) was suspected to be LSDV. The common clinical signs of infected cattle were fever (40°C–41.5°C), depression, enlargement of superficial lymph nodes, circumscribed localized, and generalized nodules in different parts of the body, some intradermal nodules ulcerated and ruptured to form a “sitfast” (Fig. 1a–e), dyspnea, lacrimation, ocular and nasal discharges, and loss of body weight (Fig. 1c and d). A conventional PCR was performed using specific primers that targeted the LSDV ORF103 gene at the 570 bp region to confirm the presence of LSDV viral DNA from suspected clinical cases. Of 61 samples obtained from suspected clinical cases, 46 (75.4%) tested positive by PCR (Table 2). This is likely because only oculonasal swabs were taken from some animals and a few swab samples tested negative. All DNAs extracted from positive samples yielded a clear band in the gel after amplification of the target gene, except those obtained from oculonasal swabs. Skin nodules and scabs had more obvious bands than swabs (Fig. 2). Of these, all scab and nodular samples (26/26, 100%) were positive, while only 20 oculonasal swabs of 35 tested samples (57%) were positive. There were seven sequenced samples (three in 2020, two in 2019, and two in 2018). These sequenced samples were translated to amino acid and registered in GenBank under accession number MW357655-MW357661. Phylogenetic construction was performed to determine the genetic relationship among seven sequenced LSDV isolates by ORF103 gene primers from diseased cattle. These sequenced LSDV isolates were clustered with other LSDV from Egypt and different regions in the same clade and other separated clade to SPPV and GTPV isolates retrieved from the GenBank (Fig. 3 and Table 3) to detect the identity and near region of the disease transmission responsible for frequent and continuing outbreaks. The field LSDV isolates from Egypt were more closely related to other LSDV sequences from Africa (Kenya), Asia (Israel, Russia, Kazakhstan, India, and China), Europe (Belgium and Greece), and the United States. The isolated outbreak samples showed nucleotide sequence identities of 100% with MW357655, MW357658, and MW357660 (outbreak in 2020) and 98.60% with MW357656, MW357657, MW357659, and MW357661 (outbreaks in 2018 and 2019) when compared with other sequences of LSDV from Israel in 2012, Greece in 2015, Russia in 2015, and Serbia in 2016. Furthermore, when compared with LSDV vaccine strains, nucleotide identities were 0.2%–1.6% lower than when compared to outbreak sequences and gave identities between 98.6% and 99.82% with previously mentioned accession number. The diversity of sequences from Egypt is low when compared between each other and clusters with each other, whereas other samples of GTPV and SPPV show great diversity. The analysis of the ORF103 gene showed major sequence differences between the vaccine strain and the field isolates (Table 3). The sequence analysis of the current field isolates indicated single nucleotide mutation at position 161C (2020)/T (2018&2019) between the isolates of the outbreak in 2020 and isolates of the outbreaks in 2018 and 2019 (Fig. 4). The genetic variation between the phenotypically LSD vaccine strains, isolated and sequenced strains generated from GenBank in one and/or two single nucleotide polymorphisms (Fig. 4). The isolated sequences in 2018 and 2019 were equal to the other sequence in GenBank isolated from Egypt in 2018 with a substitute of only one nucleotide in position 161, replacing T with C when compared with the isolates in 2020. While the difference between these isolates and other LSD sequences either concerning isolates or vaccine were shown no substitute or 1 or 2 or 6 nucleotide substitutes. The current isolates differed from goatpox isolates in 10 or 11 nucleotide substitutes, while there were major substitute changes (13 nucleotides) when compared with sheeppox isolates in GenBank (Fig. 4).
Fig. 1. Cattle with characteristic of LSDV. (a and b) Generalized circumscribed active nodules covering entire body; (c) nodules on head with lacrimation; and (d and e) progression of the disease with the observation of eroded and crusted nodules and scab formation. Table 2. The data of collected LSD samples either nodules or oculonasal swabs during three outbreaks (2018, 2019, and 2020) from cattle by conventional PCR with demarcation of the incidence.
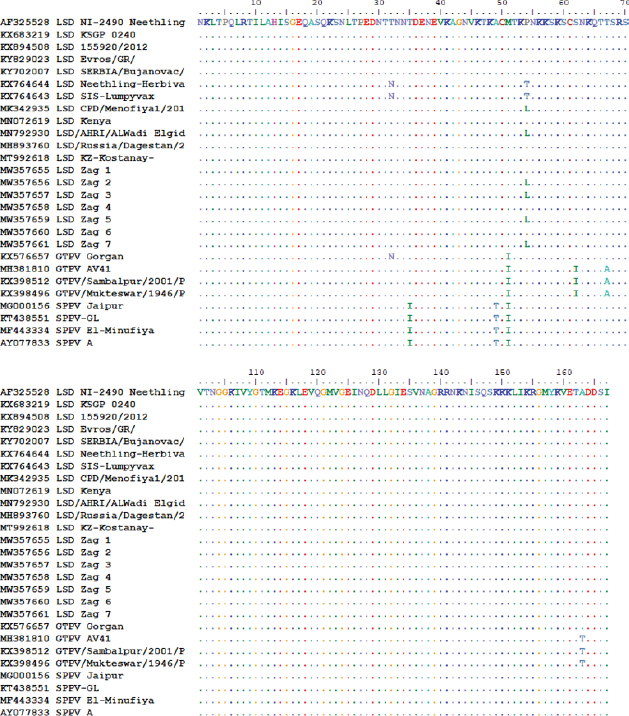
Similarly, the deduced amino acid sequences of the ORF103 gene of the isolates of the outbreak in 2020 and those in 2018 and 2019 revealed a single amino acid variation proline by leucine (P/L) at residue position 54 (Fig. 5). The protein (AA) differed from isolated strains from other LSD sequences in GenBank by substituting only 1 or 2 AA at position 54 substituted P by L (only isolates of 2018 and 2019) and 71 position substituted G by S (recorded this change on all field isolates). Whereas, vaccine strains of LSD either LSD Neethling-Herbivac or LSDsis–lumpyvax substitute 32 T by N when compared with all isolates either field isolates or those derived from GenBank. When compared with goatpox, 3 or 5 amino acids were substituted, while sheeppox was substituted by 4 AAs as illustrated in Fig. 5. The putative virion core protein was the most abundant protein found in the sequence. DiscussionLSD is endemic in several African countries, rapidly spreading throughout the Middle East, including Egypt. A prolonged debilitating clinical course, reduced weight gain, permanent damage to hides, temporary or permanent infertility or even sterility in bulls, temporary or permanent loss of milk production, and abortion of pregnant cattle have all been identified as important cattle health problems (Leliso et al., 2021).
Fig. 2. Agarose gel electrophersis picture of PCR product showing the amplification of fragment of ORF103 gene at 570 bp. P: Control positive; L: DNA ladder; Lane 1: Negative fragment of LSDV in samples taken from oculonasal swabs; Lanes 4,5,9: Positive fragment of LSDV in samples taken from oculonasal swabs; Lanes 2,3,6,7,8: Positive fragment of LSDV in samples taken from scabs and skin nodules specimens.
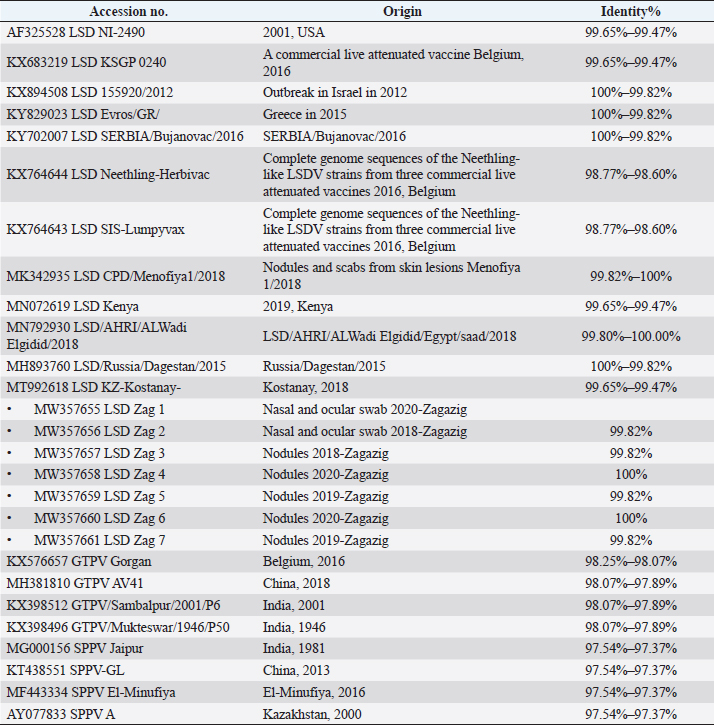
Fig. 3. Phylogentic analysis of 27 CaPVs based on nucleotide sequences of ORF103 gene derived from Genebank and drawn by MEGA6. Table 3. Details and identity % of selected CaPVs sequences based on ORF103 gene used to construct phylogenetic tree.
Regarding the three different outbreaks over 3 years (2018, 2019, and 2020) during the summer season, we investigated the abrupt emergence of dispersed skin nodules on cattle in Sharkia Governorate, Egypt. LSD is a cattle disease that can be easily identified by clinical signs that vary in severity from animal to animal, such as extensive circumscribed skin nodules that become necrotic, ulcerate, and eventually form a deep scab, in addition to fever, enlargement of superficial lymph nodes, and edema of the leg in some animals, consistent with the findings of Gari et al. (2010) and Rouby et al. (2021). In this scenario, the use of conventional PCR directly from collected samples without exposure to VI is considered to be a more sensitive and accurate test with the ability to detect the DNA of LSD in low genome quantity with slightly ease to analysis and interpret this result is supported by other authors (Tuppurainen et al., 2005; Awad et al., 2010; Al-Salihi, 2014). In addition, to facilitate rapid application of control measures and isolation of positive cases (Sharawi and Abd El-Rahim, 2011) while, VI in tissue culture takes longer time, may be exposed to bacterial and fungal contamination and require several passages (Van Rooyen et al., 1969). In addition, serological tests are time-consuming to be used as primary diagnostic methods and can’t differentiate between infected and vaccinated animals or antibodies resulting from LSDV infection caused by different members of CaPVs and those of other Poxviruses (Abd El-Rahim et al., 2002; Awad et al., 2010).
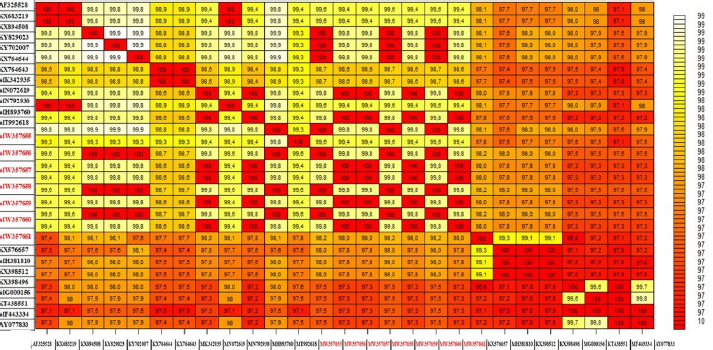
Fig. 4. The pairwise heatmap shows the distance matrix between each pair of isolates to determine degree of similarity and diversity relationship among selected CaPVs sequences. The heatmap was generated using R software (package: plot.matrix). The PCR assay detected LSDV in 26 (100%) of 26 skin nodules from the corresponding infected cattle. The PCR result was fully correlated with the field diagnosis based on clinical symptoms. Although, the presences of several other sources for virus detection, such as oral, nasal, and ocular discharges, were determined, scabs and nodules are classified as the best sample materials as they are easy to collect without the use of local anesthesia and can withstand long transport in different temperatures with more concentration of viral particles than other origins (Tuppurainen et al., 2005; El-Kholy et al., 2008). While the PCR assay detected LSDV in 20 (75%) of 35 oculonasal swabs from the corresponding infected cattle, this was attributed to intermittent virus discharge in the nasal and ocular discharges during infection with LSDV, which started on day 11 post-infection (pi) and was thought to persist until day 38 pi (Aleksandr et al., 2020). In this study, the PCR assay showed high specificity as a unique band of the expected size (approximately 570 bp) was obtained for the ORF103 gene of all positive DNA samples derived from skin scabs and nodule samples and oculonasal swabs. LSDV is closely related to other CaPVs; however, it has a unique complement of genes responsible for viral host range and virulence, according to genomic comparison (Sharawi and Abd El-Rahim, 2011). These results were consistent with the findings of El-Nahas et al. (2011); Sharawi and Abd El-Rahim (2011); and El-Khabaz (2014). This indicates that there was less nucleotide exchange through the years between the outbreaks. This finding is correlated with the known fact that CaPVs are highly conserved within and among their species (Tulman et al., 2002; Babiuk et al., 2008). Direct sequencing of PCR amplicons and comparative genetic analyses were useful to not only trace the new outbreak LSDV but also establish genetic tools for epidemiological studies and the development of novel efficient LSDV vaccines (El-Kholy et al., 2008; Varshovi et al., 2009). To prevent the introduction of LSDV from neighboring countries during importation, it is strongly recommended to apply the PCR assay in examination of live domestic and wild bovine species and bovine semen from endemic countries at quarantines (El-Kholy et al., 2008; Tuppurainen et al., 2017). Molecular analysis indicated that there was no significant nucleotide variation among the current and previous field isolates included in this study, except a single nucleotide mutation and a single amino acid variation. On the other hand, a sequence difference was observed between the vaccine strain and the field isolates. The phylogenetic analysis of the ORF103 gene sequences was able to group the CaPVs into three distinct groups: LSDV, SPPV, and GTPV. It further showed that LSDVs from the outbreaks in Sharkia, Egypt, grouped with LSDV isolates from Kenya, Belgium, Serbia, Russia, Kazakhstan, and Greece (Fig. 3 and Table 3). These sequences were closely related to the sequences from Kenya when compared according to nucleotide identity, therefore suggesting that the same LSDVs are responsible for outbreaks across borders. The BLAST analysis revealed high sequence homology of 98.6%–100% between the LSDV sequences and sequences in GenBank, while the sequence corresponding to the outbreak in 2020 showed an identity of 100% with the LSD sequences from Israel in 2012 (KX894508), Greece in 2015 (KY829023), and Serbia/Bujanovac in 2016 (KY702007). Moreover, the sequence samples corresponding to the outbreaks in 2018 and 2019 showed complete identity between two outbreaks and 100% identity with the outbreak in 2018 in ALWadi Elgidid and Menofiya, Egypt, while the identity between these two outbreaks and the outbreak in 2020 was 99.82%. From the previous analysis, no new virus circulated among different breeds of cattle in different countries of Egypt. This may be due to the small number of LSDV sequences used in this assay with the use of only a single gene and only one governorate and the highly genetic stability of LSDV (Mafirakureva et al., 2017; Ochwo et al., 2020). The variation between the wild-type LSDV and sequence corresponding to SPPV either wild or virus may be interpreted as the need for further diagnostic testing, along with the sequencing of several LSDV genes, and must be performed along with the sequence of the Romanian strain of SPPV used as a vaccine by the Egyptian authorities.
Fig. 5. Amino acid sequences alignment of ORF103 gene of current field isolates and related to LSD either virus or vaccine and SPPV and GTPV generated from database of GenBank. Despite annual mass vaccination by the Egyptian authorities with sheeppox vaccine (VSVRI, Egypt), not all vaccinated animals develop a protective level of immunity against LSDV (Tuppurainen and Oura, 2012; Gelaye et al., 2015; Rouby et al., 2021). This may be attributed to several reasons, such as the refusal of some owners to vaccinate their animals, unsuccessful vaccination, and uncontrolled animal movement together with the high stability of LSDV in the environment and presence of insect vectors (Sharawi and Abd El-Rahim, 2011; Tuppurainen and Oura, 2012; Hodhod et al., 2020). The wide route of transmission and inadequate control of the arthropod vector contribute to the difficulty in applying efficient control and eradication of LSD (Coetzer et al., 1994; Helmy et al., 2017). The sequence variation of amino acids between the outbreaks analyzed in this study with SPPV from either field isolates or vaccine derived from GenBank revealed 13 variations of nucleotides, indicating the failure of sheeppox vaccine to protect cattle against circulating isolates that cause LSD in Egypt. This finding is consistent with the reports in Israel, Ethiopia, Sudan, and Oman on LSD vaccines providing insufficient protection and causing severe reactions in cattle after immunization (Khalafalla et al., 1993; Tamam, 2006; Brenner et al., 2009; Somasundaram, 2011; Ayelet et al., 2013; Eeva et al., 2014; Gelaye et al., 2015). LSDV is believed to be introduced during imports of cattle from neighboring countries, like Kenya, as the lack of strict quarantine measures results in the rapid spread of the disease. Homologues between local isolated strains and the strain used in vaccines to control infection may be necessary to reduce the recurrent outbreaks of LSD in Egypt, instead of using the Romanian SPPV vaccine; these concurrent with rapid detection and prompt culling of infected one are effective control measures of the infection in cattle. ConclusionsTo the best of authors’ knowledge, this is the first study on molecular detection and phylogenetic analysis of LSDV in Sharkia, Egypt, using the ORF103 gene for three successive years. Molecular analysis indicated that there were no significant nucleotide mutation and amino acid variation among the current isolates during the three successive years between each other and with previous field isolates, except for a single nucleotide mutation and a single amino acid variation; this indicates that LSDV has a genetically conserved nature. These data point to the presence of genetically similar LSDVs circulating in the Egypt, and this emphasizes the transboundary nature of LSDV from a nearby nation. Furthermore, we note that outbreak viruses differ from vaccine strain viruses based on single gene sequence analysis, implying that additional assays should be performed to determine the effect of nucleotide and amino acid changes on virus pathogenicity and vaccine immunogenicity by sequencing multiple genes. AcknowledgmentsThe authors would like to thank the owner of the cases admitted to Zagazig Veterinary Clinic for their facility and collection of samples. The authors also would like to thank the workers in the Department of Animal Medicine, Zagazig University, Egypt, for their help in sample collection. Conflict of interestThe authors declare that there is no conflict of interest. Authors contributionsAll authors contributed to the concept planning, the study’s execution, the analysis of data, and the interpretation. EMF and EBA prepared the initial draft of the manuscript. All authors had complete access to all the data in the study, took responsibility for data reliability and analysis accuracy, and approved the final submission. Data availabilityData used in this article are included within the article. ReferencesAbd El-Rahim, I.H.A., El-Ballal, S. and Hussein, M. 2002. An outbreak of lumpy skin disease among cattle in upper Egypt (El-Menia governorate). Minufyia Vet. J. 2(1), 185–200. Abdallah, F.M., El Damaty, H.M. and Kotb, G.F. 2018. Sporadic cases of lumpy skin disease among cattle in Sharkia province, Egypt: genetic characterization of Lumpy skin disease virus isolates and pathological findings. Vet. World 11(8), 1150–1158. Aleksandr, K., Olga, B., David, W.B., Pavel, P., Yana, P., Svetlana, K., Alexander, N., Vladimir, R., Dmitriy, L. and Alexander, S. 2020. Non-vector-borne transmission of Lumpy skin disease virus. Sci. Rep. 10, 7436. Allam, A.M., Elbayoumy, M.K., Abdel-Rahman, E.H., Hegazi, A.G. and Farag, T.K. 2020. Molecular characterization of the 2018 outbreak of lumpy skin disease in cattle in Upper Egypt. Vet. World 13(7), 1262–1268. Al-Salihi, K. 2014. Lumpy skin disease: review of literature. Mirror Res. Vet. Sci. Anim. 3(3), 6–23. Awad, W.S., Ibrahim, A.K., Mahran, K., Fararh, K.M. and Moniem, M.I.A. 2010. Evaluation of different diagnostic methods for diagnosis of lumpy skin disease in cows. Trop. Anim. Health Prod. 42(4), 777–783. Awadin, W., Hussein, H., Elseady, Y., Babiuk, S. and Furuoka, H. 2011. Detection of Lumpy skin disease virus antigen and genomic DNA in formalin-fixed paraffin-embedded tissues from an Egyptian outbreak in 2006. Transbound. Emerg. Dis. 58(5), 451–457. Ayelet, G., Abate, Y., Sisay, T., Nigussie, H., Gelaye, E., Jemberie, S. and Asmare, K. 2013. Lumpy skin disease: preliminary vaccine efficacy assessment and overview on outbreak impact in dairy cattle at DebreZeit, central Ethiopia. Anti. Res. 98(2), 261–265. Babiuk, S., Bowden, T.R., Boyle, D.B., Wallace, D.B. and Kitching, R.P. 2008. Capripoxviruses: an emerging worldwide threat to sheep, goats and cattle. Transbound. Emerg. Dis. 55(7), 263–272. Brenner, J., Bellaiche, M., Gross, E., Elad, D., Oved, Z., Haimovitz, M., Wasserman, A., Friedgut, O., Stram, Y., Bumbarov, V. and Yadin, H. 2009. Appearance of skin lesions in cattle populations vaccinated against lumpy skin disease: statutory challenge. Vaccine 27(10), 1500–1503. Carn, V.M. and Kitching, R.P. 1995. An investigation of possible routes of transmission of Lumpy skin disease virus (Neethling). Epidemiol. Infect. 114(1), 219–226. Chihota, C.M., Rennie, L.F., Kitching, R.P. and Mellor, P.S. 2001. Mechanical transmission of Lumpy skin disease virus by Aedes aegypti (Diptera: Culicidae). Epidemiol. Infect. 126, 317–321. Coetzer, J.A.W. and Tustin R.C. 2004. Lumpy skin disease, 2nd ed. In Infectious diseases of livestock. Cape Town, South Africa: Oxford University Press Southern Africa, pp: 1268–1276. Coetzer, J., Thomson, G. and Tustin, R. 1994. Poxviridae. In Infectious diseases of livestock. Cape Town, South Africa: Oxford University Press, pp: 601–603. Constable, P., Hinchcliff, K., Done, S. and Grünberg, W. 2017. Clinical examination and making a diagnosis. In Veterinary Medicine, 11th ed. Elsevier Press, St. Louis, Missouri, USA. Eeva, S.M.T., Caroline, R., Katarzyna, B.B., Nick, J.K., Shadi, A., Lorraine, F., Mark, R.H., Charles, E.L. and Peter, P.C.M. 2014. Characterization of Sheeppox virus vaccine for cattle against Lumpy skin disease virus. Anti. Res. 109, 1–6. Elhaig, M.M., Selim, A. and Mahmoud, M. 2017. Lumpy skin disease in cattle: frequency of occurrence in a dairy farm and a preliminary assessment of its possible impact on Egyptian buffaloes. Onderstepoort J. Vet. Res. 84(1), e1–e6. El-Khabaz, K.A. 2014. Rapid laboratory diagnosis of lumpy skin disease by using PCR. Assiut Vet. Med. J. 60(143), 37–41. El-Kholy, A.A., Soliman, H.M. and Abdelrahman, K.A. 2008. Polymerase chain reaction for rapid diagnosis of a recent Lumpy skin disease virus incursion to Egypt. Arab J. Biotech. 11(2), 293–302. El-Nahas, E., El-Habbaa, A., El-Bagoury, G. and Radwan, M. 2011. Isolation and identification of Lumpy skin disease virus from naturally infected buffaloes at Kaluobia, Egypt. Glob. Vet. 7(3), 234–237. Gari, G., Abie, G., Gizaw, D., Wubete, A., Kidane, M., Asgedom, H., Bayissa, B., Ayelet, G., Oura, C.A., Roger, F. and Tuppurainen, E.S. 2015. Evaluation of the safety, immunogenicity and efficacy of three Capripoxvirus vaccine strains against Lumpy skin disease virus. Vaccine 33(28), 3256–3261. Gari, G., Bonnet, P., Roger, F. and WaretSzkuta, A. 2011. Epidemiological aspects and financial impact of lumpy skin disease in Ethiopia. Prev. Vet. Med. 102(4), 274–283. Gari, G., Waret-Szkuta, A., Grosbois, V., Jacquiet, P. and Roger, F. 2010. Risk factors associated with observed clinical lumpy skin disease in Ethiopia. Epidemiol. Infect. 138(11), 1657–1666. Gelaye, E., Belay, A., Ayelet, G., Jenberie, S., Yami, M., Loitsch, A., Tuppurainen, E., Grabherr, R., Diallo, A. and Lamien, C.E. 2015. Capripox disease in Ethiopia: genetic differences between field isolates and vaccine strain, and implications for vaccination failure. Anti. Res. 119, 28–35. Helmy, N.M., Ahmed, A.S. and Mohamed, Z.Y. 2017. Molecular, clinicopathological and serodiagnosis of LSDV in cattle at Sharkia and Fayoum Governorates. J. Virol. Sci. 1, 1–11. Hodhod, A., Emad, E., Mervat, I.A.M. and Madiha, S.I. 2020. Isolation and molecular characterization of Lumpy skin disease virus in Egypt during 2017-2018. Eur. J. Pharm. Med. Res. 7(1), 96–103. Irons, P.C., Tuppurainen, E.S.M. and Venter, E.H. 2005. Excretion of Lumpy skin disease virus in bull semen. Theriogenology 63(5), 1290–1297. Keshta, H., Allam, A., Fadl, S. and El Beskawy, M. 2020. Detection of Lumpy skin disease during an outbreak in summer 2019 in Menoufia governorate, Egypt using clinical, biochemical and molecular diagnosis. Zagazig Vet. J. 48(4), 378–389. Khalafalla, A.I., Gaffar Elamin, M.A. and Abbas, Z. 1993. Lumpy skin disease: observations on the recent outbreaks of the disease in the Sudan. Rev. Elev. Med. Vet. Pays Trop. 46(4), 548–550. Kitching, R.P. 2003. Vaccines for lumpy skin disease, sheeppox and goatpox. Dev. Biol. (Basel) 114, 161–167. Leliso, S.A., Bari, F.D. and Chibssa, T.R. 2021. Molecular characterization of Lumpy skin disease virus isolates from outbreak cases in cattle from Sawena district of Bale Zone, Oromia, Ethiopia. Vet. Med. Int. 2021, 1–9. Mafirakureva, P., Saidi, B. and Mbanga, J. 2017. Incidence and molecular characterisation of Lumpy skin disease virus in Zimbabwe using the P32 gene. Trop. Anim. Health Prod. 49(1), 47–54. Mikhael, C.A., Nakhla, O.E. and Mohamed, N.A. 2017. Study on the capability of a dual capripox vaccine in protection of cattle against LSD infection. J. Vet. Med. Res. 24(1), 61–70. Molla, W., de Jong, M.C.M., Gari, G. and Frankena, K. 2017. Economic impact of lumpy skin disease and cost effectiveness of vaccination for the control of outbreaks in Ethiopia. Prev. Vet. Med. 147, 100–107. Ochwo, S., VanderWaal, K., Ndekezi, C., Nkamwesiga, J., Munsey, A., Witto, S.G., Nantima, N., Mayanja, F., Okurut, A., Atuhaire, D.K. and Mwiine, F.N. 2020. Molecular detection and phylogenetic analysis of Lumpy skin disease virus from outbreaks in Uganda 2017-2018. BMC Vet. Res. 16(1), 16–66. OIE. 2018. Manual of diagnostic tests and vaccines for terrestrial animals. Lumpy skin disease: aetiology, epidemiology, diagnosis, prevention and control, references. Paris, France: OIE. Rouby, S.R., Safwat, N.M., Hussein, K.H., Abdel- Ra’ouf, A.M., Madkour, B.S. and Abdel-Moneim, A.S. 2021. Lumpy skin disease outbreaks in Egypt during 2017-2018 among sheeppox vaccinated cattle: epidemiological, pathological, and molecular findings. PLoS One 16(0), e0258755. Salib, F.A. and Osman, A.H. 2011. Incidence of lumpy skin disease among Egyptian cattle in Giza Governorate, Egypt. Vet. World 4(4), 162–167. Sharawi, S.S.A. and Abd El-Rahim, I.H.A. 2011. The utility of polymerase chain reaction for diagnosis of lumpy skin disease in cattle and water buffaloes in Egypt. Rev. Sci. Tech. 30(3), 821–830. Shen, Y.J., Shephard, E., Douglass, N., Johnston, N., Adams C., Williamson, C. and Williamson, A.L. 2011. A novel candidate HIV vaccine vector based on the replication deficient Capripoxvirus, Lumpy skin disease virus (LSDV). Virol. J. 8, 265. Somasundaram, M.K. 2011. An outbreak of lumpy skin disease in a Holstein dairy herd in Oman: a clinical report. Asian J. Anim. Vet. Adv. 6, 851–859. Tamam, S.M. 2006. Isolation of Lumpy skin disease virus form naturally infected cattle previously vaccinated with live attenuated Sheeppox virus vaccine. Beni-Suef Vet. Med. J. 16(1), 27–31. Tamura, K., Stechner, G., Petersun, D., Filipski, A. and Kumar, S. 2013. MEGA6: molecular evolutionary genetics analysis version 6.0. Mol. Biol. Evol. 30(12), 2725–2729. Thompson, J.D., Higgins, D.G. and Gibson, T.J. 1994. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22(22), 4673–4680. Tian, H., Chen, Y., Wu, J., Shang, Y. and Liu, X. 2010. Serodiagnosis of Sheeppox and Goatpox using an indirect ELISA based on synthetic peptide targeting for the major antigen P32. Virol. J. 7, 245. Tulman, E.R., Afonso, C.L., Lu, Z.E.R., Zsak, L., Kutish, G.F. and Rock, D.L. 2001. Genome of lumpy skin disease virus. J. Virol. 75(5), 7122–7130. Tulman, E.R., Afonso, C.L., Lu, Z.E.R., Zsak, L., Sur, J.H., Sandybaev, N.T., Kerembekova, U.Z., Zaitsev, V.L., Kutish, G.F. and Rock, D.L. 2002. The genomes of Sheeppox and Goatpox viruses. J. Virol. 76(12), 6054–6061. Tuppurainen, E.S. and Oura, C.A. 2012. Review: lumpy skin disease: an emerging threat to Europe, the Middle East and Asia. Transbound. Emerg. Dis. 59(1), 40–48. Tuppurainen, E.S., Stoltsz, W.H., Troskie, M., Wallace, D.B., Oura, C., Mellor, P.S., Coetzer, J.A. and Venter, E.H. 2011. A potential role for ixodid (hard) tick vectors in the transmission of Lumpy skin disease virus in cattle. Transbound. Emerg. Dis. 58, 93–104. Tuppurainen, E., Venter, E. and Coetzer, J. 2005. The detection of Lumpy skin disease virus in samples of experimentally infected cattle using different diagnostic techniques. Onderstepoort J. Vet. Res. 72(2), 153–164. Tuppurainen, E.S.M., Venter, E.H., Shisler, J.L., Gari, G., Mekonnen, G.A., Juleff, N., Lyons, N.A., De Clercq, K., Upton, C., Bowden, T.R., Babiuk, S. and Babiuk, L. 2017. Review: Capripoxvirus diseases: current status and opportunities for control. Transbound. Emerg. Dis. 64(3), 729–745. Van Rooyen, P.J., Munz, E.K. and Weiss, K.E. 1969. The optimal conditions for the multiplication of the Neethling-type Lumpy skin disease virus in embryonated eggs. Onderstepoort J. Vet. Res. 36(2), 165–174. Varshovi, H.R., Keyvanfar, H., Aghaiypour, K., Pourbakhsh, S.A., Shooshtari, A.H. and Aghaebrahimian, M. 2009. Capripoxvirus identification by PCR based on P32 gene. Arch. Razi Inst. 64(1), 19–25. Zhou, T., Jia, H., Chen, G., He, X., Fang, Y. and Wang, X. 2012. Phylogenetic analysis of Chinese Sheeppox and Goatpox virus isolates. Virol. J. 9, 1–8. Zhu, X.L., Yang, F., Li, H.X., Dou, Y.X., Meng, X.L., Li, H., Luo, X.N. and Cai, X.P. 2013. Identification and phylogenetic analysis of a Sheeppox virus isolated from the Ningxia Hui Autonomous Region of China. Genet. Mol. Res. 12(2), 1670–1678. | ||
| How to Cite this Article |
| Pubmed Style Fawzi EM, Morsi AM, Abd-elfatah EB. Molecular Diagnosis of Three Outbreaks During 3 Successive Years (2018, 2019, and 2020) of Lumpy Skin Disease Virus in Cattle in Sharkia Governorate, Egypt. Open Vet. J.. 2022; 12(4): 451-462. doi:10.5455/OVJ.2022.v12.i4.6 Web Style Fawzi EM, Morsi AM, Abd-elfatah EB. Molecular Diagnosis of Three Outbreaks During 3 Successive Years (2018, 2019, and 2020) of Lumpy Skin Disease Virus in Cattle in Sharkia Governorate, Egypt. https://www.openveterinaryjournal.com/?mno=101852 [Access: January 25, 2026]. doi:10.5455/OVJ.2022.v12.i4.6 AMA (American Medical Association) Style Fawzi EM, Morsi AM, Abd-elfatah EB. Molecular Diagnosis of Three Outbreaks During 3 Successive Years (2018, 2019, and 2020) of Lumpy Skin Disease Virus in Cattle in Sharkia Governorate, Egypt. Open Vet. J.. 2022; 12(4): 451-462. doi:10.5455/OVJ.2022.v12.i4.6 Vancouver/ICMJE Style Fawzi EM, Morsi AM, Abd-elfatah EB. Molecular Diagnosis of Three Outbreaks During 3 Successive Years (2018, 2019, and 2020) of Lumpy Skin Disease Virus in Cattle in Sharkia Governorate, Egypt. Open Vet. J.. (2022), [cited January 25, 2026]; 12(4): 451-462. doi:10.5455/OVJ.2022.v12.i4.6 Harvard Style Fawzi, E. M., Morsi, . A. M. & Abd-elfatah, . E. B. (2022) Molecular Diagnosis of Three Outbreaks During 3 Successive Years (2018, 2019, and 2020) of Lumpy Skin Disease Virus in Cattle in Sharkia Governorate, Egypt. Open Vet. J., 12 (4), 451-462. doi:10.5455/OVJ.2022.v12.i4.6 Turabian Style Fawzi, Elshaima Mohamed, Abdelkarem Mansour Morsi, and Eman Beshry Abd-elfatah. 2022. Molecular Diagnosis of Three Outbreaks During 3 Successive Years (2018, 2019, and 2020) of Lumpy Skin Disease Virus in Cattle in Sharkia Governorate, Egypt. Open Veterinary Journal, 12 (4), 451-462. doi:10.5455/OVJ.2022.v12.i4.6 Chicago Style Fawzi, Elshaima Mohamed, Abdelkarem Mansour Morsi, and Eman Beshry Abd-elfatah. "Molecular Diagnosis of Three Outbreaks During 3 Successive Years (2018, 2019, and 2020) of Lumpy Skin Disease Virus in Cattle in Sharkia Governorate, Egypt." Open Veterinary Journal 12 (2022), 451-462. doi:10.5455/OVJ.2022.v12.i4.6 MLA (The Modern Language Association) Style Fawzi, Elshaima Mohamed, Abdelkarem Mansour Morsi, and Eman Beshry Abd-elfatah. "Molecular Diagnosis of Three Outbreaks During 3 Successive Years (2018, 2019, and 2020) of Lumpy Skin Disease Virus in Cattle in Sharkia Governorate, Egypt." Open Veterinary Journal 12.4 (2022), 451-462. Print. doi:10.5455/OVJ.2022.v12.i4.6 APA (American Psychological Association) Style Fawzi, E. M., Morsi, . A. M. & Abd-elfatah, . E. B. (2022) Molecular Diagnosis of Three Outbreaks During 3 Successive Years (2018, 2019, and 2020) of Lumpy Skin Disease Virus in Cattle in Sharkia Governorate, Egypt. Open Veterinary Journal, 12 (4), 451-462. doi:10.5455/OVJ.2022.v12.i4.6 |