| Research Article | ||
Open Vet. J.. 2023; 13(6): 732-741 Open Veterinary Journal, (2023), Vol. 13(6): 732-741 Original Research The use of RT-PCR in the diagnosis and differentiation of vaccine strains of chicken infectious bronchitis and Newcastle diseaseNigmetulla Assanov1*, Ryskeldi Bazarbayev1, Assilbek Mussoyev1, Bauyrzhan Otarbayev1 and Kairat Iskhan21Faculty of Veterinary Science, Kazakh National Agrarian Research University, Almaty, Republic of Kazakhstan 2Faculty of Bioresources and Technology, Kazakh National Agrarian Research University, Almaty, Republic of Kazakhstan *Corresponding Author: Nigmetulla Assanov. Faculty of Veterinary Science, Kazakh National Agrarian Research University, Almaty, Republic of Kazakhstan. Email: assanovnigmetulla [at] gmail.com Submitted: 11/01/2023 Accepted: 11/05/2023 Published: 10/06/2023 © 2023 Open Veterinary Journal
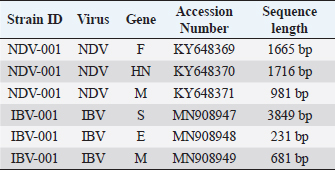
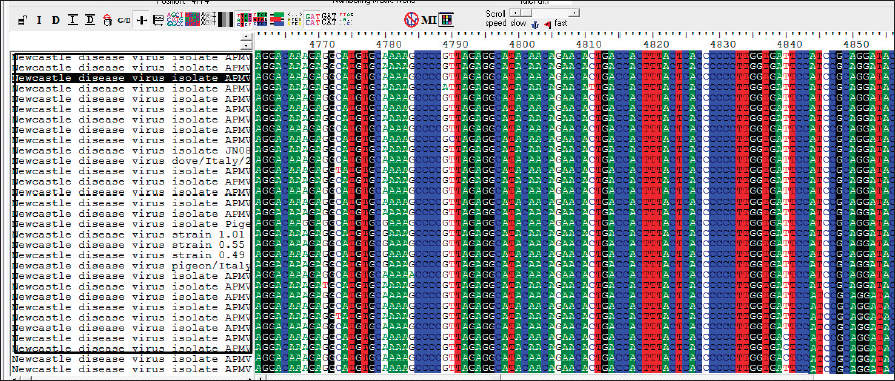
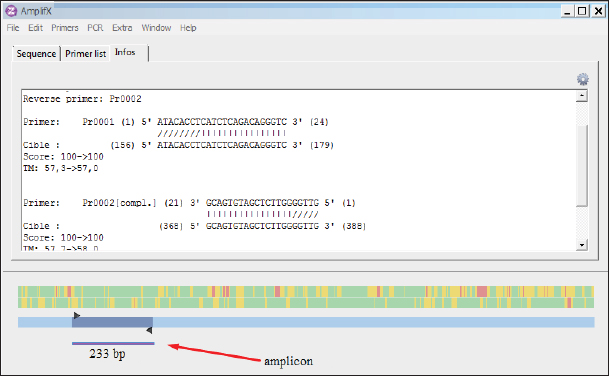
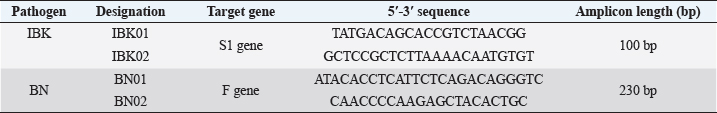
AbstractBackground: Infectious diseases of young and adult birds with respiratory syndrome are a significant deterrent to the development of industrial poultry farming due to decreased productivity and significant mortality. The only effective method of combating viral diseases is timely and targeted vaccination, which largely depends on laboratory diagnostic results. Aim: This article aims to study the real-time reverse transcription polymerase chain reaction, (RT-PCR) which has the prospect of more effective diagnosis of vaccine strains of chicken infectious bronchitis and Newcastle disease. Methods: The fastest and most accurate method for the differential diagnosis of pathogens in an associative viral infection is RT-PCR. The method proposed in the article for selecting primers for amplification made it possible to use this method for the simultaneous interspecies differential diagnosis of two or more viral agents, significantly accelerating their diagnosis. Results: The correlation of the nucleotide sequence obtained from sequencing to a specific virus strain is complicated by the lack of a single nomenclature mechanism for separating genetic groups. Conclusion: The results of this study will allow easy and fast typing of sequences into known and databased virus strains and avoid further confusion in the nomenclature of genetic groups in the future. Keywords: Amplification, Nucleotide sequence, Phylogenetic tree, Primer, Sequence. IntroductionSeveral studies indicated that viral diseases are the most common in industrial poultry farms (Haji-Abdolvahab et al., 2019; Vishchur et al., 2020; Dukhnitskyi and Tyshkivska, 2021). A review article by Ike et al. (2021) also focuses on significant economic losses in broiler and egg flocks due to the development of Newcastle disease (ND) and infectious bronchitis (IB) due to the significant mortality of young birds and increased losses during rearing. The primary method of preventing the incidence of poultry remains active vaccination of the entire population against pathogens of these diseases. There are many live, attenuated, inactivated, and recombinant, both monovalent and combined vaccines against Newcastle disease virus (NDV) and infectious bronchitis virus (IBV) in the world, which stimulate the development of both cellular and humoral responses of the body. Unfortunately, due to a significant mutation of the capsid proteins of the causative agent of IB in chickens (Tan et al. , 2016) and a considerable diversity of NDV genetic variants (Huang and Wang, 2006) the effectiveness of vaccines is insufficient because cross-infection with different strains does not guarantee the protection of chickens after vaccination. The classification of virus serotypes is difficult due to the need for a worldwide system for determining mutations. As a result, the same virus types in different regions are called differently, which also introduces a certain destructiveness. Molecular genetic studies showed that the primary source of modification of the IBV genotype is a single nucleotide substitution in the S1 gene, while other parts of the genome remain unchanged (Tan et al., 2016). Therefore, the only effective combating of these diseases is constantly monitoring circulating strains in a particular area and selecting vaccines with the desired genotypes (Lim et al., 2015). In addition, a significant part of vaccine viruses can become a new source of infection due to genome recombination (Gallardo, 2021). With viruses exhibiting high variability and constantly giving rise to new variants and genotypes, there is a need for a rapid and sensitive method of diagnosing and distinguishing viral virions. This allows for better control of the genetic and phenotypic variability within the local population of pathogens that cause infectious diseases in poultry. ND and IBV cause a significant number of respiratory diseases in young poultry. Since both pathogens are RNA viruses, their identification may be difficult (Mo et al., 2022). Various diagnostic tests have been developed worldwide to detect IBV and NDV particles; these are serological and molecular techniques (Brown and Bevins, 2017). Serological methods include hemagglutination tests and enzyme immunoassay. A publication by Wu et al. (2019) concluded that serological methods can quickly and relatively cheaply detect the presence of a virus. Still, low specificity does not make it possible to differentiate between genotypes. This is particularly important in the case of IBV, as cross-reactions between serotypes make serological tests uninformative (Miller et al., 2010). Gallardo (2021) reports that, in most cases, serological methods are used to determine the level of immunoglobulins (antibodies) after the vaccination of birds. Molecular genetic studies have a higher sensitivity, specificity, and reliability of the results obtained compared to serological methods (Legnardi et al., 2020). The primary molecular genetic method for studying the viral genome is the polymerase chain reaction (PCR) in its various modifications. Several PCR options allow you to highly reliably determine the genotypic structure of the virus and attribute it to a specific strain. Most often, for the quantitative and qualitative determination of the incidence in birds caused by RNA-containing viruses, the real-time reverse transcription polymerase chain reaction (qRT-PCR or RT-PCR) method is used (Shanmuganathan et al., 2017; Okino et al., 2018; Al-Jallad et al., 2020; Mo et al., 2020; Salem et al., 2020; Ongor et al., 2021; Worku et al., 2022) for the typing of the chicken IBV as well as for the isolation of the NDV. The difference between the methods of different researchers was using various primers and probes for typing viral strains. The susceptible reverse transcription method allows the detection of deficient levels of viral copies 102 −04. Valastro’s et al. (2016) identified 6 IBV genotypes containing 32 viral lines based on point mutations in the S1 gene sequence. Dey et al. (2014) study results showed significant differentiation of NDV strains. To a lesser extent, the method of loop isothermal amplification is used for genetic typing, which also uses a PCR followed by specific primers to target the 5-untranslated region of the IBV genome and the conserved region of the ND large polymerase gene. In research by Wu et al. (2019), the specificity of this method was inferior to the classical real-time RT-PCR method, and with the combined detection of IBV and ND viruses, the result was on the same level as RT-PCR. Other methods for diagnosing and differentiating viral particles for the typing of pathogens of respiratory diseases of birds also use the technique of sequence-independent, single-primer amplification (SISPA) (Bahador et al., 2021; Brinkmann et al., 2021). This method has recently been used to identify RNA-containing viruses and is the most effective in differentiating their strains. It is easier to conduct research but requires further sequencing of PCR products, which is associated with laboriousness and the high cost of research. Consequently, the objective of this study was to enhance the technique for simultaneous detection and differentiation of IBVs and NDVs genomes in chickens through the use of RT-PCR. Additionally, the study aimed to identify the molecular genetic characteristics of IBV and ND viruses. Materials and MethodsThe studies were carried out based on the Laboratory of Molecular Genetics using the information on the nucleotide sequence of strains of infectious chicken bronchitis and NDVs in GenBank, as well as by testing methods using vaccine strains of these viruses with polyvalent attenuated vaccines. This study used materials of methodological recommendations, scientific publications describing the stages of molecular genetic analysis of IBV and ND viruses, and bioinformatic methods for identifying their genomes. Development of specific primers. When developing primer systems for detecting the ND virus, the sequences of the F gene were used, while for the IBV, the sequences of the S1 gene were used. When developing specific oligonucleotides, local bases of nucleotide sequences of pathogen genes obtained from GenBank were created (The National Center for Biotechnology Information, 2022). The analysis of their conservatism is carried out using the BioEdit program (7.2.0.). After that, specific oligonucleotides were developed in the areas of conservative regions using the AmplifX program. RNA extraction. Genetic material was isolated using a commercial kit RNeasy mini kit (Qiagen, Hilden, Germany) for extraction, stabilization, and subsequent use of RNA from animal cells and tissues. RNA extraction was performed according to the manufacturer’s instructions. The extracted viral RNAs were used in following reverse transcription for Complementary deoxyribonucleic acid (cDNA) (deoxyribonucleic acid) synthesis or stored at −70°C for later use. cDNA was obtained from extracted RNA using the reverse transcriptase enzyme. For duplex PCR, two sets of universal primers were used in one reaction. cDNA was obtained after reverse transcription as a template for further PCR amplification. Libraries, fragmented IB, and BN sequences were prepared using the SISPA protocol ‒ non-selective logarithmic amplification of heterogeneous populations of complementary DNA obtained from templates purified with RNA. Sequencing using the SISPA method consists of a single primer with random hexamers at the end for amplification. With primers, SISPA from RNA gets double-stranded complementary DNA according to the method of Chrzastek et al. (2017). Identification of IB and ND strains based on the analysis of the primary nucleotide sequence. The primary nucleotide sequence analysis was performed in the GenBank database using the built-in BLASTn (Basic Local Alignment Search Tool) software. BLAST is a software package designed to match a nucleotide sequence with a translated amino acid chain, compare and align them. Alignment refers to the graphic placement of homologous sequences to search for similar regions. Programmatic exon analysis with Blastx (“Translated query versus protein database”) makes it possible to convert a nucleotide sequence according to the principle of complementarity into amino acid molecules and then search for homologous sequences in protein databases. Blastx allows for a complete analysis of possible variants of the nucleotide sequence of the analyzed genome. This allows efficient analysis of unknown sequences or sequences containing sequencing errors that could lead to frameshifts or other translational errors. Such an algorithm is often used to analyze the resulting sequenced nucleotide sequences and fragmented sequences. The completeness of the sequence analysis is evaluated with the available sequences from the BLAST database with the definition of such indicators as the maximum identity, the area of query overlap, and the validity of this alignment. Building a phylogenetic tree. When conducting phylogenetic and evolutionary analysis, multiple alignments of the virus genotype sample were carried out according to the studied genes from the GenBank database. Then, with the help of special programs, a tree was built, and the results were presented as graphical information. To build a phylogenetic tree, a selection of nucleotide sequences was compiled in the FASTA format using the Neighbour-joining (NJ) method; this is a fast method suitable for working with many sequences. For this, the molecular evolutionary genetic analysis (MEGA) 11 program was used. MEGA 11 is a computer program used for molecular evolutionary genetics analysis. It is a software suite for conducting various molecular biology analyses, including phylogenetic analysis, sequence alignment, evolutionary rate estimation, and ancestral sequence reconstruction. ResultsIn the work, several methodological approaches were used in molecular genetic studies with the aim of simultaneous detection and possible differentiation of the causative agents of two respiratory diseases in poultry ‒ RNA viruses of ND and IB. Such methods in industrial poultry farming, when the probability of infection of a large number of livestock is very high, will reduce the time to determine the pathogen and the costs of treatment and prevention measures on infected farms. The first method used to determine the simultaneous presence of respiratory viruses was classical RT-PCR using primers whose amplification products are easy to differentiate. Using primers for specific species genes made it possible to detect the presence of viruses during joint (duplex or triplex) amplification. Development of synthetic primersThis stage of work aimed to create specific primers for iteration and identification of the viral genome in the reaction mixture. At the same time, they were guided by particular regions of the virus genome inherent in all strains of a specific species. To do this, the complete gene sequences of different strains of chicken IB and NDVs were downloaded from the GenBank database for further analysis (Table 1). To find gene-conserved regions in the virus genome, viral nucleotide sequences were aligned in the BioEdit program (7.2.0.) using the Clustal W function (Fig. 1). When analyzing the available database from GenBank with genome sequences of IBV and ND viruses, target regions were obtained in the most variable genes, S1 (IBV) and F (ND). When aligning the nucleotide sequences, conservative areas were found and subsequently used to create primers. Alignment, analysis, and primer selection were performed using the AmplifX software. This is one of the software elements for PCR experiments. The program contains all information about primers in a single database and can create them by analyzing the target region of the nucleotide sequence of the target gene. After loading the information into AmplifX, primer systems for the genes of interest with detailed characterizations were designed to perform theoretical amplification (Fig. 2). When processing an array of information about the S1 target genes for IB of chickens and the F gene for the NDV, primers for each were determined. Thus, (Figure 2) shows the window of the AmplifX program with information on the main characteristics of the recommended primer for the iteration of the F gene of the NDV. The software, in addition to the nucleotide sequence of the primer, indicates the length of the synthesized amplicon (233 bp), as well as the recommended conditions for the PCR reaction, in particular, and the annealing temperature. The selected primers were also checked for the purine and pyrimidine bases ratio. A similar analysis was carried out for the S1 gene of the chicken IBV. Simultaneously with the independent selection of primers, the primers used in previous studies, described in articles by authors from other countries, were analyzed. As a result of this combined approach, two pairs of primers were selected for detecting IB and BN by target genes (Table 2). Table 1. The sequence information of NDV and IBV strains.
Fig. 1. BioEdit program (7.2.0.) with aligned F1 sequences of the NDV gene (The National Center for Biotechnology Information, 2022).
Fig. 2. AmplifX program. Carrying out theoretical amplification on the target gene F1 of the NDV (The National Center for Biotechnology Information, 2022). One of the main advantages of the selected primers was that they differed in the size of the nucleotide sequence of the target amplicons and could be used in the reaction to identify the species of the virus during PCR simultaneously and at the same time, it was easy to distinguish the amplification products. This approach made it possible to carry out the identification of viruses with minimal costs due to the visualization of the result using the electrophoretic separation of amplicons. Table 2. Primer systems for detection of target genes of infectious chicken bronchitis and NDV (The National Center for Biotechnology Information, 2022).
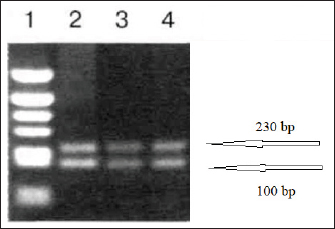
To test the performance of the primers, vaccine strains of ND and IBV viruses from a polyvalent live attenuated vaccine were used. Viral material was prepared and set up a PCR using reagents and equipment from Qiagen (Germany). After the extraction of viral RNA, duplex RT-PCR was performed in a total reaction volume of 25 µl with 0.5 µl of each primer. The temperature profile for duplex RT-PCR included an initial denaturation at 94°C for 2 minutes, followed by 35 cycles of 94°C for 45 seconds, 55°C for 45 seconds, and 72°C for 45 seconds. The results of amplifying the reaction mixture are presented as an electropherogram (Fig. 3). The performance of a method on vaccine strains does not necessarily guarantee its success in detecting the viruses in actual samples. The technique would need to be further validated on a larger number of clinical samples to assess its sensitivity and specificity in detecting the target viruses. Additionally, the method should be evaluated for its ability to see other strains or subtypes of the viruses and any potential cross-reactivity with other viruses or pathogens that may be present in the samples. As a result of electrophoresis of the amplification products in polyacrylamide gel, two products were obtained that had sizes of 100 and 230 nucleotide sequences and which corresponded to the target amplicons of the genes of vaccine strains of infectious chicken bronchitis and NDVs. These results indicate the possibility of performing species-specific detection of viral target genes by real-time duplex reverse transcription PCR. The experiment was carried out in three repetitions when setting up the reaction under the same conditions. The results of all reactions were at the same level, which indicates the efficiency of the proposed primers when used together to diagnose possible simultaneous infection of poultry with two respiratory viruses of ND and IB of chickens. The following molecular genetic method used for the joint identification and possible differentiation of viral strains was obtaining fragmented sequences of infectious chicken bronchitis and NDVs using a modified SISPA protocol. This method involves some modification of the standard RT-PCR procedure using a specific K-8N primer and enzyme. This approach is also used to obtain full-sized viral genomes with the possibility of subsequent virus identification at its low concentration in the material.
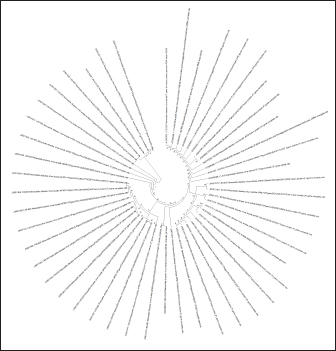
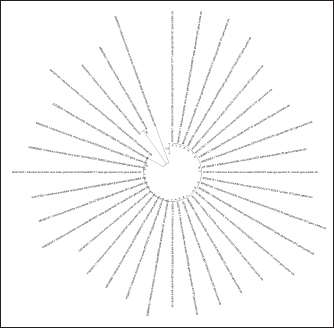
Fig. 3. Results of detection of vaccine strains of infectious chicken bronchitis (100 bp) and Newcastle disease (230 bp) viruses after duplex RT-PCR. A simplified genome iteration method using the SISPA protocol allowed for replicating viral genome sequences from ND and IBV vaccine strains. In contrast to the classical reverse transcription PCR, the modified SISPA protocol is more straightforward and does not require multiple cycles of amplification of the RNA viral genome and virus propagation, thus significantly reducing the time and costs for virus isolation. Vaccine viral genomes were extracted using a commercial RNeasy mini kit (Qiagen, Hilden, Germany). Then, using RT-PCR and primer K-8N (GAC CAT CTA GCG ACC TCC CAN NNN NNN NNN N), primary viral cDNAs were isolated. Subsequent amplification was carried out using maple polymerase at 37°C for 60 minutes. The synthesized DNA was subsequently used for sequencing. Samples for analysis were prepared using the SQK-LSK109 kit (Oxford Nanopore Technologies). PCR reaction products were sequenced for 3 hours on Oxford Nanopore MinION SpotON Flow Cells Mk I, R9.4 (Oxford Nanopore Technologies). After sequencing cDNA obtained by amplification by the SISPA method, information about the nucleotide sequence was identified in the GenBank database using multiple alignments in the BLASTn program. As a result of the study, it was possible to locate both types of viruses, while only the NDV could determine the belonging to the strain. These results confirm the possibility of using the SISPA method to identify respiratory viruses in associated poultry infections. To determine the strain identification of viruses, the approach turned out to be ineffective and needs further improvement. Phylogenetic analysis of the samplesPhylogenetic analysis, based on the comparison of nucleotide sequences, makes it possible to determine the relationship and group different groups of viruses behind common gene pools, both at the interspecific and intraspecific levels. Given the significant mutagenicity of individual genes of RNA-containing avian viruses, a phylogenetic tree was constructed according to the most variable genes of IBV and ND viruses to simplify further differentiation of strains. This approach will make it possible to quickly compare the nucleotide sequence of the virus obtained during sequencing with that presented in the phylogenetic tree and determine its taxonomic and other characteristics. This will simplify the differentiation of wild and vaccine strains of IB of chickens and ND when analyzing the causes of morbidity in poultry in industrial herds. For phylogenetic analysis of genomes, information on possible variants of the nucleotide sequence in the S1 spike gene of the IBV (about 1600 nt) and the F gene of the NDV was loaded from the GenBank database. Nucleotide sequences were aligned and analyzed to search for homologous regions using the Megablast method of the BLAST software. The obtained analysis results were processed with the subsequent construction of a phylogenetic tree using the MEGA 11 software product. The software for MEGA contains a large set of methods and tools for computational molecular evolution, including constructing a phylogenetic tree. In this case, the NJ method was used, making it possible to combine both phylogenetic and evolutionary approaches simultaneously. The results of this approach are shown in (Figs. 4 and 5). Evolutionary relationships of taxaA bootstrap file derived from 500 iterations was used to represent the evolutionary history of the analyzed taxa. Branches corresponding to genotypes reproduced in less than 50% of bootstrap replicas are collapsed. Evolutionary distances were calculated using the maximum composite likelihood method and expressed in units of base substitutions per site. This analysis involved 48 nucleotide sequences. All positions that contained errors or missing nucleotides were not considered when compiling the tree. In total, there were 537 positions in the final dataset. A similar approach was implemented for the S1 gene of the IBV since this fragment is the most frequently used in the international classification system for the causative agent of the disease. The classification system contains 32 lines that belong to 6 genotypes. When creating a phylogenetic tree, the original scaling was used, which corresponded to the evolutionary distance. These distances were calculated using maximum likelihood estimation and are expressed in units of mutations (nucleotide substitutions) per gene. This analysis involved 35 nucleotide sequences. The use of a phylogenetic and evolutionary approach makes it possible to establish the sources and dynamics of the spread of infection, and an in-depth study of the genetic relationships between viruses isolated in various farms provided important information about the causes and routes of infection.
Fig. 4. Ring phylogenetic tree of sequences of the F gene of the NDV (The National Center for Biotechnology Information, 2022).
Fig. 5. Ring phylogenetic tree of the S1 gene of the infectious chicken bronchitis virus (The National Center for Biotechnology Information, 2022). Regarding the relationship between evolutionary trees and PCR products, it is important to note that PCR products can be used as input for phylogenetic analysis, a method for reconstructing evolutionary relationships between different organisms or strains based on their genetic sequences. By comparing the sequences of the PCR products from different strains, researchers can construct evolutionary trees that illustrate the relationships between the strains. Valastro et al. (2016) conducted a study on the phylogeny of IBV, which identified different subtypes or clades of IBV that PCR primers could target. This finding is particularly relevant to the current research on PCR-based detection of IBV, as the authors of this study aimed to evaluate the potential of real-time RT-PCR for the more effective diagnosis of vaccine strains of chicken IBV and ND. By optimizing the RT-PCR method to detect and differentiate these viruses in poultry simultaneously, the authors were able to identify the genetic and phenotypic variability of local pathogens causing infectious diseases in poultry. The results of this study demonstrate that RT-PCR is a fast and accurate method for the simultaneous detection and differentiation of RNA-containing viruses that cause respiratory diseases in poultry. Overall, the findings of this study have important implications for developing more efficient diagnostic methods for IBV and ND in poultry, which will ultimately help protect poultry health and improve the efficiency of poultry farming. Further research is needed to refine the intraspecific classification of virus strains and develop phylogenetic schemes for other genes characterized by high variability. DiscussionThe rapid detection and differentiation of viruses can play a significant role in their control since the pathogens of IB of chickens and ND affect the respiratory tract of chickens, are characterized by considerable lethality, and, therefore, cause significant economic damage to the industry. There are several other viral diseases with a similar clinical picture and high mortality in young and adult birds that occur with the intensive development of poultry farming. These reasons force the constant search and development of reliable and fast methods of laboratory differential diagnosis for such diseases. The same opinion is shared by Salem et al. (2020) conducted studies on the incidence of respiratory diseases in chickens in Egypt. The conducted literature review, presented within the framework of this article, indicates that the fastest and most reliable diagnostic method is molecular genetic approaches in the typing of pathogens at the interspecific and intraspecific levels. Whereas serological diagnostic methods (ELISA, neutralization tests and hemagglutination retention, etc.) behind the message of Sakai et al. (2006) have not always been informative. Due to the high sensitivity and specificity of the PCR method, it is widely used as an indispensable method for diagnosing and detecting viruses. For the simultaneous detection of different types of infectious agents, the most suitable tool is the real-time RT-PCR. Carrying out by several authors of a duplex reaction (Saba and Mardani, 2014; Shirvan and Mardani, 2014) and triplex reaction (Zhang et al., 2020) RT-PCR for the simultaneous species identification of avian respiratory RNA viruses confirmed the possibility of multi-vector simultaneous amplification and visualization of PCR results for diagnosis. This method of rapid detection of viral particles in the system of industrial poultry farming is critical and will be in demand in case of combined infection of birds with two or more pathogens and will speed up diagnostic measures. One-step RT-PCR is time-consuming, but duplex RT-PCR can significantly reduce this time. The primary condition for conducting a duplex PCR reaction is to select primers that would be both species-specific and not overlap during amplification. Another of the requirements for the joint determination of pathogens of several diseases is that the amplification products (primers) must differ significantly so that they can be easily distinguished when visualizing the PCR results. The article describes in detail the mechanism of primer selection for carrying out the RT-PCR, and the software products used in the selection and verification of primers are considered. This approach made it possible to identify vaccine strains of IBV and ND viruses with selected primers at minimal cost and in a short time. In the study, the main focus was on the interspecies identification of viruses that cause respiratory diseases in birds. Duplex RT-PCR can also be used for intraspecific differentiation of virus strains. This is confirmed in the work of Shirvan and Mardani (2014), who carried out simultaneous amplification of the H5, H7, and H9 subtypes of the avian influenza virus. Unfortunately, the intraspecific determination of virus strains is associated with high costs for continuous monitoring of possible mutations in the virus genome and the development of primers to identify these serotypes. Therefore, using RT-PCR for interspecies differentiation of pathogens in developing an associative infection will be the fastest and most effective diagnostic method. No less effective in the detection and interspecies differentiation of pathogens was the method of amplifying the virus genome using the SISPA protocol. This approach also succeeded in identifying both vaccine strain viruses from the live attenuated vaccine. Despite some simplification of the formulation of the first stage of the polymerase chain reaction ‒ the use of the universal primer K-8N and a specific polymerase enzyme, they allow quick obtaining amplified particles of the virus genomes present in the reaction mixture. Further visualization of the obtained amplification products is associated with a more expensive method ‒ sequencing and subsequent analysis of the nucleotide sequence using software products for its identification in the international GenBank database. This diagnostic method is longer and more expensive than the classical RT-PCR method, with visualization of results using electrophoretic separation of amplification products. But on the other hand, sequencing of the sequences obtained as a result of the reaction also makes it possible to determine whether the virus belongs to a particular strain, which makes this method more informative. In the studies carried out, it was possible to accurately determine the strain of the NDV, which corresponded to the one declared by the vaccine manufacturer, while the intraspecific matching of the IBV using alignment programs and analysis of the nucleotide sequence did not bring a positive result, and the strain was not identified. Perhaps the reason for this was the low concentration of viral RNA in the vaccine since Chrzastek et al. (2017) in his studies also pointed out the dependence of intraspecific detection of viruses on their concentration in the material during analysis using the SISPA protocol. Therefore, the technique for determining the intraspecific affiliation of viruses using this method still needs to be further improved to obtain the maximum and reliable result in most cases. Using the sequencing technique for target gene regions made it possible to map the nucleotide sequences of almost all known strains at the intraspecific level. The exchange of such information between scientific laboratories, institutes, and other genetic organizations made it possible to create a GenBank scientific database on the site of the biotechnological information center with unlimited access to everyone. Unfortunately, a unified nomenclature mechanism for separating genetic groups at the site has not yet been developed, and this greatly complicates the classification of strains. Thus, the lack of standard rules in genotyping the S1 gene region of the infectious chicken bronchitis virus makes the interpretation of possible virus strains very difficult. The same situation is observed in several other viruses that are not associated with developing diseases in humans. Therefore, based on this situation, a phylogenetic tree method is proposed, taking into account the evolutionary component, to form a taxonomic classification of chicken IBVs and ND for genes that are characterized by the most frequent replacement (mutations, insertions, deletions, etc.) in the nucleotide sequence. Such genes are the S1 spike gene of the IBV virus and the F gene of the ND virus. The algorithm and principle of constructing a phylogenetic tree, described in detail in this article, made it possible to create a classifier of known virus strains and discard information from the general information base of GenBank, which contained genome variants with errors and did not correspond to the maximum likelihood statistical method. The resulting phylogenetic trees for the target genes of the studied IBV and ND viruses, presented in the article, will subsequently allow easy and quick typing of the nucleotide sequences obtained as a result of sequencing into known virus strains that are in the database and to avoid further confusion in the nomenclature of genetic groups in the future. In addition, the use of this approach will make it possible to detect possible sources of infection in poultry and the dynamics of the spread of infection in herds. And the analysis of genetic relationships between strains of viruses isolated in different farms will provide important information about the routes of infection and will help in developing a system for preventing the disease in industrial herds. The approaches considered in diagnosing and differentiating viral diseases in poultry in industrial poultry farming are already partially used by various genetic laboratories around the world. But combining all the proposed activities into a single system for determining viral strains and their classification will bring the biosafety of poultry farming to a new level. ConclusionThis study aimed to evaluate the potential of RT-PCR for the more effective diagnosis of vaccine strains of chicken IB and ND. By optimizing the RT-PCR method to detect and differentiate these viruses in poultry simultaneously, the authors were able to identify the genetic and phenotypic variability of local pathogens causing infectious diseases in poultry. The results of this study demonstrate that RT-PCR is a fast and accurate method for the simultaneous detection and differentiation of RNA-containing viruses that cause respiratory diseases in poultry. The proposed method for selecting primers and performing RT-PCR simplifies the study and provides a more efficient approach to controlling the spread of these diseases in poultry populations. Overall, the findings of this study have important implications for developing more efficient diagnostic methods for IBV and ND in poultry, which will ultimately help protect poultry health and improve the efficiency of poultry farming. Further research is needed to refine the intraspecific classification of virus strains and develop phylogenetic schemes for other genes characterized by high variability. ReferencesAl-Jallad, T., Kassouha, M., Salhab, M., Alomar, A., Al-Masalma, M. and Abdelaziz, F. 2020. Molecular characterization of isolated infectious bronchitis viruses from affected vaccinated broiler flocks in Syria. BMC Vet. Res. 16, 449. Bahador, D., Mohammadi, A., Foroughi, A. and Alirezaie, B. 2021. Sequence-independent single-primer-amplification (SISPA) as a screening technique for detecting unexpected RNA viral adventitious agents in cell cultures. Arch. Biotechnol. Biomed. 5, 8–12. Brinkmann, A., Uddin, S., Krause, E., Surtees, R., Dinçer, E., Kar, S., Hacıoğlu, S., Özkul, A., Ergünay, K. and Nitsche, A. 2021. Utility of a sequence-independent, single-primer-amplification (SISPA) and nanopore sequencing approach for detection and characterization of tick-borne viral pathogens. Virus 13(2), 203. Brown, V.R. and Bevins, S.N. 2017. A review of virulent newcastle disease viruses in the United States and the role of wild birds in viral persistence and spread. Vet. Res. 48, 68. Chrzastek, K., Lee, D.H., Smith, D., Sharma, P., Suarez, D.L., Pantin-Jackwood, M. and Kapczynski, D.R. 2017. Use of sequence-independent, single-primer-amplification (SISPA) for rapid detection, identification, and characterization of avian RNA viruses. Virology 509, 159–166. Dey, S., Chellappa, M.M., Gaikwad, S., Kataria, J.M. and Vakharia, V.N. 2014. Genotype characterization of commonly used Newcastle disease virus vaccine strains of India. PLoS One. 9(6), e98869. Dukhnitskyi, V.B. and Tyshkivska, A.M. 2021. Tilmicosin intake and distribution in the body of broiler chickens with ornithobacteriosis. Ukrainian J. Vet. Sci. 12(2), 46–58. Gallardo, R.A. 2021. Infectious bronchitis virus variants in chickens: evolution, surveillance, control and prevention. Aust. J. Vet. Sci. 53, 55–62. Haji-Abdolvahab, H., Ghalyanchilangeroudi, A., Bahonar, A., Ghafouri, S.A., Marandi, M.V., Mehrabadi, M.H.F. and Tehrani, F. 2019. Prevalence of avian influenza, Newcastle disease, and infectious bronchitis viruses in broiler flocks infected with multifactorial respiratory diseases in Iran, 2015-2016. Trop. Anim. Health Prod. 51, 689–695. Huang, Y.P. and Wang, C.H. 2006. Development of attenuated vaccines from Taiwanese infectious bronchitis virus strain. Vaccine 24, 785–791. Ike, A.C., Ononugbo, C.M., Obi, O.J., Onu, C.J., Olovo, C.V., Muo, S.O., Chukwu, O.S., Reward, E.E. and Omeke, O.P. 2021. Towards improved use of vaccination in the control of infectious bronchitis and Newcastle disease in poultry: understanding the immunological mechanisms. Vaccines 9, 20. Legnardi, M., Tucciarone, C.M., Franzo, G. and Cecchinato, M. 2020. Infectious Bronchitis virus evolution, diagnosis, and control. Vet. Sci. 7(2), 79. Lim, T.H., Youn, H.N., Yuk, S.S., Kwon, J.H., Hong, W.T., Gwon, G.B., Lee, J.A., Lee, J.B., Lee, S.W. and Song, C.S. 2015. Successful cross-protective efficacy induced by heat-adapted live attenuated nephropathogenic infectious bronchitis virus derived from a natural recombinant strain. Vaccine 33, 7370–7374. Miller, P.J., Decanini, E.L. and Afonso. C.L. 2010. Newcastle disease: evolution of genotypes and the related diagnostic challenges. Inf. Gen.Evolut. 10, 26–35. Mo, J., Angelichio, M., Gow, L., Leathers, V. and Jackwood, M.W. 2022. Quantitative real-time PCR assays for the concurrent diagnosis of infectious laryngotracheitis virus, Newcastle disease virus and avian metapneumovirus in poultry. J. Vet. Science. 23(2), e21. Mo, J., Angelichio, M., Gow, L., Leathers, V. and Jackwood, M.W. 2020.Validation of specific quantitative real-time RT-PCR assay panel for infectious bronchitis using synthetic DNA standards and clinical specimens. J. Virolog. Met. 276, 113773. Okino, C.H., Montassier, M.F.S., Oliveira, A.P. and Montassier, H.J. 2018. Rapid detection and differentiation of avian infectious bronchitis virus: an application of mass genotype by melting temperature analysis in RT-qPCR using SYBR Green I. J. Vet. Med. Sci. 80(4), 725–730. Ongor, H., Timurkaan, N., Coven, F., Karabulut, B., Eroksuz, H., Cetinkaya, B. and Carli, T. 2021. Detection of Israel variant 2 (IS/1494/06) genotype of Infectious bronchitis virus in a layer chicken flock. Vet. Fakült. Derg. 68, 167–172. Saba, A. and Mardani, K. 2014. Molecular detection of infectious bronchitis and Newcastle disease viruses in broiler chickens with respiratory signs using duplex RT-PCR. Vet. Res. Forum Int. Quar. J. 5, 319–323. Sakai, K., Yada, K., Sakabe, G., Tani, O., Miyaji, K., Nakamura, M. and Takehara, K. 2006. Serological and virological studies of Newcastle disease and avian influenza in slaughter-age ostriches (Struthio camelus) in Japan. J. Vet. Med. Sci. 68(5), 491–494. Salem, M.H.I., Hagag, N.M., Ali, A.A.H. and El-Shahidy, M.S.M. 2020. Validation of one-step multiplex RT-PCR for diagnosis of respiratory viruses coinfections in chickens. Adv. Anim. Vet. Sci. 8(1), 62–67. Shanmuganathan, L., Anggoro, D. and Wibowo, M. 2017. Newcastle disease virus detection from chicken organ samples using reverse transcriptase polymerase chain reaction. J. Sain Vet. 35(1), 127. Shirvan, A.S. and Mardani, K. 2014. Molecular detection of infectious bronchitis and Newcastle disease viruses in broiler chickens with respiratory signs using duplex RT-PCR. Vet. Res. Forum. 5(4), 319. Tan, L., Zhang, Y., Liu, F., Yuan, Y., Zhan, Y., Sun, Y., Qui, X., Meng, C., Song, C. and Ding, C. 2016. Infectious bronchitis virus poly-epitope- based vaccine protects chickens from acute infection. Vaccine 34, 5209–5216. The National Center for Biotechnology Information. 2022. Available via https://www.ncbi.nlm.nih.gov/genbank/. Valastro, V., Holmes, E.C., Britton, P., Fusaro, A., Jackwood, M.W., Cattoli, G. and Monne, I. 2016. S1 gene-based phylogeny of infectious bronchitis virus: an attempt to harmonize virus classification. Infect. Genet. Evol. 39, 349–364. Vishchur, O.I., Romanovych, L.V. and Kurtyak, B.M. 2020. Influence of vitamins E and C on the quantity and functional activity of τ- ι β-lymphocytes of blood-chicken broilers. Ukrainian J. Vet. Sci. 11(1), 59–69. Worku, T., Dandecha, M., Shegu, D., Aliy, A. and Negessu, D. 2022. Isolation and molecular detection of Newcastle disease virus from field outbreaks in chickens in Central Ethiopia. Vet. Med. 13, 65–73. Wu, X., Song, Z., Zhai, X., Zuo, L., Mei, X., Xiang, R., Kang, Z., Zhou, L. and Wang, H. 2019. Simultaneous and visual detection of infectious bronchitis virus and Newcastle disease virus by multiple LAMP and lateral flow dipstick. Poult. Sci. 98(11), 5401–5411. Zhang, X., Yao, M., Tang, Z., Xu, D., Luo, Y., Gao, Y. and Yan, L. 2020. Development and application of a triplex real-time PCR assay for simultaneous detection of avian influenza virus, Newcastle disease virus, and duck Tembusu virus. BMC Vet. Res. 16(1), 203. | ||
| How to Cite this Article |
| Pubmed Style Assanov N, Bazarbayev R, Mussoyev A, Otarbayev B, Iskhan K. The use of RT-PCR in the diagnosis and differentiation of vaccine strains of chicken infectious bronchitis and Newcastle disease. Open Vet. J.. 2023; 13(6): 732-741. doi:10.5455/OVJ.2023.v13.i6.8 Web Style Assanov N, Bazarbayev R, Mussoyev A, Otarbayev B, Iskhan K. The use of RT-PCR in the diagnosis and differentiation of vaccine strains of chicken infectious bronchitis and Newcastle disease. https://www.openveterinaryjournal.com/?mno=138508 [Access: November 22, 2025]. doi:10.5455/OVJ.2023.v13.i6.8 AMA (American Medical Association) Style Assanov N, Bazarbayev R, Mussoyev A, Otarbayev B, Iskhan K. The use of RT-PCR in the diagnosis and differentiation of vaccine strains of chicken infectious bronchitis and Newcastle disease. Open Vet. J.. 2023; 13(6): 732-741. doi:10.5455/OVJ.2023.v13.i6.8 Vancouver/ICMJE Style Assanov N, Bazarbayev R, Mussoyev A, Otarbayev B, Iskhan K. The use of RT-PCR in the diagnosis and differentiation of vaccine strains of chicken infectious bronchitis and Newcastle disease. Open Vet. J.. (2023), [cited November 22, 2025]; 13(6): 732-741. doi:10.5455/OVJ.2023.v13.i6.8 Harvard Style Assanov, N., Bazarbayev, . R., Mussoyev, . A., Otarbayev, . B. & Iskhan, . K. (2023) The use of RT-PCR in the diagnosis and differentiation of vaccine strains of chicken infectious bronchitis and Newcastle disease. Open Vet. J., 13 (6), 732-741. doi:10.5455/OVJ.2023.v13.i6.8 Turabian Style Assanov, Nigmetulla, Ryskeldi Bazarbayev, Assilbek Mussoyev, Bauyrzhan Otarbayev, and Kairat Iskhan. 2023. The use of RT-PCR in the diagnosis and differentiation of vaccine strains of chicken infectious bronchitis and Newcastle disease. Open Veterinary Journal, 13 (6), 732-741. doi:10.5455/OVJ.2023.v13.i6.8 Chicago Style Assanov, Nigmetulla, Ryskeldi Bazarbayev, Assilbek Mussoyev, Bauyrzhan Otarbayev, and Kairat Iskhan. "The use of RT-PCR in the diagnosis and differentiation of vaccine strains of chicken infectious bronchitis and Newcastle disease." Open Veterinary Journal 13 (2023), 732-741. doi:10.5455/OVJ.2023.v13.i6.8 MLA (The Modern Language Association) Style Assanov, Nigmetulla, Ryskeldi Bazarbayev, Assilbek Mussoyev, Bauyrzhan Otarbayev, and Kairat Iskhan. "The use of RT-PCR in the diagnosis and differentiation of vaccine strains of chicken infectious bronchitis and Newcastle disease." Open Veterinary Journal 13.6 (2023), 732-741. Print. doi:10.5455/OVJ.2023.v13.i6.8 APA (American Psychological Association) Style Assanov, N., Bazarbayev, . R., Mussoyev, . A., Otarbayev, . B. & Iskhan, . K. (2023) The use of RT-PCR in the diagnosis and differentiation of vaccine strains of chicken infectious bronchitis and Newcastle disease. Open Veterinary Journal, 13 (6), 732-741. doi:10.5455/OVJ.2023.v13.i6.8 |