| Review Article | ||
Open Vet. J.. 2024; 14(4): 941-951 Open Veterinary Journal, (2024), Vol. 14(4): 941–951 Review Article African swine fever virus proteins against host antiviral innate immunity and their implications for vaccine developmentFredmoore L. Orosco1, 2*1Virology and Vaccine Institute of the Philippines Program, Industrial Technology Development Institute, Department of Science and Technology, Taguig, Philippines 2Department of Biology, College of Arts and Sciences, University of the Philippines Manila, Metro Manila, Philippines *Corresponding Author: Fredmoore L. Orosco. Virology and Vaccine Institute of the Philippines Program, Industrial Technology Development Institute, Department of Science and Technology, Taguig, Philippines. Email: orosco.fredmoore [at] gmail.com; florosco [at] up.edu.ph Submitted: 03/12/2023 Accepted: 13/03/2024 Published: 30/04/2024 © 2024 Open Veterinary Journal
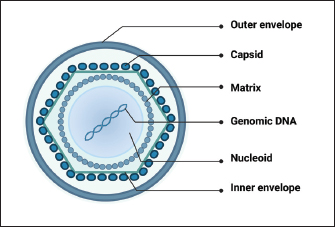
AbstractAfrican swine fever virus (ASFV) poses a significant threat to global swine populations, necessitating a profound understanding of viral strategies against host antiviral innate immunity. This review synthesizes current knowledge regarding ASFV proteins and their intricate interactions with host defenses. Noteworthy findings encompass the modulation of interferon signaling, manipulation of inflammatory pathways, and the impact on cellular apoptosis. The implications of these findings provide a foundation for advancing vaccine strategies against ASFV. In conclusion, this review consolidates current knowledge, emphasizing the adaptability of ASFV in subverting host immunity. Identified research gaps underscore the need for continued exploration, presenting opportunities for developing targeted vaccines. This synthesis provides a roadmap for future investigations, aiming to enhance our preparedness against the devastating impact of ASFV on global swine populations. Keywords: African swine fever virus, Antiviral innate immune response, Virulence factors, Pathogenicity, Vaccine development. IntroductionAfrican swine fever (ASF), caused by the African swine fever virus (ASFV), is a potent viral infection affecting swine (Zhao et al., 2019). Characteristics of this contagious ailment encompass heightened body temperature, bleeding, ataxia, and a state of depression in affected pigs (Wang et al., 2019). Since its initial identification in Kenya in 1921, ASF has swiftly disseminated across diverse regions, including Africa, Europe, South America, Latin America, and Asia (Zhao et al., 2019). The deleterious consequences of ASF outbreaks have led to substantial economic setbacks and posed a formidable challenge to the sustainable development of the global pig industry (Wang et al., 2020). In the era of accelerated globalization, traditional methods such as animal removal and restricting movements prove ineffective against ASFV. Consequently, the primary solution lies in creating a dependable, secure, and efficient vaccine to combat the ASFV threat. Despite the virus being recognized for more than a century, there is currently no authorized vaccine accessible (Wang et al., 2020; Orosco, 2023a). The limited understanding of how ASFV-specific proteins govern the host innate immune system has significantly impeded the progress of reliable and safe ASFV vaccines (Zheng et al., 2022). Recent studies have revealed that ASFV has evolved diverse mechanisms to evade host immune responses, resulting in severe disease and high mortality rates (He et al., 2022; Zheng et al., 2022). This review aims to provide an overview of the different ASFV proteins that suppress host-directed antiviral innate immune responses and their implications for ASFV virulence and vaccine development. Overview of ASFVThe DNA genome of ASFV is double-stranded and linear, exhibiting variable sizes typically between 170 and 193 kbp, contingent on the isolate (Chapman et al., 2008). Within the ASFV genome, there are 151 to 167 open reading frames (ORFs). Variations in genome length and gene count primarily arise from the acquisition or loss of ORFs within the virus-encoded multigene families (MGFs). In addition, discrepancies in length can be attributed to variations in the quantity of short tandem repeats in genes or intergenic regions (Lubisi et al., 2007). The viral genetic material is tightly arranged on both DNA strands, with each gene possessing a brief upstream sequence housing a promoter region acknowledged by the viral RNA polymerase complex. These promoters are characterized by short lengths and a prevalence of A+T nucleotides, interacting with virus-encoded transcription factors tailored for distinct virus gene expression stages—early, intermediate, and late gene categories (Almazán et al., 1993; Rodríguez et al., 1996). ASFV exhibits a complex structure with an overall size ranging from 175 to 215 nm (Salas and Andrés, 2013). Its virion comprises various elements, such as a nucleoprotein core, an internal lipid layer encasing a core shell, an icosahedral capsid, and an optional lipid envelope. However, insights into the virus’s structure and architecture have been limited until recent times (Salas and Andrés, 2013). Recent advancements in single-particle cryo-EM analyses have provided a three-dimensional view of the ASFV particle’s structure (Fig. 1). This analysis disclosed that the nucleoid is enveloped by two distinct icosahedral protein capsids and two lipoprotein membranes. One membrane aligns with the inner capsid’s icosahedral symmetry, while the other surrounds the outer capsid, originating from the budding process (Andrés et al., 2020; Blome et al., 2020). Several ASFV proteins have been recognized as immunogens, with p72, p54, p30, and CD2v identified as the primary ones (Jancovich et al., 2018). In vitro expression of the CD2v protein has exhibited partial safeguarding against infection by closely related virulent strains. Despite the capability of p30, p54, and p72 proteins to prompt neutralizing antibodies, they lack effectiveness in shielding pigs from potent ASFV challenges. Investigations indicate that eliminating DP71L, DP96R, B119L, or DP148R significantly diminishes viral virulence, offering defense against virulent ASFV strain infection (Bosch-Camós et al., 2020) but necessitates further examination. Hence, it is imperative to explore the biological functions of diverse ASFV-encoded proteins to enhance comprehension of their involvement in immune evasion. Such insights are pivotal for advancing novel ASF vaccines.
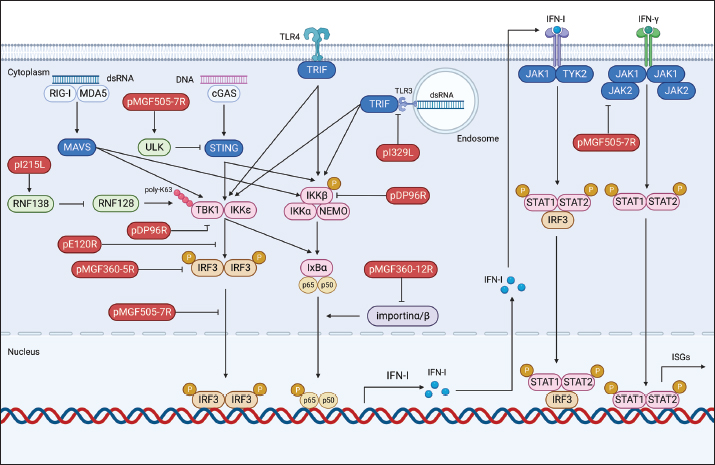
Fig. 1. Schematic representation of ASFV structure. Modulation of host antiviral innate immunityUpon viral invasion, the host immune system depends on early detection of pathogen-associated molecular patterns (PAMPs) through pattern recognition receptors (PRRs) to initiate antiviral innate immune and inflammatory reactions (Hu and Shu, 2020). PRR activation initiates a downstream signaling cascade, culminating in the synthesis of interferons (IFNs) and proinflammatory cytokines (Takeuchi and Akira, 2010). These antiviral effectors play a crucial role in coordinating efficient innate and adaptive immune responses to combat viral infections. Comprehending the intricate interaction between viral evasion tactics and host immune reactions is essential for innovating novel vaccines and antiviral treatments (He et al., 2022; Orosco, 2024a). The global pig industry has suffered notable harm from ASFV over an extended period, despite the intricate nature of the antiviral immune system. This implies that ASFV has developed an extensive immune evasion system in balance with its hosts. The expansive ASFV genome contains multiple proteins that aid in efficient infection, replication, and evasion of the immune response. Recently, specific ASFV proteins implicated in undermining the host’s antiviral innate immunity have been recognized (He et al., 2022; Orosco, 2024b). In this review, we will discuss the molecular mechanisms by which ASFV regulates the production of IFNs, inflammatory responses, and induction of apoptosis. Regulation of IFNsDespite differences in structure, receptor distribution, and tissue-specific activities, all types of IFNs can induce an antiviral state (van Boxel-Dezaire et al., 2006; Orosco, 2023b). Specifically, type I IFNs (IFN-I) serve as an initial defense against viral infections. They curtail viral replication and spread by triggering the production of multiple antiviral proteins that disrupt various stages of the viral life cycle. The initiation of the IFN-I response involves the recognition of viral PAMPs, such as Toll-like receptors (TLRs), RIG-I-like receptors, and cytoplasmic DNA sensors, by PRRs. This recognition activates innate immune signaling pathways, leading to the synthesis of proinflammatory cytokines and IFN-I (Fig. 2) (Zhu et al., 2019). In recent years, a small number of ASFV proteins, including multigene family 360 (MGF360), MGF530/505, pI329L, pDP96R, pE120R, pI215L, and pI267L have been identified to inhibit IFN-I responses, thus facilitating ASFV pathogenesis. MGF360 and MGF530/505The extensive genome of ASFV encodes a diverse array of proteins, including MGFs, implicated in both immune evasion and viral replication. Specifically, MGF100, MGF110, MGF300, MGF360, and MGF505/530 are recognized as pivotal contributors to ASFV pathogenesis (Dixon et al., 2019). MGF360 encompasses 11–15 members, while MGF530/505 comprises 9 or 10 members. Both MGF360 and MGF530/505 have been demonstrated to hinder IFN-I responses and facilitate viral replication by prolonging the survival of infected cells (O’Donnell et al., 2015; Reis et al., 2016).
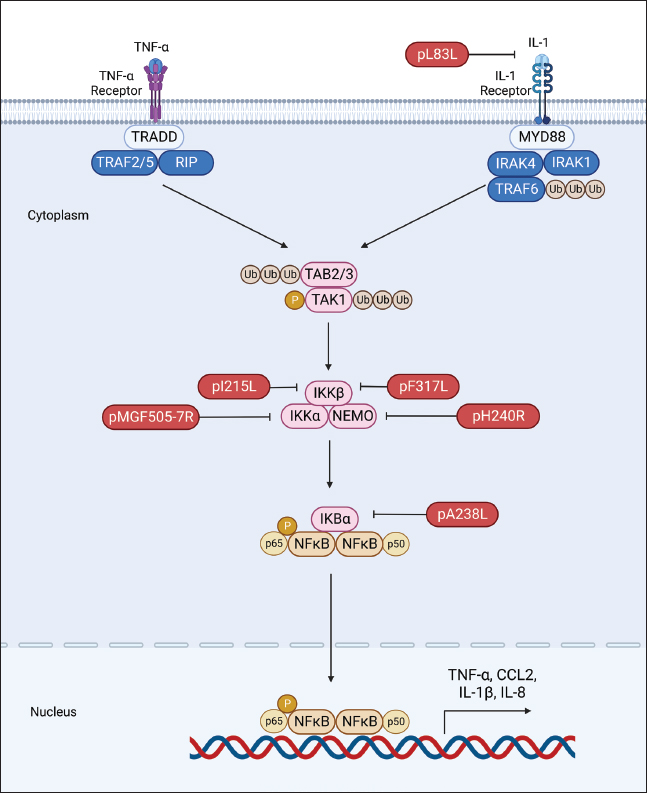
Fig. 2. Regulation of IFN production by ASFV proteins. The ASFV OURT88/3 variant, lacking specific genes from MGF360 or MGF505, has demonstrated heightened induction of IFN-I compared to its parent strain. This indicates the involvement of MGF360 and MGF505 members in regulating IFN expression (O’Donnell et al., 2015; Golding et al., 2016). Beyond IFN expression regulation, certain ASFV MGF family members directly influence IFN production. For instance, A276R, a MGF360 member, impedes IFN-β induction in an IRF3-dependent manner, with the underlying mechanism warranting further investigation (Correia et al., 2013). Similarly, MGF360-12L significantly hinders poly (I:C)-induced activation of the host immune response by disrupting the importin α-NF-κB interaction and inhibiting p65 nuclear translocation, resulting in diminished host antiviral response activation (Zhuo et al., 2021). Within the ASFV multigene family, the early-expressed MGF505-7R emerges as a crucial regulator of the cGAS-STING pathway. Research indicates that an ASFV strain lacking MGF505-7R prompts elevated levels of type I IFN and exhibits diminished virulence in piglets compared to the wild-type ASFV. MGF505-7R’s modulation of the cGAS-STING pathway involves its interaction with STING, resulting in reduced STING expression through heightened ULK1, an autophagy-related protein. In addition, MGF505-7R hinders the nuclear translocation of IRF3 and NF-κB by interacting with IRF3 and IKKα, ultimately leading to decreased production of IFN-β and proinflammatory cytokines (Yang et al., 2022a). MGF505-11R, an MGF family member, impedes cGAS-STING-driven immune responses by selectively interacting with STING. It promotes STING degradation through diverse pathways, including lysosomal, ubiquitin-proteasome, and autophagy routes (Yang et al., 2021a). In addition, MGF360-11L forms associations with TBK1 and IRF7 to hinder IFN-I production. Subsequently, TBK1 and IRF7 undergo degradation via the cysteine, ubiquitin-proteasome, and autophagy pathways (Yang et al., 2022b; Orosco and Espiritu, 2024). Furthermore, MGF360-14L inhibits type I IFN production by binding to the E3 ubiquitin ligase TRIM21. This interaction facilitates K63-linked ubiquitination of IRF3, destabilizing IRF3, and suppressing type I IFN production (Wang et al., 2022). In a recent investigation, it was disclosed that ASFV MGF360-9L functions as a suppressive regulator of IFN-β signaling. Precisely, MGF360-9L induces the decay of signal transducer and activator of transcription (STAT) 1 via apoptotic pathways and STAT2 through the ubiquitin-proteasome pathway. These revelations shed light on how ASFV eludes host antiviral responses and could potentially guide the creation of live attenuated ASFV vaccines (Zhang et al., 2022). pI329LASFV’s pI329L, a late protein encompassing 329 amino acids, exhibits a noteworthy degree of conservation. Intriguingly, aligning the pI329L sequence with TLR proteins unveils a 35% similarity between the intracellular domain of pI329L and the BOX1 and BOX2 regions within the human Toll-interleukin-1 receptor-like domain of TLR3 (de Oliveira et al., 2011). Prior research indicates that the ASFV protein pI329L functions as a viral antagonist to TLR3, hindering the activation of IFN-I by dsRNA or LPS. Structural analysis suggests pI329L targets TRIF, an adaptor protein in the TLR3 pathway. Overexpressing TRIF eliminates pI329L’s impact on IRF3 and NF-κB activation and IFN expression (Henriques et al., 2011; de Oliveira et al., 2011). In addition, the attenuated ASFV OURT88/3 strain, lacking I329L, induces heightened IFN-I expression in infected macrophages compared to its parent virus. However, it demonstrates reduced protection against challenges with the virulent ASFV OURT88/1 strain in pigs. Intriguingly, I329L deletion does not attenuate the ASFV Georgia 2007/1 strain (Reis et al., 2020). pDP96RpDP96R, an early expressed protein of ASFV, is encoded in the genome’s right variable region, with a molecular weight of around 10.7 kDa (Zsak et al., 1998). Recent research demonstrates that removing double genes 9GL (B119L) and UK (DP96R) in the Georgia 2007/1 mutant (ASFV-G-Δ9GL/ΔUK) significantly diminishes virus replication and pathogenicity in domestic pigs (O’Donnell et al., 2017; Orosco, 2023c). In addition, pDP96R has been identified as a negative regulator of IFN-I production via the cGAS-STING signaling pathway and influences NF-κB signaling by obstructing TBK1 and IKKβ activation (Wang et al., 2018). This underscores the crucial role of pDP96R in ASFV immune evasion. pE120RpE120R, a highly conserved capsid constituent of ASFV, links with the principal capsid protein p72 and encompasses about 120 amino acids (Liu et al., 2021). Recent investigations unveil pE120R’s pivotal role in inhibiting cGAS-STING-mediated immune response. It achieves this by binding to the C-terminal domain of IRF3, hindering IRF3 phosphorylation and impeding its recruitment to TBK1. This ultimately results in diminished IFN production (Liu et al., 2021; He et al., 2022). In addition, the inhibitory effect of pE120R relies on specific amino acids at positions 72 and 73. Deleting these residues annuls the IRF3-pE120R interaction, amplifies IFN-β and various ISG expressions, and suppresses ASFV replication in porcine alveolar macrophages (Liu et al., 2021). pI215LpI215L stands as the sole E2-ubiquitin conjugating enzyme of ASFV. Its conserved N-terminal region exhibits 31%–45% identity and 52%–66% similarity with other ubiquitin-conjugating enzymes (UBC) (Hingamp et al., 1992; Orosco, 2023d). Recent studies affirm pI215L as a potent inhibitor of IFN-I. Silencing pI215L hampers ASFV HLJ/18 strain replication while elevating IFN-β production. Further analysis reveals that pI215L curtails K63-linked polyubiquitination of TBK1, a pivotal post-translational modification for TBK1 activation, consequently impeding IFN-β production (Huang et al., 2021). pI267LThe ASFV protein I267L is identified as a key player in suppressing innate antiviral responses via the RNA Pol-III-RIG-I axis. I267L’s interaction with the E3 ubiquitin ligase Riplet disrupts Riplet’s binding with RIG-I, impeding K63-polyubiquitination and RIG-I activation. This results in decreased IFN-β levels and sustains wild-type ASFV replication. Conversely, I267L-deficient ASFV exhibits heightened IFN-β levels and diminished replication in primary macrophages and pigs, underscoring I267L’s pivotal role in ASFV virulence and pathogenesis (Ran et al., 2022). Regulation of inflammatory responsesASFV infection induces the release of diverse pro-inflammatory cytokines, including tumor necrosis factor (TNF)-α, interleukin (IL)-1β, IL-6, and IL-8. Notably, TNF-α plays a substantial role in ASF pathogenesis, exerting pro-inflammatory, pro-apoptotic, and pro-coagulant effects (Gómez-Villamandos et al., 2013). TNF-α and IL-1β constitute essential elements of the host’s innate antiviral immune response, critical for regulating the replication of various viruses, such as classical swine fever virus (Liniger et al., 2021) and Japanese encephalitis virus (Das et al., 2008). At present, there is broad consensus that ASFV possesses the capability to induce a robust inflammatory reaction in pigs, leading to severe inflammatory lesions and fatality. Contrary to expectations, numerous investigations have revealed that low-virulence ASFV strains can provoke markedly higher levels of inflammatory cytokines in comparison to their high-virulence counterparts. This hints at the complexity of the inflammatory response to ASFV, suggesting possible interference with host responses by viral proteins (Fig. 3) (Zheng et al., 2022). A growing body of research has pinpointed various ASFV proteins, such as pA238L, pMGF505-7R, pL83L, pF317L, pS273R, and pH240R, as agents inhibiting inflammatory responses. The mechanisms of action of these proteins will be described in detail below. pA238LpA238L, an early ASFV protein, is distributed in both the nucleus and cytoplasm of infected cells. Post-translational modifications yield two protein forms, with molecular weights of 28 and 32 kDa (Silk et al., 2007). Research involving cells overexpressing pA238L reveals its capability to impede the expression of genes regulated by the NF-κB transcription factor (Powell et al., 1996).
Fig. 3. Regulation of inflammatory responses by ASFV proteins. Evidence indicates that pA238L engages with the catalytic subunit of the serine-threonine protein phosphatase calcineurin (CaN) to regulate its phosphatase activity (Miskin et al., 2000). CaN plays a pivotal role in various pathways, including the activation of NFAT family transcription factors that stimulate cytokine production, such as IL-2, IL-4, and GM-CSF (Hogan, 2017). Through its interaction with CaN, pA238L obstructs NFAT activation by modulating its phosphatase activity (Miskin et al., 2000). In addition, pA238L has been demonstrated to repress COX-2 transcription, a potent lipid mediator of inflammation, in an NFAT-dependent manner (Granja et al., 2004). pMGF505-7RIL-1β, a versatile proinflammatory cytokine with diverse effects, is primarily synthesized by monocytes, macrophages, and lymphocytes, playing a pivotal role in regulating innate and adaptive immune responses (Aarreberg et al., 2018). Within ASFV-encoded proteins, pMGF505-7R emerges as a potent inhibitor of inducer-mediated IL-1β production. Research indicates that pMGF505-7R interacts with IKKα, impeding IκBα phosphorylation, and NF-κB nuclear translocation, and subsequently diminishing pro-IL-1β transcription. In addition, pMGF505-7R engages with NLRP3, hindering NLRP3 inflammasome assembly and thus suppressing mature IL-1β secretion. Importantly, the removal of MGF505-7R from the ASFV HLJ/18 strain is associated with attenuated ASFV virulence and heightened IL-1β production in vivo compared to its parental ASFV strain (Li et al., 2021b). pL83LThe L83L gene encodes pL83L, a protein highly conserved across ASFV strains and anticipated to undergo prenylation (Marakasova et al., 2017). Positioned near the left end of the ASFV genome, pL83L, an early-expressed protein, exhibited binding to IL-1β in a yeast two-hybrid assay. Although lacking in vivo evidence regarding its impact on IL-1β activity, there is speculation about pL83L potentially influencing this proinflammatory cytokine. Notably, studies involving the deletion of the L83L gene observed no reduction in virus replication in macrophages or a decrease in virulence in domestic pigs (Borca et al., 2018). Comprehensive investigations are required to unveil the precise functional role of pL83L. pF317LpF317L protein is a novel, poorly characterized protein of ASFV, consisting of 317 amino acids with an unknown function (Dixon et al., 2013). Recent studies have revealed that pF317L interacts with IKKβ and negatively regulates its activation. This leads to the stabilization of IκBα and inhibition of NF-κB activation, thereby reducing the expression of various proinflammatory cytokines and facilitating viral replication (Yang et al., 2021b; Mahedi et al., 2023). It has been demonstrated that deletion of F317L is lethal to ASFV, while ectopic expression of pF317L can enhance ASFV replication efficiency in porcine alveolar macrophages (iPAMs) (Yang et al., 2021b). However, whether the knockdown of F317L affects the inflammatory response has yet to be confirmed. pS273RpS273R, expressed in the late phases of ASFV infection, is situated within the cytoplasmic viral factories. This 273-amino acid protein, identified as a cysteine proteinase in the SUMO-1-specific protease family, has been studied extensively (Andrés et al., 2001). Recent investigations underscore the pivotal role of pS273R in modulating the host’s inflammatory reactions by impeding ASFV-induced pyroptosis (Zhao et al., 2022), a programmed cell death crucial for eliminating pathogens and instigating inflammatory responses to enhance protective host immunity (Kesavardhana et al., 2020). pS273R engages with GSDMD and cleaves it at G107–A108, generating a truncated N-terminal fragment, GSDMD-N1–107. Unlike the conventional GSDMD-N1–279 induced by caspase-1, GSDMD-N1–107 lacks the capacity to induce pyroptosis. In addition, pS273R further processes GSDMD-N1-279 into GSDMD-N1-107, which, in turn, hinders GSDMD-N1-279-triggered pyroptosis and facilitates ASFV replication (Zhao et al., 2022). pH240RASFV pH240R serves as a robust suppressor of IL-1β transcription, maturation, and secretion, along with restraining NLRP3 inflammasome activation in inflammatory responses (Huang et al., 2023). Its inhibitory actions extend beyond NF-κB signaling to encompass NLRP3 inflammasome activation. In this process, pH240R interacts with NF-κB essential modulator (NEMO), a component of the inhibitor of kappa B kinase (IKK) complex, leading to diminished phosphorylation of IκBα and p65. Moreover, pH240R binds to NLRP3, impeding NLRP3 inflammasome activation and subsequently reducing IL-1β production. Noteworthy is the observation that the removal of the H240R gene amplifies antiviral inflammatory responses, evident through heightened inflammatory cytokine expression both in vitro and in vivo, ultimately mitigating viral pathogenicity in pigs compared to its parental strain. These findings underscore the pivotal role of pH240R in ASFV virulence, indicating its potential as a subject for in-depth exploration into ASFV immune evasion mechanisms. Such investigations may pave the way for the development of attenuated live vaccines and therapeutic interventions for effective ASF prevention and control (Huang et al., 2023). Regulation of apoptosisApoptosis, a programmed cell death mechanism vital for maintaining normal tissue homeostasis in multicellular organisms, serves as a significant host defense against virus infections, curbing virus replication and clearing infected cells (D’Arcy, 2019; Simbulan et al., 2024). Given its pivotal role, it is unsurprising that numerous viruses employ diverse strategies to impede or prolong apoptosis, ensuring their survival and replication (D’Arcy, 2019; Fan, 2019). A distinctive characteristic of acute ASF is the initiation of apoptosis in infected macrophages in vivo, as evidenced by earlier investigations (Dixon et al., 2017). In its quest to enhance virus replication and elude the host immune system, ASFV has developed diverse tactics, such as the expression of specific proteins dedicated to impeding apoptosis (Zheng et al., 2022). This section will delve into the distinct functions of ASFV proteins in apoptosis regulation and elucidate their molecular mechanisms. pA224LThe ASFV pA224L protein, belonging to the inhibitor of apoptosis (IAP) family, features a BIR motif distinctive to this family. A224L expression effectively impedes apoptosis triggered by stimuli like TNF-α. Deletion of the A224L gene in ASFV-infected cells resulted in heightened caspase 3 activity, indicating A224L’s role in inhibiting caspase 3-mediated apoptosis (Nogal et al., 2001). While the precise mechanism of pA224L’s caspase 3 inhibition remains unclear, it likely involves direct binding to caspase 3. Furthermore, A224L activates NF-κB, a transcription factor regulating anti-apoptotic genes like IAP and Bcl-2 family members. NF-κB activation induces cFLIP expression, an inactive caspase 8 homolog that inhibits caspase 8 activity, effectively blocking apoptosis (Rodríguez et al., 2002). pA179LThe protein pA179L, a small and highly conserved molecule with a 21 kDa molecular mass and 179 amino acids, is consistently expressed throughout the virus infection cycle (Hernaez et al., 2013). As a member of the Bcl-2 family, pA179L possesses BH1, BH2, BH3, and BH4 domains but lacks corresponding transmembrane domains (Afonso et al., 1996). Its interaction profile includes BH3-only proteins like active truncated Bid, alongside proapoptotic factors such as Bax and Bak (Galindo et al., 2008; Banjara et al., 2017). Demonstrated to hinder the initiation of apoptosis, pA179L achieves this by directly engaging with proapoptotic factors (Galindo et al., 2008; Banjara et al., 2017). The remarkable conservation of pA179L, with 94%–99% amino acid identity among various ASFV strains, underscores its significance in ASFV replication and survival (Hernaez et al., 2013). pDP71LpDP71L assumes a pivotal role in ASFV virulence and maintains a substantial level of conservation across ASFV strains (Dixon et al., 2017). Through sequence analysis, it is evident that pDP71L shares homology with Myd116, GADD34 (a host DNA damage-inducible protein), and ICP34.5 (a neurovirulence-associated gene of HSV-1). Recent investigations underscore pDP71L’s direct interaction with eIF2α, where it recruits PP1α to dephosphorylate eIF2α. This interaction amplifies protein translation and concurrently suppresses the eIF2α-ATF4-CHOP apoptosis pathway. Intriguingly, even in the absence of the DP71L gene, ASFV-infected cells exhibit CHOP induction, implying the potential presence of compensatory mechanisms within ASFV for apoptosis regulation (Zhang et al., 2010). pEP153RThe pEP153R glycoprotein is a transmembrane protein featuring a C-type lectin-like region and a cell attachment (RGD) sequence crucial for its biological functions. Removal of the EP153R gene from the ASFV BA71V strain does not impact virulence in vitro but eliminates the hemadsorption phenomenon in infected cells (Galindo et al., 2000). The absence of pEP153R correlates with increased caspase-3 activity and heightened apoptosis compared to the parental BA71V strain. When expressed in Vero or COS cells, pEP153R partially inhibits apoptosis triggered by actinomycin, staurosporine, or ASFV, indicating its anti-apoptotic nature. In addition, pEP153R is demonstrated to dampen the transactivation activity of the cellular protein p53, a key player in the apoptotic cascade (Hurtado et al., 2004). p54The ASFV E183L gene encodes the p54 protein, a late-stage virus protein situated in the inner envelope of mature virions (Rodríguez et al., 2004). Through a shared amino acid sequence with Bim, a pro-apoptotic BH3-only protein, p54 interacts with DLC8, potentially influencing apoptosis by relocating Bim from microtubules to the mitochondrion. Overexpressing p54 triggers caspase-3 activation and cell apoptosis, with the 13-amino-acid domain essential for DLC8 binding also being crucial for caspase-3 activation. Despite being packaged in virions, p54’s apoptosis-inducing capability is hindered in the early infection stage by the presence of the apoptosis inhibitor protein pA179L. Nevertheless, in the later infection stage, heightened p54 expression may induce apoptosis, facilitating virus transmission (Hernáez et al., 2004). pE199LThe ASFV E199L gene encodes the pE199L protein, a late-stage viral protein found in the inner envelope of ASFV-infected cells (Sun et al., 1996). This protein, with a molecular mass of 19-20 kDa, is highly conserved among various ASFV strains. pE199L, characterized as a type I transmembrane protein, comprises a cysteine-rich extended segment at the N-terminus, a putative transmembrane domain, and a brief C-terminal tail (Matamoros et al., 2020). Prior investigations propose that pE199L triggers complete autophagy in Vero and HEK293T cells by suppressing pyrroline-5-carboxylate reductase 2 (PYCR2) (Chen et al., 2021). Recent findings indicate that pE199L also functions as an apoptosis initiator. Specifically, pE199L competes with Bak for Bcl-XL, resulting in the activation of the pro-apoptotic factor Bak. Furthermore, pE199L facilitates the activation of another pro-apoptotic factor, Bax, and its relocation to the mitochondria, thus instigating mitochondrial-dependent apoptosis (Li et al., 2021a). ConclusionThis comprehensive review has delved into the intricate interactions between ASFV proteins and the host’s antiviral innate immunity. Our exploration of key viral proteins has illuminated the multifaceted strategies employed by ASFV to subvert host defenses. The evidence presented underscores the pivotal roles of these viral proteins in orchestrating a sophisticated evasion of host innate immune responses. Notably, the diverse mechanisms involved, such as interference with IFN signaling, modulation of inflammatory pathways, and manipulation of apoptosis, highlight the adaptability of ASFV in overcoming the host’s defenses. Despite the strides made in understanding ASFV-host interactions, notable research gaps persist. Deeper insights into the precise molecular mechanisms employed by specific viral proteins, especially in modulating cellular apoptosis and inflammasome activation, merit further investigation. Unraveling the intricate crosstalk between viral proteins and host factors remains a challenging but essential avenue for future exploration. Looking ahead, the identified gaps pave the way for future studies aimed at elucidating novel targets for antiviral interventions. The development of vaccines against ASFV hinges on a comprehensive understanding of viral immune evasion strategies. Exploring innovative vaccine platforms, considering the identified protein targets, holds promise for enhancing vaccine efficacy and broadening our arsenal against this devastating virus. AcknowledgmentsThe author would like to thank the DOST S&T Fellows Program, the Philippine Council for Agriculture, Aquatic and Natural Resources Research and Development (DOST-PCAARRD) for funding this research project, and the Industrial Technology Development Institute (DOST-ITDI) for hosting this research project. Conflict of interestThe author declares that there is no conflict of interest. FundingDOST S&T Fellows Program, the Philippine Council for Agriculture, Aquatic and Natural Resources Research and Development (DOST-PCAARRD). Data availabilityAll data are provided in the manuscript. ReferencesAarreberg, L.D., Wilkins, C., Ramos, H.J., Green, R., Davis, M.A., Chow, K. and Gale, M. 2018. Interleukin-1β signaling in dendritic cells induces antiviral interferon responses. mBio 9, e00342–18. Afonso, C.L., Neilan, J.G., Kutish, G.F. and Rock, D.L. 1996. An African swine fever virus Bc1-2 homolog, 5-HL, suppresses apoptotic cell death. J. Virol. 70, 4858–4863. Almazán, F., Rodríguez, J.M., Angulo, A., Viñuela, E. and Rodriguez, J.F. 1993. Transcriptional mapping of a late gene coding for the p12 attachment protein of African swine fever virus. J. Virol. 67, 553–556. Andrés, G., Alejo, A., Simón-Mateo, C. and Salas, M.L. 2001. African swine fever virus protease, a new viral member of the SUMO-1-specific protease family. J. Biol. Chem. 276, 780–787. Andrés, G., Charro, D., Matamoros, T., Dillard, R.S. and Abrescia, N.G.A. 2020. The cryo-EM structure of African swine fever virus unravels a unique architecture comprising two icosahedral protein capsids and two lipoprotein membranes. J. Biol. Chem. 295, 1–12. Banjara, S., Caria, S., Dixon, L.K., Hinds, M.G. and Kvansakul, M. 2017. Structural insight into African swine fever virus A179L-mediated inhibition of apoptosis. J. Virol. 91, e02228–16. Blome, S., Franzke, K. and Beer, M. 2020. African swine fever – A review of current knowledge. Virus Res. 287, 198099. Borca, M.V., O’Donnell, V., Holinka, L.G., Ramírez-Medina, E., Clark, B.A., Vuono, E.A., Berggren, K., Alfano, M., Carey, L.B., Richt, J.A., Risatti, G.R. and Gladue, D.P. 2018. The L83L ORF of African swine fever virus strain Georgia encodes for a non-essential gene that interacts with the host protein IL-1β. Virus Res. 249, 116–123. Bosch-Camós, L., López, E. and Rodriguez, F. 2020. African swine fever vaccines: a promising work still in progress. Porc. Health Manag. 6, 17. https://doi.org/10.1186/s40813-020-00154-2. van Boxel-Dezaire, A.H.H., Rani, M.R.S. and Stark, G.R. 2006. Complex modulation of cell type-specific signaling in response to type I interferons. Immunity 25, 361–372. Chapman, D.A.G., Tcherepanov, V., Upton, C. and Dixon, L.K. 2008. Comparison of the genome sequences of non-pathogenic and pathogenic African swine fever virus isolates. J. Gen. Virol. 89, 397–408. Chen, S., Zhang, X., Nie, Y., Li, H., Chen, W., Lin, W., Chen, F. and Xie, Q. 2021. African swine fever virus protein E199L promotes cell autophagy through the interaction of PYCR2. Virol. Sin. 36, 196–206. Correia, S., Ventura, S. and Parkhouse, R. M. 2013. Identification and utility of innate immune system evasion mechanisms of ASFV. Virus Res. 173, 87–100. D’Arcy, M.S. 2019. Cell death: a review of the major forms of apoptosis, necrosis and autophagy. Cell Biol. Int. 43, 582–592. Das, S., Mishra, M.K., Ghosh, J. and Basu, A. 2008. Japanese encephalitis virus infection induces IL-18 and IL-1beta in microglia and astrocytes: correlation with in vitro cytokine responsiveness of glial cells and subsequent neuronal death. J. Neuroimmunol. 195, 60–72. Dixon, L.K., Chapman, D.A.G., Netherton, C.L. and Upton, C. 2013. African swine fever virus replication and genomics. Virus Res. 173, 3–14. Dixon, L.K., Sánchez-Cordón, P.J., Galindo, I. and Alonso, C. 2017. Investigations of pro- and anti-apoptotic factors affecting African swine fever virus replication and pathogenesis. Viruses 9, 241. Dixon, L.K., Sun, H. and Roberts, H. 2019. African swine fever. Antiviral Res. 165, 34–41. Fan, L. 2019. Signaling pathways involved in regulating apoptosis induction in host cells upon PRRSV infection. Virus Genes 55, 433–439. Galindo, I., Almazán, F., Bustos, M.J., Viñuela, E. and Carrascosa, A.L. 2000. African swine fever virus EP153R open reading frame encodes a glycoprotein involved in the hemadsorption of infected cells. Virology 266, 340–351. Galindo, I., Hernaez, B., Díaz-Gil, G., Escribano, J.M. and Alonso, C. 2008. A179L, a viral Bcl-2 homologue, targets the core Bcl-2 apoptotic machinery and its upstream BH3 activators with selective binding restrictions for Bid and Noxa. Virology 375, 561–572. Golding, J.P., Goatley, L., Goodbourn, S., Dixon, L.K., Taylor, G. and Netherton, C.L. 2016. Sensitivity of African swine fever virus to type I interferon is linked to genes within multigene families 360 and 505. Virology 493, 154–161. Gómez-Villamandos, J.C., Bautista, M.J., Sánchez-Cordón, P.J. and Carrasco, L. 2013. Pathology of African swine fever: the role of monocyte-macrophage. Virus Res. 173, 140–149. Granja, A.G., Nogal, M.L., Hurtado, C., Vila, V., Carrascosa, A.L., Salas, M.L., Fresno, M. and Revilla, Y. 2004. The viral protein A238L inhibits cyclooxygenase-2 expression through a nuclear factor of activated T cell-dependent transactivation pathway. J. Biol. Chem. 279, 53736–53746. He, W.R., Yuan, J., Ma, Y.-., Zhao, C.Y., Yang, Z.Y., Zhang, Y., Han, S., Wan, B. and Zhang, G.P. 2022. Modulation of host Aativiral innate immunity by African swine fever virus: a review. Animals 12, 2935. Henriques, E.S., Brito, R.M.M., Soares, H., Ventura, S., de Oliveira, V.L. and Parkhouse, R.M.E. 2011. Modeling of the toll-like receptor 3 and a putative toll-like receptor 3 antagonist encoded by the African swine fever virus. Protein Sci. 20, 247–255. Hernaez, B., Cabezas, M., Muñoz-Moreno, R., Galindo, I., Cuesta-Geijo, M.A. and Alonso, C. 2013. A179L, a new viral Bcl2 homolog targeting Beclin 1 autophagy related protein. Curr. Mol. Med. 13, 305–316. Hernáez, B., Dı́az-Gil, G., Garcı́a-Gallo, M., Ignacio Quetglas, J., Rodrı́guez-Crespo, I., Dixon, L., Escribano, J.M. and Alonso, C. 2004. The African swine fever virus dynein-binding protein p54 induces infected cell apoptosis. FEBS Lett. 569, 224–228. Hingamp, P.M., Arnold, J.E., Mayer, R.J. and Dixon, L.K. 1992. A ubiquitin conjugating enzyme encoded by African swine fever virus. EMBO J. 11, 361–366. Hogan, P.G. 2017. Calcium–NFAT transcriptional signalling in T cell activation and T cell exhaustion. Cell Calcium 63, 66–69. Hu, M.M. and Shu, H.B. 2020. Innate immune response to cytoplasmic DNA: mechanisms and diseases. Annu. Rev. Immunol. 38, 79–98. Huang, L., Liu, H., Ye, G., Liu, X., Chen, W., Wang, Z., Zhao, D., Zhang, Z., Feng, C., Hu, L., Yu, H., Zhou, S., Zhang, X., He, X., Zheng, J., Bu, Z., Li, J. and Weng, C. 2023. Deletion of African swine fever virus (ASFV) H240R gene attenuates the virulence of ASFV by enhancing NLRP3-mediated inflammatory responses. J. Virol. 97, e0122722. Huang, L., Xu, W., Liu, H., Xue, M., Liu, X., Zhang, K., Hu, L., Li, J., Liu, X., Xiang, Z., Zheng, J., Li, C., Chen, W., Bu, Z., Xiong, T. and Weng, C. 2021. African swine fever virus pI215L negatively regulates cGAS-STING signaling pathway through recruiting RNF138 to inhibit K63-linked Ubiquitination of TBK1. J. Immunol. 207, 2754–2769. Hurtado, C., Granja, A.G., Bustos, M.J., Nogal, M.L., González de Buitrago, G., de Yébenes, V.G., Salas, M.L., Revilla, Y. and Carrascosa, A.L. 2004. The C-type lectin homologue gene (EP153R) of African swine fever virus inhibits apoptosis both in virus infection and in heterologous expression. Virology 326, 160–170. Jancovich, J.K., Chapman, D., Hansen, D.T., Robida, M.D., Loskutov, A., Craciunescu, F., Borovkov, A., Kibler, K., Goatley, L., King, K., Netherton, C.L., Taylor, G., Jacobs, B., Sykes, K. and Dixon, L.K. 2018. Immunization of pigs by DNA prime and recombinant vaccinia virus boost to identify and rank African swine fever virus immunogenic and protective proteins. J. Virol. 92, e02219–17. Kesavardhana, S., Malireddi, R.K.S. and Kanneganti, T.D. 2020. Caspases in cell death, inflammation, and pyroptosis. Annu. Rev. Immunol. 38, 567–595. Li, J., Jin, P., Zhu, J., Zou, H., Xu, X., Tang, M., Zhou, M., Gan, Y., He, J., Ling, Y. and Su, Y. 2021a. Multi-scale GCN-assisted two-stage network for joint segmentation of retinal layers and discs in peripapillary OCT images. Biomed. Opt. Express 12, 2204–2220. Li, J., Song, J., Kang, L., Huang, L., Zhou, S., Hu, L., Zheng, J., Li, C., Zhang, X., He, X., Zhao, D., Bu, Z. and Weng, C. 2021b. pMGF505-7R determines pathogenicity of African swine fever virus infection by inhibiting IL-1β and type I IFN production. PLOS Pathog. 17, e1009733. Liniger, M., Gerber, M., Renzullo, S., García-Nicolás, O. and Ruggli, N. 2021. TNF-mediated inhibition of classical swine fever virus replication is IRF1-, NF-κB- and JAK/STAT signaling-dependent. Viruses 13, 2017. Liu, H., Zhu, Z., Feng, T., Ma, Z., Xue, Q., Wu, P., Li, P., Li, S., Yang, F., Cao, W., Xue, Z., Chen, H., Liu, X. and Zheng, H. 2021. African swine fever virus E120R protein inhibits interferon beta production by interacting with IRF3 To block its activation. J. Virol. 95, e0082421. Lubisi, B.A., Bastos, A.D.S., Dwarka, R.M. and Vosloo, W. 2007. Intra-genotypic resolution of African swine fever viruses from an East African domestic pig cycle: a combined p72-CVR approach. Virus Genes 35, 729–735. Mahedi, M.R.A., Rawat, A., Rabbi, F., Babu, K.S., Tasayco, E.S., Areche, F.O., Pacovilca-Alejo, O.V., Flores, D.D.C., Aguilar, S.V., Orosco, F.L., Syrmos, N., Mudhafar, M., Afrin, S. and Rahman, M.M. 2023. Understanding the global transmission and demographic distribution of Nipah virus (NiV). Res. J. Pharm. Technol. 16, 3588–3594. Marakasova, E.S., Eisenhaber, B., Maurer-Stroh, S., Eisenhaber, F. and Baranova, A. 2017. Prenylation of viral proteins by enzymes of the host: virus-driven rationale for therapy with statins and FT/GGT1 inhibitors. BioEssays 39, 1700014. Matamoros, T., Alejo, A., Rodríguez, J.M., Hernáez, B., Guerra, M., Fraile-Ramos, A. and Andrés, G. 2020. African swine fever virus protein pE199L mediates virus entry by enabling membrane fusion and core penetration. mBio 11, e00789–20. Miskin, J.E., Abrams, C.C. and Dixon, L.K. 2000. African swine fever virus protein A238L interacts with the cellular phosphatase calcineurin via a binding domain similar to that of NFAT. J. Virol. 74, 9412–9420. Nogal, M.L., González de Buitrago, G., Rodríguez, C., Cubelos, B., Carrascosa, A.L., Salas, M. L. and Revilla, Y. 2001. African swine fever virus IAP homologue inhibits caspase activation and promotes cell survival in mammalian cells. J. Virol. 75, 2535–2543. O’Donnell, V., Holinka, L.G., Gladue, D.P., Sanford, B., Krug, P.W., Lu, X., Arzt, J., Reese, B., Carrillo, C., Risatti, G.R. and Borca, M.V. 2015. African swine fever virus georgia isolate harboring deletions of MGF360 and MGF505 genes is attenuated in swine and confers protection against challenge with virulent parental virus. J. Virol. 89, 6048–6056. O’Donnell, V., Risatti, G.R., Holinka, L.G., Krug, P.W., Carlson, J., Velazquez-Salinas, L., Azzinaro, P.A., Gladue, D.P. and Borca, M.V. 2017. Simultaneous deletion of the 9GL and UK genes from the African swine fever virus georgia 2007 isolate offers increased safety and protection against homologous challenge. J. Virol. 91, e01760–16. de Oliveira, V.L., Almeida, S.C.P., Soares, H.R., Crespo, A., Marshall-Clarke, S. and Parkhouse, R.M.E. 2011. A novel TLR3 inhibitor encoded by African swine fever virus (ASFV). Arch. Virol. 156, 597–609. Orosco, F. L. 2023a. Current progress in diagnostics, therapeutics, and vaccines for African swine fever virus. Vet. Integr. Sci. 21, 751–781. Orosco, F. 2023b. Advancing the frontiers: revolutionary control and prevention paradigms against Nipah virus. Open Vet. J. 13, 1056–1056. Orosco, F. L. 2023c. Breaking the chains: advancements in antiviral strategies to combat Nipah virus infections. Int. J. One Health 9, 122–133. Orosco, F. L. 2023d. Host immune responses against African swine fever virus: insights and challenges for vaccine development. Open Vet. J. 13, 1517–1535. Orosco, F. 2024a. Recent advances in peptide-based nanovaccines for re-emerging and emerging infectious diseases. J. Adv. Biotechnol. Exp. Ther. 7, 106. Orosco, F. L. 2024b. Immune evasion mechanisms of porcine epidemic diarrhea virus: a comprehensive review. Vet. Integr. Sci. 22, 171–192. Orosco, F.L. and Espiritu, L.M. 2024. Navigating the landscape of adjuvants for subunit vaccines: recent advances and future perspectives. Int. J. Appl. Pharm. 18–32. Powell, P.P., Dixon, L.K. and Parkhouse, R.M. 1996. An IkappaB homolog encoded by African swine fever virus provides a novel mechanism for downregulation of proinflammatory cytokine responses in host macrophages. J. Virol. 70, 8527–8533. Ran, Y., Li, D., Xiong, M.G., Liu, H.N., Feng, T., Shi, Z.W., Li, Y.H., Wu, H.N., Wang, S.Y., Zheng, H.X. and Wang, Y.Y. 2022. African swine fever virus I267L acts as an important virulence factor by inhibiting RNA polymerase III-RIG-I-mediated innate immunity. PLOS Pathog. 18, e1010270. Reis, A.L., Abrams, C.C., Goatley, L.C., Netherton, C., Chapman, D.G., Sanchez-Cordon, P. and Dixon, L.K. 2016. Deletion of African swine fever virus interferon inhibitors from the genome of a virulent isolate reduces virulence in domestic pigs and induces a protective response. Vaccine 34, 4698–4705. Reis, A.L., Goatley, L.C., Jabbar, T., Lopez, E., Rathakrishnan, A. and Dixon, L.K. 2020. Deletion of the gene for the type I interferon inhibitor I329L from the attenuated African swine fever virus OURT88/3 strain reduces protection induced in pigs. Vaccines 8, 262. Rodríguez, J.M., García-Escudero, R., Salas, M.L. and Andrés, G. 2004. African swine fever virus structural protein p54 is essential for the recruitment of envelope precursors to assembly sites. J. Virol. 78, 4299–1313. Rodríguez, C.I., Nogal, M.L., Carrascosa, A.L., Salas, M.L., Fresno, M. and Revilla, Y. 2002. African swine fever virus IAP-like protein induces the activation of nuclear factor kappa B. J. Virol. 76, 3936–3942. Rodríguez, J.M., Salas, M.L. and Viñuela, E. 1996. Intermediate class of mRNAs in African swine fever virus. J. Virol. 70, 8584–8589. Salas, M.L. and Andrés, G. 2013. African swine fever virus morphogenesis. Virus Res. 173, 29–41. Silk, R.N., Bowick, G.C., Abrams, C.C. and Dixon, L.K. 2007. African swine fever virus A238L inhibitor of NF-κB and of calcineurin phosphatase is imported actively into the nucleus and exported by a CRM1-mediated pathway. J. Gen. Virol. 88, 411–419. Simbulan, A.M., Banico, E.C., Sira, E.M.J.S., Odchimar, N.M.O. and Orosco, F.L. 2024. Immunoinformatics-guided approach for designing a pan-proteome multi-epitope subunit vaccine against African swine fever virus. Sci. Rep. 14, 1354. Sun, H., Jenson, J., Dixon, L.K. and Parkhouse, R.M.E. 1996. Characterization of the African swine fever virion protein j18L. J. Gen. Virol. 77, 941–946. Takeuchi, O. and Akira, S. 2010. Pattern recognition receptors and inflammation. Cell 140, 805–820. Wang, Y., Cui, S., Xin, T., Wang, X., Yu, H., Chen, S., Jiang, Y., Gao, X., Jiang, Y., Guo, X., Jia, H. and Zhu, H. 2022. African swine fever virus MGF360-14L negatively regulates type I interferon signaling by targeting IRF3. Front. Cell. Infect. Microbiol. 11, 818969. Wang, T., Sun, Y., Huang, S. and Qiu, H.J. 2020. Multifaceted immune responses to African swine fever virus: implications for vaccine development. Vet. Microbiol. 249, 108832. Wang, X., Wu, J., Wu, Y., Chen, H., Zhang, S., Li, J., Xin, T., Jia, H., Hou, S., Jiang, Y., Zhu, H. and Guo, X. 2018. Inhibition of cGAS-STING-TBK1 signaling pathway by DP96R of ASFV China 2018/1. Biochem. Biophys. Res. Commun. 506, 437–443. Wang, N., Zhao, D., Wang, J., Zhang, Y., Wang, M., Gao, Y., Li, F., Wang, J., Bu, Z., Rao, Z. and Wang, X. 2019. Architecture of African swine fever virus and implications for viral assembly. Science 366, 640–644. Yang, K., Huang, Q., Wang, R., Zeng, Y., Cheng, M., Xue, Y., Shi, C., Ye, L., Yang, W., Jiang, Y., Wang, J., Huang, H., Cao, X., Yang, G. and Wang, C. 2021a. African swine fever virus MGF505-11R inhibits type I interferon production by negatively regulating the cGAS-STING-mediated signaling pathway. Vet. Microbiol. 263, 109265. Yang, J., Li, S., Feng, T., Zhang, X., Yang, F., Cao, W., Chen, H., Liu, H., Zhang, K., Zhu, Z. and Zheng, H. 2021b. African swine fever virus F317L protein inhibits NF-κB activation to evade host immune response and promote viral replication. mSphere 6, e0065821. Yang, K., Xue, Y., Niu, T., Li, X., Cheng, M., Bao, M., Zou, B., Shi, C., Wang, J., Yang, W., Wang, N., Jiang, Y., Yang, G., Zeng, Y., Cao, X. and Wang, C. 2022a. African swine fever virus MGF505-7R protein interacted with IRF7and TBK1 to inhibit type I interferon production. Virus Res. 322, 198931. Yang, K., Xue, Y., Niu, H., Shi, C., Cheng, M., Wang, J., Zou, B., Wang, J., Niu, T., Bao, M., Yang, W., Zhao, D., Jiang, Y., Yang, G., Zeng, Y., Cao, X. and Wang, C. 2022b. African swine fever virus MGF360-11L negatively regulates cGAS-STING-mediated inhibition of type I interferon production. Vet. Res. 53, 7. Zhang, F., Moon, A., Childs, K., Goodbourn, S. and Dixon, L. K. 2010. The African swine fever virus DP71L protein recruits the protein phosphatase 1 catalytic subunit to dephosphorylate eIF2alpha and inhibits CHOP induction but is dispensable for these activities during virus infection. J. Virol. 84, 10681–10689. Zhang, K., Yang, B., Shen, C., Zhang, T., Hao, Y., Zhang, D., Liu, H., Shi, X., Li, G., Yang, J., Li, D., Zhu, Z., Tian, H., Yang, F., Ru, Y., Cao, W. J., Guo, J., He, J., Zheng, H. and Liu, X. 2022. MGF360-9L Is a Major virulence factor associated with the african swine fever virus by antagonizing the JAK/STAT signaling pathway. mBio 13, e0233021. Zhao, G., Li, T., Liu, X., Zhang, T., Zhang, Z., Kang, L., Song, J., Zhou, S., Chen, X., Wang, X., Li, J., Huang, L., Li, C., Bu, Z., Zheng, J. and Weng, C. 2022. African swine fever virus cysteine protease pS273R inhibits pyroptosis by noncanonically cleaving gasdermin D. J. Biol. Chem. 298. Zhao, D., Liu, R., Zhang, X., Li, F., Wang, J., Zhang, J., Liu, X., Wang, L., Zhang, J., Wu, X., Guan, Y., Chen, W., Wang, X., He, X. and Bu, Z. 2019. Replication and virulence in pigs of the first African swine fever virus isolated in China. Emerg. Microbes Infect. 8, 438–447. Zheng, X., Nie, S. and Feng, W.H. 2022. Regulation of antiviral immune response by African swine fever virus (ASFV). Virol. Sin. 37, 157–167. Zhu, Y., Deng, J., Nan, M.L., Zhang, J., Okekunle, A., Li, J.Y., Yu, X.Q. and Wang, P.H. 2019. The interplay between pattern recognition receptors and autophagy in inflammation. In autophagy regulation of innate immunity. Cui, J., ed. Advances in experimental medicine and biology. Singapore: Springer, pp: 79–108. Zhuo, Y., Guo, Z., Ba, T., Zhang, C., He, L., Zeng, C. and Dai, H. 2021. African swine fever virus MGF360-12L inhibits type I interferon production by blocking the interaction of importin α and NF-κB signaling pathway. Virol. Sin. 36, 176–186. Zsak, L., Caler, E., Lu, Z., Kutish, G. F., Neilan, J. G. and Rock, D. L. 1998. A nonessential African swine fever virus gene UK Is a significant virulence determinant in domestic swine. J. Virol. 72, 1028–1035. | ||
| How to Cite this Article |
| Pubmed Style Fredmoore L. Orosco. African swine fever virus proteins against host antiviral innate immunity and their implications for vaccine development. Open Vet. J.. 2024; 14(4): 941-951. doi:10.5455/OVJ.2024.v14.i4.1 Web Style Fredmoore L. Orosco. African swine fever virus proteins against host antiviral innate immunity and their implications for vaccine development. https://www.openveterinaryjournal.com/?mno=179757 [Access: January 08, 2026]. doi:10.5455/OVJ.2024.v14.i4.1 AMA (American Medical Association) Style Fredmoore L. Orosco. African swine fever virus proteins against host antiviral innate immunity and their implications for vaccine development. Open Vet. J.. 2024; 14(4): 941-951. doi:10.5455/OVJ.2024.v14.i4.1 Vancouver/ICMJE Style Fredmoore L. Orosco. African swine fever virus proteins against host antiviral innate immunity and their implications for vaccine development. Open Vet. J.. (2024), [cited January 08, 2026]; 14(4): 941-951. doi:10.5455/OVJ.2024.v14.i4.1 Harvard Style Fredmoore L. Orosco (2024) African swine fever virus proteins against host antiviral innate immunity and their implications for vaccine development. Open Vet. J., 14 (4), 941-951. doi:10.5455/OVJ.2024.v14.i4.1 Turabian Style Fredmoore L. Orosco. 2024. African swine fever virus proteins against host antiviral innate immunity and their implications for vaccine development. Open Veterinary Journal, 14 (4), 941-951. doi:10.5455/OVJ.2024.v14.i4.1 Chicago Style Fredmoore L. Orosco. "African swine fever virus proteins against host antiviral innate immunity and their implications for vaccine development." Open Veterinary Journal 14 (2024), 941-951. doi:10.5455/OVJ.2024.v14.i4.1 MLA (The Modern Language Association) Style Fredmoore L. Orosco. "African swine fever virus proteins against host antiviral innate immunity and their implications for vaccine development." Open Veterinary Journal 14.4 (2024), 941-951. Print. doi:10.5455/OVJ.2024.v14.i4.1 APA (American Psychological Association) Style Fredmoore L. Orosco (2024) African swine fever virus proteins against host antiviral innate immunity and their implications for vaccine development. Open Veterinary Journal, 14 (4), 941-951. doi:10.5455/OVJ.2024.v14.i4.1 |