| Research Article | ||
Open Vet. J.. 2024; 14(12): 3213-3218 Open Veterinary Journal, (2024), Vol. 14(12): 3213-3218 Research Article Isolation and PCR-based detection of parvovirus in dogshuha Ismaeel Abdul Allmjeed, Rawaa Saladdin Jumaa* and Osamah Mohammed IbrahimDepartment of Microbiology, College of Veterinary Medicine, University of Baghdad, Baghdad, Iraq *Corresponding Author: Rawaa Saladdin Jumaa. Department of Microbiology, College of Veterinary Medicine, University of Baghdad, Baghdad, Iraq. Email: rawaa.saladdin [at] covm.uobaghdad.edu.iq Submitted: 22/07/2024 Accepted: 15/11/2024 Published: 31/12/2024 © 2024 Open Veterinary Journal
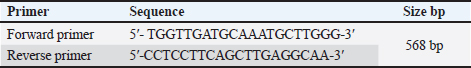
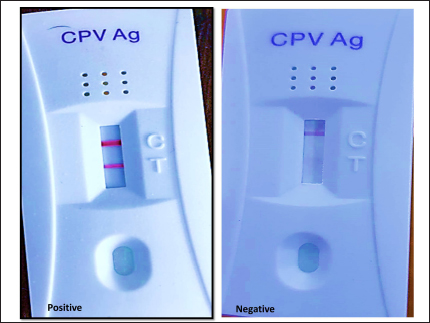
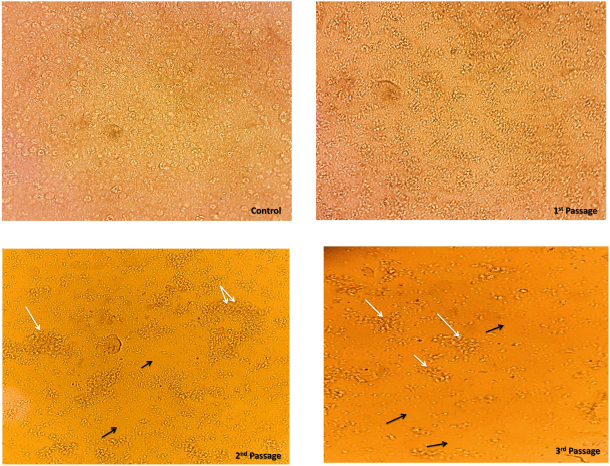
AbstractBackground: The Canine parvovirus type 2 is an acute viral disease in puppies and is considered a worldwide disease. Aim: This study was performed to identify the Canine parvovirus (CPV) throughout 2022 and 2023 in Iraq. The novelty of this study was to isolate this virus in two types of cell cultures (diploid and fibroblast cell cultures) and confirm the isolated virus via PCR technique. Methods: Sixty fecal samples were taken from suspected infected dogs in the Baghdad Governorates who were clinically sick and thought to have CPV. The first performed qualitative antigen detection on the samples with a rapid test, then the positive samples were propagated on the feline kidney (FK) and embryonated chicken fibroblast cells and finally identified the isolated virus on six randomly selected samples by amplifying the VP2 gene with a conventional polymerase chain reaction. Results: Rapid tests found 50% of fecal swabs as positive results. Additionally, the results of the cytopathic effect (CPE) at the second and third passages on chicken embryo and FK cell cultures revealed rounder and more detached cells. Furthermore, the isolated virus was confirmed through amplified the Vp2 gene of the virus by polymerase chain reaction. Conclusion: The focus of this study was to isolate of parvovirus that causes diarrhea in puppies, which is quite frequent in Iraq. The virus was isolated using both primary and secondary cell cultures. The solitary virus causes cells to become more differentiated and rounded due to CPEs. The virus was confirmed to be real using a polymerase chain reaction experiment. This study also suggests doing more epidemiological research and sequencing analysis to determine the prevalence of CPV serotypes in Iraq and the efficacy of trade in and locally made vaccinations in preventing against viral infection. Keywords: Parvovirus, Isolation, PCR, Pups, Iraq. IntroductionCanine parvovirus type 2 (CPV2) is a virus that affects the dog population globally, mainly in puppies. This disease is considered acute hemorrhagic enteritis and its clinical signs include diarrhea, vomiting, myocarditis, and immunosuppression. The CPV2 virus is encircled in an icosahedral capsid structure and has a very small diameter of about 25 nm (Ahmed et al., 2018). CPV2 is categorized under the taxonomic classification of the family Parvoviridae, more specifically the subfamily Parvovirinae, genus protoparvovirus, and carnivore protoparvovirus 1 species (Cotmore et al., 2019). The genetic material of this virus is a single stranded, linear DNA and has a length of about 5,200 nucleotides. The genome encodes for four different proteins. Two structural proteins, (VP1 and VP2), VP2 is the main protective antigen and forms the viral capsid. Two non-structural proteins (NSs) are (NS1 and NS2), the NS1 which is important to viral replication, and NS2, which is essential for the viral capsid to assemble (Nur-Farahiyah et al., 2021). This virus was first recognized in the 1970s due to spreading among dogs. It was also called CPV2 since a similar one, CPV1, is seen in dogs (Chen et al., 2021). Moreover, the genetic variations in the virus during the 1980s, the original CPV2 virus gave two distinct antigenic subtypes (CPV2a and CPV2b). Additionally, the first of a new antigenic subtype, CPV2c, in 2000 led to the phase out of the prototype CPV2 in Italy (Clark et al., 2018). As well as, several viruses (RNA and DNA viruses) were diagnosed using PCR (Ali et al., 2015; Allawe, 2016; Mhemid, 2016; Jumaa et al., 2019, 2020, and 2021; Ahmed, 2020; Taha and Alhankawe, 2023; Jbr and Jumaa, 2024). This study aims to isolate the circulating Canine parvovirus (CPV) in Baghdad (Iraq) by isolation of this virus on several types of cell cultures and confirm the isolated virus using the PCR method. Materials and MethodsSamples collection and its preparationSixty fecal swabs were collected from suspected infected dogs in Baghdad governorates through 2022. Between the ages of (2–6) months, these dogs showed moderate to severe gastrointestinal symptoms, including (vomiting, lethargy, in appetence, diarrhea, and dehydration). These samples were prepared according to Soliman et al. (2018). Each fecal swab was put in a container with labels that had phosphate-buffered saline (PBS) and a (10%) antibiotic solution. After that, after two cycles of freezing and thawing, the samples were centrifuged for (10 minutes) at 2,000 relative centrifugal force. Finally, the supernatant was separated and kept at (−80°C) until it was needed for viral isolation and molecular identification. Rapid test for viral detectionThis kit is a commercial Kit (Rapid test kit, BIONOTE) and a diagnostic tool that can identify the presence of CPV antigen in fecal swabs. Viral isolation on feline kidney (FK) and embryonated chicken fibroblast (ECF) cellsFirst, these cells were grown in minimal essential medium (MEM) supplemented with fetal calf serum (10%) at 37°C in an environment of CO2 (5%) until they reached full coverage as monolayers. Afterward, the growth medium was withdrawn from the monolayers in a 25 cm2 cell culture flask and replaced with (0.1 ml) of viral inoculum. Then, by concurrently inoculating similar flasks with an equivalent volume of sterile PBS, negative culture controls were created. Subsequently, for 1 hour, the inoculum was allowed to absorb at (37°C) in temperature. After an hour, the MEM with 1% fetal calf serum was added to every monolayer including the control culture (negative) and infected culture, and incubated at 37°C. Finally, the monolayer was diagnosed using an inverted microscope every day to look for the appearance of cytopathic effects (CPEs) (Sharma et al., 2016; Kurucay et al., 2023). Extraction of DNAThe DNA Mini Kit (Promega USA) was used to extract DNA from samples according to the manufacturer’s instructions. The steps for extraction of DNA include, 200 μl of the sample suspension was incubated for ten minutes at 56°C with 10 μl of proteinase K and 200 μl of lysis buffer. After the incubation period, the lysate was mixed with 200 μl of 100% ethanol. After that, washing and centrifugation of the samples. Finally, the kit included 100 μl of elution buffer, which was used to extract the nucleic acid. PrimersThe primer of Vp2 gene was designed using NCBI described in Table 1. PCR amplificationThe primers were employed in a 25 μl reaction volume. First, 1 μl of each primer, 16.5 μl of water, 1.5 μl of DNA template, and 5 μl of PCR Master Mix from Biolab, New England made up this reaction mixture. A thermal cycler made by Applied Biosystems was used in this procedure. The protocol of amplification included the following steps: the first step is denaturation for 5 minutes at 95°C for one cycle; a second step is denaturation for 40 seconds at 95°C, annealing for 1 minute at 58°C, and extension for 1 minute at 72°C for 38 cycles; and a final step is extension for 5 minutes at 72°C for one cycle. Finally, for 1.30 hours, the PCR products were electrophoresed on a 1.5% agarose gel at a voltage of 5 volts. It was potential to see the DNA bands by utilizing a UV transilluminator. Ethical approvalAll procedures in this study followed the guidelines of the local Ethics Committee at the College of Veterinary Medicine, University of Baghdad, and approved under no. P.G./3120 on 29 April 2023. ResultsResults of rapid testThirty out of sixty samples (fecal swabs) were positive by Rapid test (Fig. 1). Result of virus propagation on FK and chicken embryo fibroblast cell culturesAfter the processing of the samples, the positive samples of the fast test were reproduced in a FK cell culture for three passes. The CPEs were readily apparent during the first passage at the 72-hour mark, as shown by the presence of rounded cells. Subsequent passages, i.e., the second and third, exhibited a further manifestation of CPE, with the presence of both rounded and detached cells (Fig. 2). Additionally, the virus underwent propagation in a culture of chicken embryo fibroblast cells for a duration of two passages. The CPEs of the first passage were seen 3 days post-infection and were defined by the presence of rounded cells. In contrast, the second passage exhibited CPE characterized by both rounded and detached cells, which occurred after 2 days (Fig. 3). These observations were contrasted to the control group (non-infected), as shown in Figures 2 and 3. Results of PCRSix samples of isolated virus were positive by PCR (Fig. 4). Table 1. Primers of VP2 gene.
Fig. 1. Positive results of rapid CPV Ag test kit.
Fig. 2. The CPEs induced by CPV on FK cells were observed at different time points. The control cell monolayer appeared normal. However, at the first passage, 72 hours post-inoculation, the cells exhibited rounding (White arrow). At the second passage, 48 hours post-inoculation, the cells showed both rounding and detachment (Black arrow). Finally, at the third passage, 24 hours post-inoculation, a greater degree of cell detachment was observed.
Fig. 3. The ECF cells were subjected to CPV, resulting in CPEs. The control cell monolayer remained unaffected. However, at the first passage, the cell monolayer exhibited cell rounding (White arrow) after 3 days of inoculation. Furthermore, at the second passage, the cell monolayer displayed both cell rounding and detachment (Black arrow) after 2 days of inoculation.
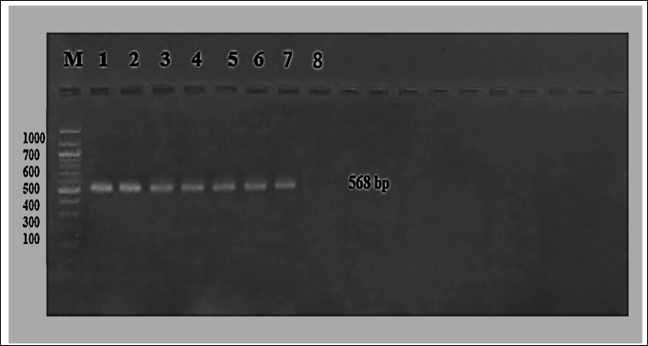
Fig. 4. The product of isolated virus was electrophoresis on 1.5% agarose at 5 volt/cm2. 1× TBE buffer for 1:30 hours. M: DNA ladder (100), 1–6 lines (isolated virus), 7 and 8 lines (positive and negative control). DiscussionThis research aimed to evaluate probable instances of CPV among individuals aged 2–6 months, including several breeds. A total of 60 fecal swabs were subjected to analysis using the rapid CPV Ag Test Kit, resulting in the identification of 30 positive samples, i.e., the positive samples about 50%. This test is widely used as a field diagnostic technique due to its expeditious nature, straight for wardens, and accessibility to both veterinarians and pet owners. Also, some studies revealed the test’s sensitivity does not surpass 50% as a result of the substantial quantity of viral antigen necessary to generate a prominent visible band. This observation corroborates the previously reported characteristics of the test, namely its high specificity and poor sensitivity, as documented by Soliman et al. (2018). The positive samples from the rapid kit were effectively cultivated on FK and ECF cells for three passages, exhibiting characteristic CPE such as cell rounding and detachment on days 2–3 and 4–6 after inoculation. These observations were made in comparison to a negative control. However, the research conducted by Sharma et al. (2016), showed that the CPV exhibited adaptation on FK cells, resulting in the manifestation of typical CPEs only after the fifth passage. Furthermore, revealed the CPE seen in the seventh passage was characterized by the shortening and rounding of cells (Sharma et al., 2016). These cuytopathic effects especially the rounding of cells may be due to the toxic effect of viral proteins but the detachment of these cells may be due to the viral replication and release of the virus from the cell to infect another cell. Also, the study of Raj et al. (2010) indicated the CPE of CPV is rounding and detached cells that are seen after 3–4 passages after infection in CRFK cell culture. CPV isolation on cell culture is a standard technique for both clinical and laboratory studies. Replicating and propagating the virus in cultured cells facilitates its research and diagnosis. Isolating CPV requires the use of canine-derived cell lines. In order to isolate CPV, FK cells are often employed as a cell line. These cells are selected because they are more likely to get infected by CPV. CPE are observable alterations brought on by a CPV infection of the cells. Examples of these impacts include cell rounding and separation from the surface of the culture flask. As well, these infected cells confirmed the presence of CPV using PCR. This assay and other virus-specific diagnostics fall within this category. But the PCR is a common molecular biology method for identifying viral infections like parvovirus because it can detect and amplify certain DNA sequences. Conventional PCR was used to amplify the six randomly chosen samples from the isolated positive samples, using specific primers targeting the Vp2 gene. All of them exhibited positivity at the 568 base pair (bp) position. This antigen was used to confirm the existence of CPV in the aforementioned samples. Also, the study of Raj et al. (2010) confirmed the isolated virus using PCR. The affirmation of the selected samples’ positive provides evidence supporting the higher sensitivity of the molecular-based technique, polymerase chain reaction, in comparison to the immunochromatographic approach, often known as the rapid test (Kurucay et al., 2023). ConclusionThe prevalence of parvovirus infection is high in Iraq. The virus was isolated in two distinct kinds of cell culture, namely primary and secondary. The isolated virus has a cytopathic impact characterized by the rounding and detaching of cells. The primary cell culture is faster than the secondary cell culture in isolation of this virus and produces a CPE. Furthermore, the presence of the isolated virus was verified by the use of a polymerase chain reaction test. AcknowledgmentsThe authors would like to express their appreciation to all members of Virology Department, College of Veterinary Medicine, University of Baghdad, for their support and help during this work. Conflict of interestThe authors declare that they have no competing interests. FundingThis research received no external funding. Authors’ contributionsAll authors contributed to this work equally. Data availabilityAll data were provided in the manuscript. ReferencesAhmed, A.I. 2020. Molecular characterization of infectious bursal disease virus isolated from naturally infected broiler chickens in Erbil, Iraq. Iraqi J. Vet. Med. 44, 21–27. Ahmed, N., Riaz, A., Zubair, Z., Saqib, M., Ijaz, S., Nawaz- Ul-Rehman, M.S., Al-Qahtani, A. and Mubin, M. 2018. Molecular analysis of partial VP-2 gene amplified from rectal swab samples of diarrheic dogs in Pakistan confirms the circulation of Canine parvovirus genetic variant CPV-2a and detects sequences of feline panleukopenia virus (FPV). Virol. J. 15(1), 45. Ali, L.F., Al-Suhail, R.G. and Naser, F.G. 2015. RT PCR detection and propagation of respiratory syncytial virus in human lung carcinoma cells (A549) cell line. Iraqi J. Vet. Sci. 56(1A), 69–74. Allawe, A.B. 2016. Molecular characterization of field isolates of avian infectious laryngeotracheitis virus from different farms in Iraq. Iraqi J. Vet. Med. 40(2), 31–35. Chen, Y., Wang, J., Bi, Z., Tan, Y., Lv, L., Zhao, H. and Qian, J. 2021. Molecular epidemiology and genetic evolution of Canine parvovirus in East China, during 2018-2020. Infect. Genet. Evol. 90, 104–780. Clark, N.J., Seddon, J.M., Kyaw-Tanner, M., Al-Alawneh, J., Harper, G., McDonagh, P. and Meers, J. 2018. Emergence of Canine parvovirus subtype 2b (CPV-2b) infections in Australian dogs. Infect. Genet. Evol. 58, 50–55. Cotmore, S.F., Agbandje-McKenna, M., Canuti, M., Chiorini, J.A., Eis-Hubinger, A.M., Hughes, J., Mietzsch, M., Modha, S., Ogliastro, M., Penzes, J.J., Pintel, D.J., Qiu, J., Soderlund-Venermo, M., Tattersall, P. and Tijssen, P. 2019. ICTV virus taxonomy profile: Parvoviridae. J. Gen. Virol. 100(3), 367–368. Jbr, A. and Jumaa, R. 2024. Sequencing analysis of the N gene of canine distemper virus from infected dogs in Baghdad City. Iraqi J. Vet. Med. 48(1), 41–47. Jumaa, R.S., Abdulmajeed, D.I. and Karim, A.J. 2021. Evaluation of secondary metabolites of herbal plant extracts as an antiviral effect on infectious bursal disease virus isolates in embryonated chicken eggs. Vet. World 14(11), 2971–2978. Jumaa, R.S., Allawe, A.B. and Jabbar, R.N. 2019. Isolation of infectious bursal disease virus and immuno histochemstry of CD4+ and CD8+ for infected Iraqi chickens. Biochem. Cell Arch. 19(2), 3375. Jumaa, R.S., Allawe, A.B. and Jabbar, R.N. 2020. Genetic analysis of field isolates of infectious bursal disease virus in Iraqi farms. Iraqi J. Vet. Med. 44(1), 18–28. Kurucay, H.N., Tamer, C., Muftuoglu, B., Elhag, A.E., Gozel, S., Cicek-Yildiz, Y. and Yazici, Z. 2023. First isolation and molecular characterization of Canine parvovirus-type 2b (CPV-2b) from red foxes (Vulpes vulpes) living in the wild habitat of Turkey. Virol. J. 20(1), 27. Mhemid, K.M. 2016. Molecular detection of lumpy skin disease virus in cattle by polymerase chain reaction in Iraq: Khitam Mahdi Mhemid and Ibtesam Qasim Hassan. Iraqi J. Vet. Med. 40(1), 83–88. Nur-Farahiyah, A.N., Kumar, K., Yasmin, A.R., Omar, A.R. and Camalxaman, S.N. 2021. Isolation and genetic characterization of Canine parvovirus in a Malayan tiger. Front. Vet. Sci. 8, 660046. Raj, J., Mukhopadhyay, H., Thanislass, J., Antony, P. and Pillai, R. 2010. Isolation, molecular characterization and phylogenetic analysis of Canine parvovirus. Infect. Genet. Evol. 10(8), 1237–1241. Sharma, S., Dhar, P., Thakur, A., Sharma, V. and Sharma, M. 2016. First detection of Canine parvovirus Type 2b from diarrheic dogs in Himachal Pradesh. Vet. World 9(9), 964–969. Soliman, R.M., Baker, N.M., Nasr, M.Y. and Khodeir, M.H. 2018. Clinical, virological and molecular characterization of Canine parvovirus in dogs. Eur. J. Pharm. Med. Res. 5(4), 525–535. Taha, F.Y. and Alhankawe, O.K. 2023. Molecular evidence of schmallenberg virus associated by ovine abortion with fetal anomalies in Nineveh province, Iraq. Iraqi J. Vet. Sci. 37(1), 115–120. | ||
| How to Cite this Article |
| Pubmed Style Allmjeed DIA, Jumaa RS, Ibrahim OM. Isolation and PCR-based detection of parvovirus in dogs. Open Vet. J.. 2024; 14(12): 3213-3218. doi:10.5455/OVJ.2024.v14.i12.6 Web Style Allmjeed DIA, Jumaa RS, Ibrahim OM. Isolation and PCR-based detection of parvovirus in dogs. https://www.openveterinaryjournal.com/?mno=208463 [Access: January 25, 2026]. doi:10.5455/OVJ.2024.v14.i12.6 AMA (American Medical Association) Style Allmjeed DIA, Jumaa RS, Ibrahim OM. Isolation and PCR-based detection of parvovirus in dogs. Open Vet. J.. 2024; 14(12): 3213-3218. doi:10.5455/OVJ.2024.v14.i12.6 Vancouver/ICMJE Style Allmjeed DIA, Jumaa RS, Ibrahim OM. Isolation and PCR-based detection of parvovirus in dogs. Open Vet. J.. (2024), [cited January 25, 2026]; 14(12): 3213-3218. doi:10.5455/OVJ.2024.v14.i12.6 Harvard Style Allmjeed, D. I. A., Jumaa, . R. S. & Ibrahim, . O. M. (2024) Isolation and PCR-based detection of parvovirus in dogs. Open Vet. J., 14 (12), 3213-3218. doi:10.5455/OVJ.2024.v14.i12.6 Turabian Style Allmjeed, Dhuha Ismaeel Abdul, Rawaa Saladdin Jumaa, and Osamah Mohammed Ibrahim. 2024. Isolation and PCR-based detection of parvovirus in dogs. Open Veterinary Journal, 14 (12), 3213-3218. doi:10.5455/OVJ.2024.v14.i12.6 Chicago Style Allmjeed, Dhuha Ismaeel Abdul, Rawaa Saladdin Jumaa, and Osamah Mohammed Ibrahim. "Isolation and PCR-based detection of parvovirus in dogs." Open Veterinary Journal 14 (2024), 3213-3218. doi:10.5455/OVJ.2024.v14.i12.6 MLA (The Modern Language Association) Style Allmjeed, Dhuha Ismaeel Abdul, Rawaa Saladdin Jumaa, and Osamah Mohammed Ibrahim. "Isolation and PCR-based detection of parvovirus in dogs." Open Veterinary Journal 14.12 (2024), 3213-3218. Print. doi:10.5455/OVJ.2024.v14.i12.6 APA (American Psychological Association) Style Allmjeed, D. I. A., Jumaa, . R. S. & Ibrahim, . O. M. (2024) Isolation and PCR-based detection of parvovirus in dogs. Open Veterinary Journal, 14 (12), 3213-3218. doi:10.5455/OVJ.2024.v14.i12.6 |