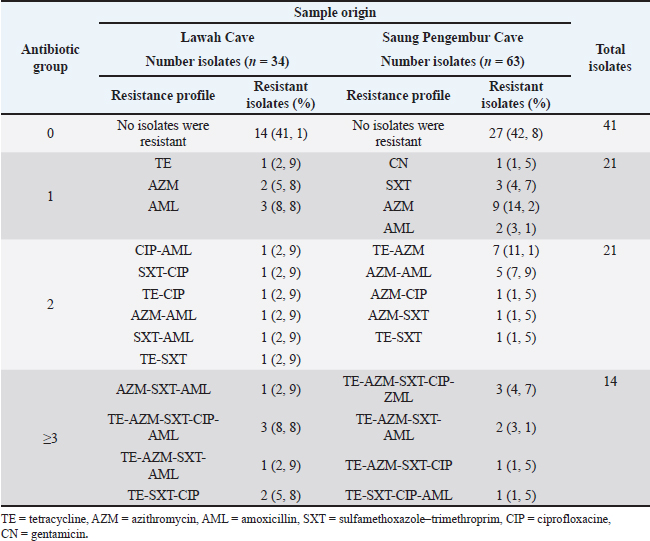
| Research Article | ||
Open Vet. J.. 2024; 14(12): 3460-3473 Open Veterinary Journal, (2024), Vol. 14(12): 3460-3473 Research Article Phylogenetic analysis blaTEM gene of Escherichia coli isolated from cave bats in West Nusa Tenggara Province, IndonesiaAlfiana Laili Dwi Agustin1, Mustofa Helmi Effendi2, Wiwiek Tyasningsih3*, Hani Plumeriastuti4, Aswin Rafif Khairullah5, Fitrine Ekawasti5, Ikechukwu Benjamin Moses6, Kurnia Nisa Kinasih7, Muhammad Khaliim Jati Kusala5, Yolla Rona Mustika7, Ima Fauziah5 and Syahputra Wibowo81Doctoral Program in Veterinary Science, Faculty of Veterinary Medicine, Universitas Airlangga, Surabaya, Indonesia 2Division of Veterinary Public Health, Faculty of Veterinary Medicine, Universitas Airlangga, Surabaya, Indonesia 3Division of Veterinary Microbiology, Faculty of Veterinary Medicine, Universitas Airlangga, Surabaya, Indonesia 4Division of Veterinary Pathology, Faculty of Veterinary Medicine, Universitas Airlangga, Surabaya, Indonesia 5Research Center for Veterinary Science, National Research and Innovation Agency (BRIN), Bogor, Indonesia 6Department of Applied Microbiology, Faculty of Science, Ebonyi State University, Abakaliki, Nigeria 7Master Program in Veterinary Science and Public Health, Faculty of Veterinary Medicine, Universitas Airlangga, Surabaya, Indonesia 8Eijkman Research Center for Molecular Biology, National Research and Innovation Agency (BRIN), Bogor, Indonesia *Corresponding Author: Wiwiek Tyasningsih. Division of Veterinary Microbiology, Faculty of Veterinary Medicine, Universitas Airlangga, Surabaya, Indonesia. Email: wiwiek-t [at] fkh.unair.ac.id Submitted: 09/09/2024 Accepted: 11/11/2024 Published: 31/12/2024 © 2024 Open Veterinary Journal
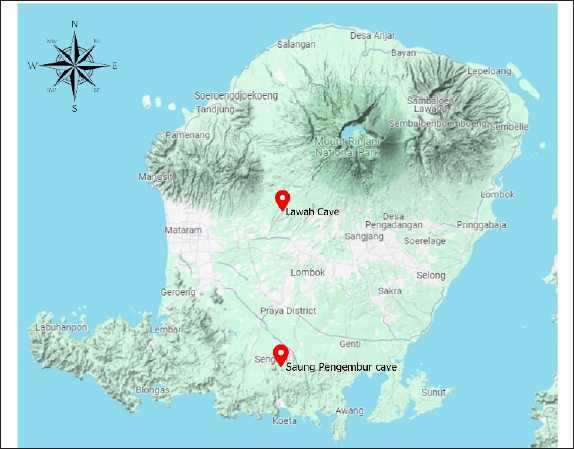
AbstractBackground: The blaTEM, blaCTX-M, and blaSHV are the result of mutations in the extended-spectrum beta-lactamases (ESBL) enzyme, which can be seen genotypically. Wild bats have never received antibiotic therapy, and the presence of antimicrobial-resistance (AMR) in bats indicates the level of AMR in the environment, the circulation of ESBL Escherichia coli between humans or animals in Indonesia and wild animals is still not widely known. Whole genome sequencing is used to determine the circulation of ESBL Escherichia coli in detail. Aim: Our research aims to determine the genetic level of relatedness of blaTEM samples of cave bat isolates in West Nusa Tenggara, Indonesia. Methods: This research is a laboratory exploration to detect ESBL genes in guano isolate samples from cave bats in West Nusa Tenggara Province, Indonesia. Samples were obtained by swabbing the rectum of live bats caught from caves on the island of West Nusa Tenggara Province, namely, Lawah Cave and Saung Pengembur Cave. A total of 50 samples from Lawah Cave and 85 samples from Saung Pengembur Cave were identified to find an E. coli bacteria using Gram-stain test and IMViC for biochemical test from isolates, which showed a metallic green color on eosin methylene blue agar media. Escherichia coli isolates were tested for sensitivity using antibiotics, followed by a polymerase chain reaction test showed positive results for the blaTEM gene and phylogenetic analysis to determine relationships. Results: Phylogenetic analysis shows a genetic closeness between the isolates from Lawah Cave and Saung Pengembur Cave is 95%. Between samples from Lawah and Saung Pengembur Cave, we found that there was a closeness of 86% to the E. coli strain A2-2 sample from Thailand with the code OR680712.1. at GenBank. Conclusion: The blaTEM gene characteristics of bat isolates from West Nusa Tenggara are close to pig isolates from Thailand. The genetic proximity between blaTEM gene samples from Lawah Cave and Saung Pengembar Cave analyzed using the phylogenetic tree shows a closeness to samples from Thailand, even though they come from samples with distant locations, which can have a negative impact on human health. Keywords: Antimicrobial resistance, Bats, blaTEM, ESBL, Human health. IntroductionBats are mammals that have a variety of environments to live, good adaptability, the ability to fly long distances, proximity to humans and livestock in urban areas, body resistance to diseases, and a relatively long lifespan making them potential as a vector for spreading various pathogens to other wild animals, humans, and animals (McDougall et al., 2021; Castelo-Branco et al., 2023). Bats have the potential to spread pathogenic bacteria such as E. coli to humans through their feces called guano (Dimkić et al., 2021). Human anthropogenic activities such as hunting bats for consumption, using caves as tourist attractions, wild animals, and livestock that eat dead bats in the human environment, using guano as fertilizer, and consuming water contaminated with guano directly without processing can increase the potential for disease transmission from bats to humans, other wild animals, and livestock (Nangoy et al., 2021). Research by Kholik et al. (2019) shows that E. coli bacteria were isolated from bats that live in Tanjung Ringgit caves East Lombok Regency, West Nusa Tenggara, the bacteria potential to risk for human and animal health. Escherichia coli bacteria are digestive tract bacteria that are often found in animals and humans and can cause pneumonia, gastroenteritis, septicemia, and cystitis (Widodo et al., 2022). Antibiotics are often used by humans to reduce mortality due to bacterial infections, antibiotics are often used as treatment resulting in an increasing antimicrobial-resistance bacteria (AMR), and E. coli often appears as a cause of multidrug resistance (MDR) among humans and animals (Wibisono et al., 2021; Putri et al., 2023). The antibiotics azithromycin, amoxicillin, tetracycline, sulfamethoxazole/trimethoprim, and ciprofloxacin are antibiotics that often appear to cause MDR of E. coli bacteria isolated from bats in Lombok, Indonesia (Agustin et al., 2024a). Escherichia coli bacteria can produce extended-spectrum beta-lactamases (ESBL) genes, which can be transmitted to other Gram-negative bacteria horizontally via bacterial plasmids (Tseng et al., 2023). The results of research by Chomkatekaew et al. (2023) conducted at Angkor Children’s Hospital, Cambodia, from several samples of mothers and children tested showed similarities between resistance plasmids beyond the relationship between mother and child, and the result of research indicates the existence of vertical transmission in the community. ESBL E. coli has increased eight times in the last decade with the highest prevalence in Asia and Africa reaching 70% (Raffelsberger et al., 2023). High mortality, long hospital stays, and increasing treatment costs are risks of ESBL-producing Enterobacteriaceae infection (Handal et al., 2023). Wild bats have never received antibiotic therapy, and the presence of AMR in bats indicates the level of AMR in the environment (Huy et al., 2023). Human anthropogenic activities result in the spread of ESBL in bats, and ESBL E. coli from animal and human origin due to anthropogenic activities can be spread through wastewater from slaughterhouses (Ugbo et al., 2023), household and hospital wastewater (Satyaprakash et al., 2024), pharmaceutical industry wastewater (Mou et al., 2023), and agriculture (Igbinosa et al., 2023). Soil harbors bacteria that have antimicrobial resistance genes (ARGs), livestock waste, and fertilizer derived from agricultural manure are a source of resistance for other animals, including wild animals (Musa et al., 2023). The circulation of ESBL E. coli between humans or animals in Indonesia and wild animals is still not widely known, and blaTEM is a gene that often appears as a cause of ESBL-produce in wild animals in Indonesia (Riwu et al., 2024; Mustika et al., 2024). The blaTEM gene is also a gene that often appears to encode ESBL in Indonesia in ducks (Prayudi et al., 2023), milk cows (Widodo et al., 2023), cats (Farizqi et al., 2023), broiler chickens (Faridah et al., 2023), and bats (Agustin et al., 2024b); however, there has been no research regarding the closeness of the ESBL E. coli kinship from West Nusa Tenggara with existing data. Whole genome sequencing (WGS) was used to properly determine the circulation of ESBL E. coli. WGS used in epidemiological investigations can be used to monitor contamination between countries provided that the available metadata must contain information such as the name of the isolated strain, sample type, isolation date, and country of origin of the sample (Rantsiou et al., 2018). WGS can be used to determine more accurate E. coli subtypes because of its high significance but is generally costly and time-consuming and requires bioinformatics expertise (Anjum et al., 2021). Research by Hayer et al. (2023) shows the results of sequencing E. coli that there is a similarity in heavy metals, antibiotic resistance genes on plasmids, and virulence genes between the feces of cows, sheep, and dogs from the same farm and rodents and wild dogs that live around the farm area the object of research. The results of bat sequencing in research by Benavides et al. (2022) in Peru show that there are similarities in genetic material from E. coli bacteria between bats and pigs. Characterization of ESBL E. coli in Australian fruit bats indicates transmission from domestic animals and/or humans to flying foxes caught in urban environments, and ExPEC in flying foxes adds evidence of the spread of pathogenic E. coli strains that have antimicrobial resistance from humans to wild animals (McDougall et al., 2019). Due to restrictions on the movement of wild bats, bats can easily spread ESBL from one environment to another, ESBL research in bats is important as a prevention for human health and antimicrobial management. Materials and MethodsType of research and study areaThis type of research is a laboratory exploration to detect ESBL genes in guano isolate samples from cave bats in West Nusa Tenggara Province, Indonesia. Samples were obtained by swabbing the rectum of live bats caught from caves on the island of West Nusa Tenggara Province, namely, Lawah Cave −8.539453, 116.276576 in West Lombok and Saung Pengembur Cave in Central Lombok −8.812872, 116.273968 (Fig. 1). Sampling was carried out once, with bats caught using a mist net placed in front of the cave mouth at 4–10 pm when the bats left the cave to look for food.
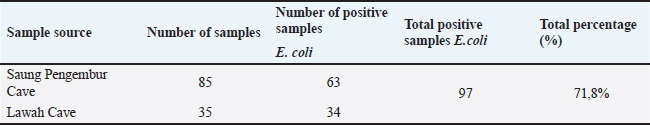
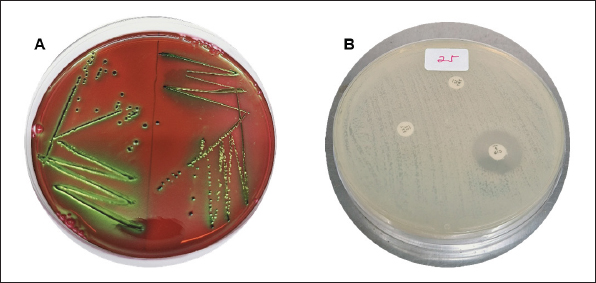
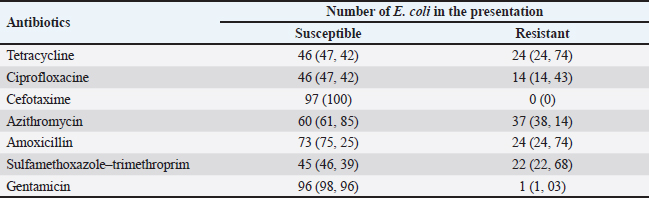
Fig. 1. Sampling locations for Lawah Cave (-8.539453, 116.276576) in West Lombok and Saung Pengembur Cave (-8.812872, 116.273968) in Central Lombok. ProceduresBefore swabbing, to avoid contamination during sample collection, the hair around the anus is cleaned first using isopropyl alcohol 70%, and a sterile cotton swab is inserted into the rectum and rotated for a few moments to ensure bacteria stick to the cotton swab (Adesiyun et al., 2009). The swab results were placed on a semi-solid medium carry Blair transport medium (Oxoid, UK) to be taken to the laboratory. Isolation and identification of samplesBrain heart infusion media (Oxoid, UK) was used to fertilize the 135 samples obtained. The samples were incubated for 24 hours at 37°C in the Bacteriology and Mycology Laboratory, Faculty of Veterinary Medicine, Airlangga University, Surabaya. Then, the samples were streaked on eosin methylene blue agar (EMBA) media (Oxoid, UK) incubated at 37°C for 24 hours, and purification was carried out on bacterial colonies, which showed a metallic green color. Bacteria that grow during purification are then identified using Gram staining and biochemical tests of indole motility (Oxoid, UK), methyl red and Voges–Proskauer (Oxoid, UK), urea (Oxoid, UK), citrate (Oxoid, UK) (IMViC), and sugars (sucrose, lactose, and glucose) (Oxoid, UK). The Gram staining test was carried out by placing the bacteria on a glass object and fixing it, dripping it with crystal violet for 2 minutes and washing it with running water, then dropping it with iodine and acetone alcohol for 1 minute, and dropping it with safranin for 2 minutes. The stained bacteria were examined using a microscope at 1000× magnification (Dameanti et al., 2023). Bacterial isolates confirmed to be E. coli were grown on nutrient agar (NA) media (Oxoid, UK). Antibiotic sensitivity test against E. coli bacteriaSensitivity testing on bacteria confirmed as E. coli was carried out using the Kirby–Bauer agar diffusion method on Mueller–Hinton agar (MHA) media (Oxoid, UK). Bacterial suspensions are made from bacteria that grow in NA media and are homogenized with NaCl liquid to match the McFarland 0.5 standard. The suspension was poured onto a plate containing MHA media and then added with seven antibiotics to determine the sensitivity of the bacteria. Antibiotics, amoxicillin 25 µg (AML) (Oxoid, UK), azithromycin 15 µg (AZM) (Oxoid, UK), cefotaxime 30 µg (CTX) (Oxoid, UK), ciprofloxacine 5 µg (CIP) (Oxoid, UK), sulfamethoxazole/trimethoprim 25 µg (SXT) (Oxoid, UK), tetracycline 30 µg (TE) (Oxoid, UK), and gentamicin 10 µg (CN) (Oxoid, UK), were used for sensitivity testing. Antibiotics that have been placed in MHA media are incubated at 37°C for 24 hours. Interpretation of results is aligned with the Clinical Laboratory Standard Institute (CLSI) standards (CLSI, 2020). Detection of blaTEM genes and phylogenetic analysis of E. coli isolates from batsEscherichia coli bacterial culture in EMBA media was continued to see the presence of the ESBL blaTEM gene using PCR with primers blaTEM (F) and blaTEM (R) (Table 1). Before extraction, E. coli bacteria were inoculated in MHA media and incubated at 37°C for 24 hours. Several bacterial colonies were tested from one isolate placed in a safe-lock Eppendorf tube containing 300 µl TE (10-mM Tris, pH8, 10-mM EDTA). The suspension was carried out by Fortex and continued with the boiling lysis method by placing it in the Eppendorf tube thermoStatTM at a temperature of 98°C for 10 minutes. After that, it was centrifuged at a speed of 10,000 rpm for 10 minutes. Then, 25-µl reaction mixture (Promega) (12.5-µl Go tag green master mix, primers blaTEM-F gene 5’-ATGAGTATTCAACATTTCCG-3’ and TEM-R 5’-CTGACAGTTACCAATGCTTA-3’ (Naelasari et al., 2018) with an amplification size of 867 bp each primer 2 µl, 0.5 µl of Dnase water, and 5 µl of DNA template) was taken and placed in an Eppendorf tube and then amplified. Thermal cycle predenaturation temperature was 96°C for 5 minutes followed by denaturation for 1 minute at 96°C, followed by 35 cycles of annealing at 58°C for 1 minute, and extension at 72°C for 1 minute. Amplification ended with a final extension at 72°C for 10 minutes. PCR sequencing results were carried out at Genetic Science Indonesia. Data analysisPhylogenetic analysis of the blaTEM gene of E. coli isolates from bats was used to determine the genetic relationship with various E. coli from animal origins from various places as reference data in GenBank using MegaX 11 software with the neighbor-joining method (Saitou and Nei, 1987). Ethical approvalThe research was approved by the Animal Care and Use Committee, Universitas Airlangga with No: 1.KEH.046.03.2023. ResultsBacteria identification as E. coliThere were 50 bats obtained from Lawah Cave and 85 bats obtained from Saung Pengembor Cave. Based on the identification results, it is known that the bats caught were Rhinolopus simplex and Hipposderos diadema. Isolates that were phenotypically confirmed as E. coli were 97 samples showing Gram-negative with biochemical test results showing indole (+) pink ring, MR (+) red, VP (−) no color change, urea (−) no color change, citrate (+) green, acid glucose, acid lactose, and acid sucrose (Fig. 2). Antibiotic for sensitivity tes from E. coli isolatesThe antibiotic sensitivity results of E. coli isolates from bats from Saung Pengembar cave showed that the highest level of antibiotic resistance was the antibiotic AZM 36.7% (29/97), followed by the antibiotic TE 15.4% (15/97), AML 13.4% (13/97), SXT 12.3% (12/97), CIP 6.1% (6/97), and gentamicin (CN) 1% (1/97), with the samples still being sensitive to cefotaxime (Fig. 3). The results of the antibiotic sensitivity test on E. coli isolates from bats from Lawah Cave showed that the highest level of antibiotic resistance was the antibiotic AML 11% (11/97) followed by the antibiotic SXT 10% (10/97); TE 9.2% (9/97), AZM, and CIP each 8.2% (8/97); and gentamicin (CN) and CTX each 0% (0/97) (Tables 2 and 3). PCR test and phylogenetic analysis blaTEM E. coliMolecular analysis shows that the blaTEM gene is the gene encoding the ESBL of Lawah Cave and Saung Pengembur Cave bat isolates (Fig. 4). We carried out a phylogenetic analysis by comparing the blaTEM gene from several countries in Southeast Asia and countries close to Indonesia with data in GenBank. The GenBank data codes that we use include JN003415.1 from Australia, OP142512.1 from Vietnam, OR095844.1 from Australia, and OR680712.1 from Thailand. Phylogenetic analysis (Fig. 5) shows a genetic closeness between the isolates from Lawah Cave (1.38) and Saung Pengembur Cave (2.70) 95%. Between samples from Lawah and Saung Pengembur Cave (1.38 and 2.70), we found that there was a closeness of 86% to the E. coli strain A2-2 sample from Thailand with the code OR680712.1. at GenBank. Isolate OR680712.1 comes from the extended-spectrum beta-lactamase gene produced by E. coli isolated from pigs. DiscussionSeveral researchers have shown that bats carry antibiotic resistance (Claudio et al., 2018; McDougall et al., 2019; Nguema et al., 2020; Obodoechi et al., 2021). Antibiotic resistance in the E. coli isolates in our study occurred in 56 samples, and 37 isolates showed the highest resistance to the antibiotic AZM. Azithromycin is a macrolide antibiotic, livestock such as cattle is often given therapy for the treatment of liver abscesses and respiratory tract infections caused by Campylobacter jejuni bacteria, and macrolide antibiotics in humans are used as supportive drugs for other antibiotics such as the β-lactam group (Trott et al., 2021). Macrolide antibiotics are the main antibiotics of choice in humans and animals for infections such as bronchitis, pneumonia, mastitis, diarrhea, chlamydia, and gonorrhea. Apart from that, it is used for the prevention of rheumatism, endocarditis, stomach ulcers, and gastritis (Kanaeva et al., 2023). Exposure to macrolides is a factor in the development of ESBL bloodstream infection (BSI) in Klebsiella pneumoniae, and macrolides are usually used in certain BSIs associated with Salmonella spp infections (Palma et al., 2017). Table 1. Prevalence of E. coli isolates from cave bats Saung Pengembur and Lawah from West Nusa Tenggara.
Fig. 2. Biochemical test results for samples that were phenotypically detected as E. coli. The biochemical test for E. coli bacteria to be positive if it shows a pink ring color on SIM media, MR media remains red, VP remains yellow, citrate is green, urea is yellow, and E. coli ferment glucose, sucrose, and lactose as indicated by the color changing from red to yellow.
Fig. 3. (A) Escherichia coli bacteria isolated from bats show metallic green colonies on EMBA media. (B) Antibiotic sensitivity test on E. coli isolates from Lawah Cave and Saung Pengempur Cave from West Nusa Tenggara Province showed that the bacteria did not die when given an antibiotic disk, which indicated that resistance had occurred in the bacteria. The TE antibiotics and AML were in second place as antibiotics that were resistant in our study (24.74%). TnpA, IS91 TE, sulfonamides, and aminoglycosides significantly spread ARGs in the environment because they are easily adsorbed in soil, and TnpA is a group of mediated transposons that mediate beta-lactam resistance genes in the cow–cow-milk food transmission chain. TnpA is a very small transposase from Y1 consisting of 140 amino acids from the HUH family, which form a single-stranded transpose ring (DNA specific 5’-TTGAT) and integrate at a new position in the transposition cycle (target 3’-TTGAT) (Yu et al., 2024). Different results were shown in research on bats in Southern Italy, and colistin sulfate showed the highest resistance at 81.6%, followed by amoxicillin at 70.7%, amoxicillin + clavulanic acid 58.1%, ampicillin 42.9%, streptomycin 40.2%, and minocycline 21.1% (Foti et al., 2023). Bacteria can be transmitted from animals to humans through various mechanisms, including the direct fecal–oral route, contaminated food products, inadequate cooking methods, improper food handling, animal scratches, and even bites (Kristianingtyas et al., 2021). Beta-lactam antibiotic from livestock manure, especially pig manure, is considered to be the main spreader of antibiotic resistance in the environment; besides being able to pollute the soil, pollutants can be carried in water over long distances; and antibiotic degradation usually occurs up to 30 days (Zalewska et al., 2023). Table 2. Antibiogram of E. coli isolates from cave bats Lawah dan Saung Pengembur, West Nusa Tenggara, Indonesia.
Table 3. Results of antibiotic sensitivity test of E. coli isolates from Saung Pengembur Cave and Lawah Cave, West Nusa Tenggara.
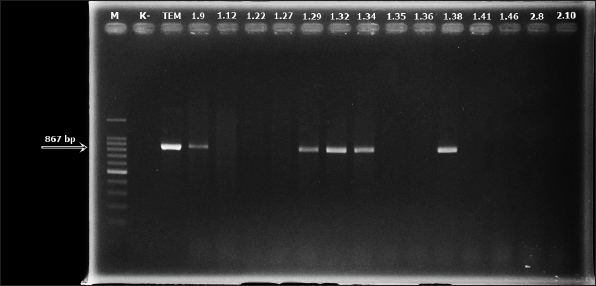
Fig. 4. blaTEM electrophoresis results of E. coli isolates from bats West Nusa Tenggara in sample numbers 1.9, 1.29, 1.32, 1.34, and 1.38. Information: M=marker and K−=negative control.
Fig. 5. blaTEM phylogenetic analysis of E. coli isolates from bats Lawah Cave and Saung Pengembur Cave with data from GenBank JN003415.1, OP142512.1, OR095844.1, and OR680712.1. Description: red circle sample from Lawah Cave and blue circle sample from Saung Pengembur Cave. Ciprofloxacin, a broad-spectrum quinolone antibiotic used to treat humans, is increasing in resistance significantly every year (Authority, 2022). In our study, TE ranks fifth with a resistance rate of 14.43%; this antibiotic is widely used in humans and animals to treat urinary tract infections; and gyrA, parC, and parE are genes that often appear as a result of mutation of quinolone-resistance determining region (Das et al., 2023). Resistance to the sulfamethoxazole–trimethoprim antibiotic in our study was not too high 22.68%, in contrast to Ssebuganda et al. (2023), which showed that E. coli bacteria from the blood, pus, and urine of patients in Uganda showed a resistance level of 85.7 % on the antibiotic sulfamethoxazole–trimethoprim. The ciprofloxacin and sulfamethoxazole–trimethoprim if given in low doses can cause residues (Stastny et al., 2024). Antibiotic residues will remain in animal waste by 60%–90% in their original form or as metabolites, and ultimately, these antibiotics will be in the soil or waterways, so they can pollute the environment which can later impact microorganisms in the soil and cause the development of antibiotic resistance ARGs in the environment (Li et al., 2023). We did not find any third-generation cephalosporin antibiotic resistance in our study (cefotaxime). The absence or low resistance of bacteria to antibiotics indicates low contamination from humans and animals, this is in line with the results of Rojas-Sereno et al. (2023) from Chile, and they did not find any resistance to cephalosporin antibiotics in isolates of E. coli and Klebsiella spp bacteria originating from bats. The location of the β-lactamase enzyme is in plasmids and transposons, which cause bacteria to be resistant to penicillin, third-generation cephalosporins such as cefotaxime, ceftazidime, ceftriaxone, and aztreonam (except carbapenem and cefamycin) (Wartiani et al., 2023). The TEM, CTX-M, and SHV genes are the result of mutations in the ESBL enzyme, which can be seen genotypically (Vanstokstraeten et al., 2023). TEM-1 and TEM-2 are derivatives of TEM and SHV-1 and the conversion of glycine from serine at position 238 is a derivative of SHV, which can hydrolyze ceftazidime; in addition, changes in glutamate from lysine position 240 of SHV derivatives can hydrolyze cefotaxime, and CTX-M-1, 2,8,9 and 25 are CTX-M ESBL mutation points originating from the Kluyvera species (Rana et al., 2022). Animal health, including pet animals, companion animals, and wild animals that have received antibiotic therapy, such as those in zoos, is an object of study that has been widely researched to determine the spread of ESBL E. coli (Naas et al., 2023). The Indonesian government has regulated the use of antibiotics by creating a national antibiotic management program and socializing appropriate antibiotic consumption; however, inappropriate use of antibiotics is still widespread in Indonesia, less than 60% of respondents understand that antibiotics are used to kill infections caused by bacteria, and there is a lack of understanding of their use. Antibiotics are precisely an important contributing factor to the emergence and increase of microorganisms that are resistant to antibiotics (Sinuraya et al., 2023). The high level of antibiotics found at the research location shows the level of antibiotic consumption for only a few types of antibiotics. This condition shows the irrational use of antibiotics so that measures are needed to control antibiotic use to reduce the risk of antibiotic resistance (Fahmi et al., 2024). In Indonesia, around 135,000 drug outlets are serving 264 million buyers, around 90,000 drug outlets are serving grocery stores, and 5,000 drug stores sell drugs without a prescription (Wulandari et al., 2021). Bacterial resistance to antibiotics arises due to excessive use of antibiotics, which will have a long-lasting impact on humans. Resistance can occur if individuals consume food sources that contain antibiotic residues (Effendi et al., 2021a). Antibiotics are used not only to treat humans but also to treat animals. Excessive use of antibiotics in livestock can pollute the environment (Effendi et al., 2021b). The release of antibiotics into the environment coincides with the release of ARG genes, and the release of ARGs into the environment can mutate when they encounter other microorganisms through mobile genetic elements and form new resistance genes or genome sets (Yanestria et al., 2022). Loss of natural habitat for wild animals and urbanization increases contact between wild animals and the environment, thereby facilitating wild animals being infected by commensal-resistant bacteria. The high and low levels of antibiotic resistance indicate contact between wild animals and bacteria that are resistant to antibiotics (Ahmed and Gulhan, 2024). One health and global health approaches are used to understand the spread of ARGs, and one health studies the role of different interconnected ecosystems, while global health studies the emergence of ARGs in a particular region, which can have an impact on other regions throughout the world. The existence of antibiotic resistance in wildlife whose relevance is not receiving antibiotics should be considered as AMR pollution, wildlife has a role in local transmission-a one health problem, and transmission between distant geographic areas is a global health problem (Laborda et al., 2022). In our study, 14 samples experienced multidrug resistance and we found blaTEM as the ESBL coding gene for E. coli. The Enterobacteriaceae family produces ESBL enzymes that encode several specific genes such as blaSHV, blaCTX-M, and blaTEM (Ansharieta et al., 2021). In line with several studies in Indonesia, blaTEM is the dominant ESBL E. coli coding gene found in animals (Effendi et al., 2022a; Effendi et al., 2022b; Tyasningsih et al., 2022). In Nigeria, the E. coli blaTEM gene was detected in 50% of isolates of the Eidolon helvum bat, which is a fruit bat, indicating that there is horizontal gene transfer in the environment (Liu et al., 2024; Modupe et al., 2022). Different results were shown in research in Portugal, and the blaTEM gene was detected in only 21.1%, while blaCTX-M was the dominant gene found as the cause of ESBL E. coli at 57.9%, followed by blaSHV at 31.6% and blaOXA at 10.5% (Garcês et al., 2019). The E coli blaTEM ESBL gene is most frequently found in humans (Sah et al., 2024). In line with this, blaTEM is the only gene that has been detected in animals on the island of Lombok, West Nusa Tenggara Indonesia, with a prevalence value of 25% in cattle (Kholik et al., 2019) and 25% in bats (Mustika et al., 2024). Escherichia coli bacteria are normal bacteria in humans and animals. However, some strains of E. coli can cause various infections, such as mastitis in cattle, colibacillosis in poultry, septicemia, meningitis, and urinary tract infections in humans (Islam et al., 2023). In our research, the bats caught were insectivorous bats, which allowed them to search for food on farms. Esmaeel et al. (2023) in their research found that Myotis emarginatus bat isolates from Iraq, which are insectivorous bats, have genetic similarities between isolates of Acinobacter spp bacteria and isolates from China. These insectivorous bats can eat mosquitoes that carry blood contaminated with bacteria resistant from Chinese travelers or imported animals, or come from migratory birds. blaTEM is a gene that is dominantly present in E. coli isolates from pigs in Thailand (Buranasinsup et al., 2018; Duy et al., 2021; Sornsenee et al., 2021; Boripun et al., 2023). It is still unclear whether our E. coli isolates were contaminated by pigs from Thailand, Indonesian tourists who were contaminated with E. coli ESBL bacteria from Thai pigs and spread in the bat habitat or from Thai tourists who carried ESBL genes spread in Lombok, West Nusa Tenggara, Indonesia. Therefore, our isolates have ESBL genetic similarities with E. coli isolates from pigs from Thailand. The similarity of ESBL genes in bat isolates from Lombok and pigs from Thailand could occur due to the high human mobility currently; in 2017 Indonesian tourists visited Bangkok; and Thailand reached 575,000 people per year and vice versa. There were 138,235 tourists from Thailand who came to Indonesia (Komalasari and Ganiarto, 2020). A total of 1012 tourists from 34 studies showed that tourists were infected with AMR after traveling to Thailand (Bokhary et al., 2021). Street food in Thailand is one of the destinations targeted by tourists, and street food in Thailand is sold in cooked, half-cooked, or even raw conditions (Jaeheng and Han, 2020; Riwu et al., 2022). ESBL E. coli contamination can occur in food to be consumed (Zurita et al., 2020), ESBL is present in pork, and pork is also known to be present in humans in the border provinces of Laos and Thailand (Pungpian et al., 2021). The results of a study conducted on fresh raw food in Thailand showed that of the 521 Enterobacter samples cultured from pork, 76.7%–100% were resistant to ampicillin antibiotics followed by co-amoxiclav, cefoxitin, and colistin and 37.1% of the highest antibiotic residues were found from enrofloxacin (Tangkoskul et al., 2021). Pig and chicken farmers in Thailand have a low level of knowledge, and pig farms in Thailand use antibiotics as much as 66.96% in animal feed (Nuangmek et al., 2020). Lombok Island, West Nusa Tenggara, has natural destinations that attract local and Thai tourists (Lesmana and Sugiarto, 2021). Anthropogenic activities carried out by tourists in natural destinations have an impact on the spread of antibiotic resistance and antibiotic resistance-carrying genes in soil and water so that they can be transmitted to wildlife (Scott et al., 2021). The existence of data on the genetic similarity of ESBL E. coli between livestock and humans (Badr et al., 2022; Puangseree et al., 2022) makes us confident that our bat isolates can be contaminated from humans traveling from West Nusa Tenggara to Thailand. ConclusionIn conclusion, our research is the first study on bats in West Nusa Tenggara that looks at genetic closeness. From the results of the phylogenetic tree, it can be seen that the E. coli isolates from Lawah Cave and Saung Pengembar Cave have the same blaTEM gene, and we assume that the bat samples came from the same source or were present that the similarity of bacterial isolates from one location to another indicates horizontal gene transfer between bats at different research locations. The genetic closeness between samples from West Nusa Tenggara bats cave with pig from Thailand from far locations could have a negative impact on human health. AcknowledgmentsResearchers would like to thank the Faculty of Veterinary Medicine, Airlangga University, for all the facilities provided during conducting research. Conflict of interestThe authors declare that there is no conflict of interest. FundingThis research was supported by funding for Doctoral Dissertation research from the Center for Higher Education Fund (BBPT) and Indonesia Endowment Funds for Education (LPDP), with grant number from keputusan kepala pusat layanan pembiayaan pendidikan kementerian pendidikan, kebudayaan, riset, and dan teknologi nomor 00087/J5.2.3./BPI.06/9/2022. BPI ID Number: 202209090573. Authors’ contributionsKNK and ALDA: collected and assembled the data. YRM and WT: collected the samples and laboratory works, and drafted the manuscript. ARK and MHE: analysis and interpretation of data. MKJK and IM: concept and design of the study. FE and HP: investigation and data curation and reviewed the manuscript critically for important intellectual content. IBM and SW: analysis and data curation and critically revised the manuscript. All authors have read, reviewed, and approved the final version of the manuscript. Data availabilityAll data are available in the manuscript. ReferencesAdesiyun, A.A., Stewart-johnson, A. and Thompson, N.N. 2009. Isolation of enteric phatogens from bats in Trinidad. J. Wildl. Dis. 45(4), 952–961. Agustin, A.L.D., Dameanti, F.N.A.E.P., Effendi, M.H., Tyasningsih, W., Khairullah, A.R., Kurniawan, S.C., Moses, I.B., Hasib, A., Mustika, Y.R. and Kinasih, K.N. 2024a. Multidrug resistance to antibiotics in Escherichia coli bacteria isolated from bats on Lombok Island, Indonesia. J. Adv. Vet. Res. 14(4), 683–686. Agustin A.L.D., Effendi, M.H., Tyasningsih, W., Khairullah, A.R., Tirtasari, K., Moses, I.B. and Ayuti, S.R. 2024b. Cave Bats as carriers of extended spectrum beta-lactamase produced by Escherichia coli from the islan of Lombok, Indonesia. Indian J. Anim. Res. 1, 9. Ahmed, N.A. and Gulhan, T. 2024. Determination of antibiotic resistance patterns and genotypes of Escherichia coli isolated from wild birds. Microbiome 12(1), 8. Anjum, M,F., Schmitt, H., Borjesson, S., Berendonk, T,U., Donner, E., Stehling, E,G. and Pedersen, K. 2021. The potential of using E. coli as an indicator for the surveillance of antimicrobial resistance (AMR) in the environment. Curr. Opinion Microbiol. 64, 152–158. Ansharieta, R., Ramandinianto, S.C., Effendi, M.H. and Plumeriastuti, H. 2021. Moleculer identification of blaCTX-M and blaTEM genes encoding extended-spestrum β-lactamase (ESBL) producing Escherichia coli isolated from raw cow’s milk in East Java, Indonesia. Biodiversitas 22(4), 1600–1605. Authority, E.F.S. 2022. The European Union summary report on antimicrobial resistance in zoonotic and indicator bacteria from humans, animals and food in 2019-2020. EFSA J. 21(3), e07867. Badr, H., Reda, R.M., Hagag, N.M., Kamel, E., Elnomrosy S.M., Mansour, A.I., Shahein, M.A., Ali, S.F. and Ali, H.R. Multidrug-resistant and genetic characterization of extended-spectrum beta-lactamase-producing E. coli recover from chickens and human in Egypt. Animals 12(3), 346. Benavides, J.A., Godreuil, S., Opazo-Capurro, A., Mahamat, O.O., Falcon, N., Oravcova, K., Streicker, D.G. and Shiva, C. 2022. Long-term maintenance of multidrug-resistant Escherichia coli carried by vampire bats and shared with livestock in Peru. Sci. Total Environ. 810(1), 152045. Bokhary, H., Pangesti, K.N.A., Rashid, H., El Ghany M.A. and Hill-Cawthorne, G.A. 2021. Travel-related antimicrobial resistance: a systematic review. Trop. Med. Infect. Dis. 6(1), 11. Boripun, R., Saengsawang, P., Intongead, S., Narinthorn, R., Wongtawan, T., Nissapatorn, V., Pereira, M.L. and Mitsuwan, W. 2023. Molecular characterization and nucleotide substitution of antibiotic resistance genes in multidrug-resistant Escherichia coli isolated from environmental swine farms. Emerg. Contam. 9(4), 100249. Buranasinsup, S., Kulpeanprasit, S., Kong-ngoen, T., Jangsangthong, A., Sookrung, N., Chaicumpa, W. and Indrawattana, N. 2018. Prevalence of the multi-drug resistance of shiga toxin-producing Escherichia coli isolated from Pigs in Central Thailand. Chiang Mai J. Sci. 45(1), 21–32. Castelo-Branco, D.S.C.M., Nobre, J.A., Souza, P.R.H., Diógenes, E.M., Guedes, G.M.M., Mesquita, F.P., Souza, P.F.N., Rocha, M.F.G., Sidrim, J.J.C., Cordeiro, R.A. and Montenegro, R.C. 2023. Role of Brazilian bats in the epidemiological cycle of potentially zoonotic pathogens. Microb. Pathog. 177(1), 106032. Chomkatekaew, C., Thaipadungpanit, J., Pasco, H., Soeng, S., Pol, S., Neou, L., Hopkins, J., Turner, P. and Btty, E.M. 2023. Detection of maternal transmission of resistant Gram-Negative bacteria in a Cambodian hospital setting. Front. Microbiol. 14(1), 1158056. Claudio, V.C., Gonzalez, I., Barbosa, G., Rocha, V., Moratelli, R. and Rassy, F. 2018. Bacteria richness and antibiotic-resistance in bats from a protected area in the Atlantic Forest of Shout Brazil. Plos One 13(9), e0203411. CLSI (Clinical and Laboratory standart Institute). 2020. Performance standards for antimicrobial suseptibility testing in CLSI supplement M100. IN. Wayne, PA: CLSI. Dameanti, F.N.A.E.P., Yanestria, S.M., Widodo, A., Effendi, M.H., Plumeriastuti, H., Tyasningsih, W., Sutrisno, R. and Akramsyah, M.A. 2023. Incidence of Escherichia coli producing extended-spectrum beta lactamase in wastewater of dairy farm in East Java, Indonesia. Biodiversitas 24(2), 1143–1150. Das, T., Nath, C., Das, P., Ghosh, K., Logno, T.A., Debnath, P., Dash, S., Devnath, H.S., Das, S. and Islam, M.Z. 2023. High prevalence of ciprofloxacin resistance in Escherichia coli isolated from chickens, humans and environment: an emerging one health issue. PLoS One 18(11), e0294043. Dimkić, I., Fira, D., Janakiev, T., Kabić, J., Satupar, M., Nenadić, M., Unković, N. and Grbić, M.L. 2021. The microbiome of bat guano: for what is this knowledge important? Appl. Microbiol. Biotechnol. 105(4), 1407–1419. Duy, D.T., Anh, N.L.L., Thoa, N.T.K., Phitsanuwattana, K., Thongratsakul, S., Carrique-Mass, J.J., Hien, L.T., Nam, N.T.T. and Toan, N.T. 2021. Identification of genotype and phenotype of antimicrobial resistance of Escherichia coli isolates from pigs in southern Vietnam. Thai J. Vet. Med. 51(1), 125–132. Effendi, M.H., Faridah, H.D., Wibisono, F.M., Wibisono, F.J., Nisa, N., Fatimah. and Ugbo, E.N. 2022a. Detection of virulence factor encoding genes on Escherichia coli isolated from broiler chicken in Blitar District, Indonesia. Biodiversitas 23(7), 3437–3442. Effendi, M.H., Hartadi, E.B., Witaningrum, A.M., Permatasari, D.A. and Ugbo, E.N. 2022b. Molecular identification of blaTEM gene of extended-spectrum beta-lactamase-producing Escherichia coli from healthy pigs in Malang district, East Java, Indonesia. J. Adv. Vet. Anim. Res. 9(3), 447–452. Effendi, M.H., Tyasningsih, W., Yurianti, Y.A., Rahmahani, J., Harijani, N. and Plumeriastuti, H. 2021a. Presence of multidrug resistance (MDR) and extended beta-spectrum beta-lactamase (ESBL) of Escherichia coli isolated from cloacal swabs of broilers in several wet market in Surabaya, Indonesia. Biodiversitas 22(1), 304–310. Effendi, M.H., Wibisono, F.J., Witaningrum, A.M. and Permatasari, D.A. 2021b. Identification of blaTEM and blaSHV genes of extended spectrum betalactamase (ESBL) producing Escherichia coli from broiler chicken in Blitar, Indonesia. Sys. Rev. Pharm. 12(1), 976–981. Esmaeel, J.R., Saleh, Z.F., Fahad, K.H. and Klaif, S.F. 2023. Phylogenetic analysis of zoonotic Acinobacter spp. Isolated from Geoffroy’s bat (Myotis emarginatus), Northern Iraq. Vet. Integr. Sci. 21(3), 843–851. Fahmi, A.G., Abidin, Z., Kusmana, C. and Noor, E. 2024. Antibiotic consumption and antibiotics occurrence into the environment: a case study of hospital in Metro, Lampung. Curr. Res. Biosci. Biotechnol. 5(2), 22–28. Faridah, H.D., Wibisono, F.M., Wibisono, F.J., Nisa, N., Fatimah, F., Effendi, M.H., Ugbo, E.N., Khairullah, A.R., Kurniawan, S.C. and Silaen, O.S.M. 2023. Prevalence of the blaCTX-M and blaTEM genes among extended-spectrum beta lactamase-producing Escherichia coli isolated from broiler chickens in Indonesia. J. Vet. Res. 67(2), 179–186. Farizqi, M.T.I., Effendi, M.H., Adikara, R.T.S., Yudaniayanti, I.S., Putra, G.D.S., Khairullah, A.R., Kurniawan, S.C., Silaen, O.S.M., Ramadhani, S., Millannia, S.K., Kaben, S.E. and Waruwu, Y.K.K. 2023. Detection of extended-spectrum β-lactamase-producing Escherichia coli genes isolated from cat rectal swabs at Surabaya Veterinary Hospital, Indonesia. Vet. World 16(9), 1917–1925. Foti, M., Grasso, R., Fisichella, V., Mascetti, A., Marco, C., Grasso, M. and Spena, M.T. 2023. Antimicrobial resistance in physiological and potentially pathogenic bacteria isolated in southern Italian Bats. Animals 13(6), 966. Garcês, A., Correia, S., Amorim, F., Pereira, J.E., Igrejas, G. and Poeta, P. 2019. First report on extended-spectrum beta-lactamase (ESBL) producing Escherichia coli from European free-tailed bats (Tadaria teniotis) in Portugal: a one-health approach of a hidden contamination problem. J. Hazard. Mater. 370(1), 219–224. Handal, N., Whitworth, J., Lyngbakken, M.N., Berda, J.E., Dalgard, O. and Jorgensen, S.B. 2023. Mortality and length of hospital stay after bloodstream infections caused by ESBL-producing compared to non-ESBL-producing E. coli. Infect. Dis. (Lond) 56(1), 19–31. Hayer, J., Salgado-Caxito, M., Opazo-Capurro, A., Munoz, P.G., Millan, J., Pineiro, A., Munita, J.M., Rivas, L. and Benavides, J.A. 2023. Multiple clonal transmissions of clinically relevant extended-spectrum beta-lactamase-producing Escherichia coli among livestock, dogs, and wildlife in Chile. J. Glob. Antimicrob. Resist. 34(1), 247–252. Huy, H.L., Koizumi, N., Nuradji, H., Susanti, Noor, S.M., Dharmayanti, N.L.P.I., Haga, T., Hirayama, K. and Miura, K. 2023. Antimicrobial resistance in Escherichia coli isolated from brown rats and house shrews in markets, Bogor, Indonesia. J. Vet. Med. Sci. 83(3), 531–534. Igbinosa, E.O., Beshiru, A., Igbinosa, I.H., Cho, G.S. and Franz, C.M.A.P. 2023. Multidrug-resistant extended spectrum β-lactamase (ESBL)-producing Escherichia coli from farm produce and agricultural environments in Edo State, Nigeria. Plos One 18(3), e0282835. Islam, M.S., Rahman, A.M.M.T., Hassan, J. and Rahman, M.T. 2023. Extended-spectrum beta-lactamase in Escherichia coli isolated from humans, animals, and environments in Bangladesh: a One Health perspective systematic review and meta-analysis. One Health 16(1), 100526. Jaeheng, Y. and Han, H. 2020. Thai street food in the fast growing global food tourism industry: preference and behaviors of food tourist. J. Hosp. Tourism Manage. 45, 641–655. Kanaeva, M.A., Yunaeva, L.L., Burevenskaya, V.Y., Bidzhieva, F.A., Ermakova, N.S. and Litvinov, A.N. 2023. Clinical and experimental substantion of the use of macrolide antibiotic in gastrointestinal and respiratory diseases. Pharmacophore 14(1), 93–99. Kholik, Agustin, A.L.D., Atma, C.D., Munawaroh, M., Ningtyas, N.S.I., Legowo, A.P. and Sukmanadi, M. 2019 Bacteria pathogens from cave-dwelling bats that are a risk to human, animal and environmental health on Lombok Island, Indonesia. Eurasia J. Biosci. 13(2), 1509–1513. Komalasari, F. and Ganiarto, E. 2020. Factore influencing Indonesian millennials travel decision: a case of Bangkok. J. Manag. Stud. 5(2), 158–179. Kristianingtyas, L., Effendi, M.H., Witaningrum, A.M., Wardhana, D.K. and Ugbo, E.N. 2021. Prevalence of extended-spectrum β-lactamase-producing Escherichia coli in companion dogs in animal clinics, Surabaya, Indonesia. Int. J. One Health 7(2), 232–236. Laborda, P., Sanz-Garcia, F., Ochoa-Sanchez, L.E., Gil-Gil, T., Hernando-Amado, S. and Martinez, J.L. 2022. Wildlife and antibiotic resistance. Front. Cell. Infect. Microbiol. 12(1), 873989. Lesmana, H. and Sugiarto, S. 2021. Formulating a competitive advantage model for tourism destinations in Indonesia. J. Asian Finance Econ. Bus. 8(3), 237–249. Li, Y., Li, Q., Ji, Z., Andom, O., Wang, X., Guo, X. and Li, Z. 2023. Current status and spatiotemporal evolution of antibiotic residues in livestock and poultry manure in China. Agriculture 13(10), 1877. Liu, T., Lee, S., Kim, M., Fan, P., Boughton, R.K., Boucher, C. and Joeng, K.C. 2024. A study at the wildlife-livestock interface unveils the potential of feral swine as a reservoir for extended-spectrum β-lactamase-producing Escherichia coli. J. Hazard. Mater. 473(1), 134694. McDougall, F., Boardman, W., Gillings, M. and Power, M. 2019. Bats a reservoirs of antibiotic resistance determinants: a survey of class 1 integrons in Grey-headed Flying Foxes (Pteropus poliocephalus). Infect. Genet. Evol. 70(1), 107–113. McDougall, F.K., Boardman, W.S.J. and Power, M.L. 2021 Characterization of beta-lactam-resistant Escherichia coli from Australian fruit bats indicates anthropogenic origins. Microb. Genom. 7(5), 000571. Modupe, A.O., Olufunke, O.A., Abike, T.O., Oluwayemisi, O.A. and Oladiran, F. 2022. Molecular characterization of resistance and virulance genes in Escherichia coli isolated from bats (Eidolon helvum) faeces in Onsu State, Nigeria. J. Adv. Microbiol. 22(2), 37–48. Mou, T.J., Nupur, N.A., Haque, F., Islam, M.F., Rahman, M.S., Huq, M.A. and Parvez. 2023 Characterization of extended spectrum beta-lactamase (ESBL) producing Escherichia coli isolates from surface water adjacent to pharmaceutical industries in Bangladesh: antimicrobial resistance and virulance pattern. Microbiol. Biotechnol. Lett. 51(3), 268–279. Musa, L., Stefanetti, V., Proietti, P.C., Grilli, G., Gobbi, M., Toppi, V., Brustenga, L., Magistrali, C.F. and Franciosini, M.P. 2023. Antimicrobial susceptibility of commensal E. coli isolated from wild bird in Umbria (Central Italy). Animals 13(11), 1776. Mustika, Y.R., Kinasih, K.N., Effendi, M.H., Puspitasari, Y., Kurniawan, S.C., Khairullah, A.R., Samodra, M.E.E., Hasib, A., Agustin, A.L.D., Moses, I.B. and Silaen, O.S.M. 2024. Molecular detection of extended-spectrum β-lactamase-producing Escherichia coli from bat cave on Lombok Island. Open Vet. J. 14(2), 699–706. Naas, T., Dabos, L. and Bonnin, R.A. 2023. β-lactamase genes without limits. Microorganism 11(5), 1200. Naelasari, D.N., Koendhori, E.B., Dewanti, L., Sulistiawati, Sarassari, R. and Kuntaman, K. 2018. The prevalence of extended spectrum β-lactamase (ESBL) producing gut bacterial floral among patients in dr. Soetomo hospital and primary health care in Surabaya. Folia Med. Indones. 54(4), 256–262. Nangoy, M., Ransaleleh, T., Lengkong, H., Koneri, R., Latinne, A. and Kyes, R.C. 2021. Diversity of fruit bats (Pteropodidae) and their ectoparasites in Batupulih Nature Tourism Park, Sulawesi, Indonesia. Biodiversitas 22(6), 3075–3082. Nguema, P.P.M., Onanga, R., Atome, G.R.N., Mbeang, J.C.O., Mabika, A.M., Yaro, M., Lounnas, M., Dumont, Y., Zohra, Z.F., Godreuil, S. and Bretagnolle, F. 2020. Characterization of ESBL-producing Enterobacteria from fruit bats in an unprotected area of Makokou, Gabon. Microorganisms 8(1), 138. Nuangmek, A., Rojanasthien, S., Yamsakul, P., Tadee, P., Thamlikitkul, V., Tansakul, N. and Patchanee, P. 2020. Perspectives on antimicrobial use in pig and layer farm in Thailand: legislation, policy, regulation and potential. Vet. Integr. Sci. 19(1), 1–21. Obodoechi, L.O., Carvalho, I., Chenouf, N.S., Martinez-Alvarez, S., Sadi, M., Nwanta, J.A., Chah, K.F. and Torres, C. 2021. Antimicrobial resistance in Escherichia coli isolates from frugivorous (Eidolon helvum) and insectivorous (Nycteris hispida) bats in Southeast Nigeria, with detection of CTX-M-15 producing isolates. Comp. Immunol. Microbiol. Infect. Dis. 75(1), 101613. Palma, N., Pons, M.J., Gomes, C., Mateu, J., Riveros, M., Garcia, W., Jacobs, J., Garcia, C., Ochoa, T.J. and Ruiz, J. 2017. Resistance to quinolones, cephalosporins and macrolides in Escherichia coli causing bacteraemia in Peruvian Children. J. Glob. Antimicrob. Resist. 11(1), 28–33. Prayudi, S.K.A., Effendi, M.H., Lukiswanto, B.S., Zahra, R.L., Benjamin, M.I., Kurniawan, S.C., Khairullah, A.R., Silaen, O.S.M., Lisnanti, E.F., Baihaqi, Z.A., Widodo, A. and Riwu, K.H.P. 2023. Detection of genes on Escherichia coli producing extended spectrum β-lactamase isolated from the small intestine of ducks in traditional markets Surabaya City, Indonesia. J. Adv. Vet. Res. 13(8), 1600–1608. Puangseree, J., Prathan, R., Srisanga, S., Angkittitrakul, S. and Chuanchuen, R. 2022. Plasmid profile analysis of Escherichia coli and Salmonella enterica isolated from pigs, pork and humans. Epidemiol. Infect. 150, e110. Pungpian, C., Sinwat, N., Angkititrakul, S., Prathan, R. and Chuanchuen, R. 2021. Presence and transver of antimicrobial resistance determinants in Escherichia coli in pigs, pork, and humans in Thailand and Lao PDR border province. Microb. Drug Resist. 27(4), 571–584. Putri, M.F.R., Kendek, I.A., Wibisono, F.J., Effendi, M.H., Rahardjo, D., Tyasningsih, W. and Ugbo, E.N. 2023. Molecular detection of iron gene on multidrug resistant avian fecal Escherichia coli isolated from broiler on traditional markets, Surabaya, Indonesia. Biodiversitas 24(12), 6454–6460. Raffelsberger, N., Buczek, D.J., Svendsen, K., Smabrekke, L., Pontinen, A.K., Lohr, I.H., Andreassen, L.L.E., Simonsen, G.S., Sundsfjord, A., Graviningen, K. and Samuelsen, Ø. 2023. Community carriage of ESBL-producing Escherichia coli and Klebsiella pneumoniae: a cross-sectional study of risk factor and comparative genomics of carriage and clinical isolates. mSphere 8(4), e00025–23. Rana, C., Rajput, S., Behera, M., Gautam, D., Vikas, V., Vats, A., Roshan, M., Ghorai, S.M. and De, S. 2022. Global epidemiology of CTX-M-type β-lactam in human and animal. Comp. Immunol. Microbiol. Infect. Dis. 86(1), 101815. Rantsiou, K., Kathariou, S., Winkler, A., Skandamis, P., Saint-Cry, M.J., Rouzeau-Szynalski, K. and Amezquita, A. 2018. Next generation microbiological risk assessment: opportunities of whole genome sequencing (WGS) for foodborne pathogen surveillance, source tracking and risk assessment. Microbiology. 287, 3–9. Riwu, K.H.P., Effendi, M.H., Rantam, F.A., Khairullah, A.R., Kurniawan, S.C., Kurniawan, A., Mosem, I.B., Hasib, A., Widodo, A., Yanestria, S.M., Silaen, O.S.M. and Shehzad, A. 2024. Molecular detection of blaTEM gene for encoding extended spectrum beta-lactamase (ESBL) on Escherichia coli isolated from deer feces in Indonesia. J. Adv. Vet. Res. 14(4), 722–726. Riwu, K.H.P., Effendi, M.H., Rantam, F.A., Khairullah, A.R. and Widodo, A. 2022. A review: virulence factors of Klebsiella pneumonia as emerging infection on the food chain. Vet. World 15(9), 2172–2179. Rojas-Sereno, Z.E., Streicker, D.G., Suarez-Yana, T., Lineros, M., Yung, V., Godreuil, S. and Benavides, J.Z. 2023. Detection of antimicrobial-resistant Enterobacterales in insectivorous bats from Chile. R. Soc. Open Sci. 10(11), 231177. Sah, R.K., Dahal, P., Parajuli, R., Giri, G.R. and Tuladhar, E. 2024. Prevalence of blaCTX-M and blaTEM genes in cefotaxime-resistant Escherichia coli recovered from tertiary care at Central Nepal: a descriptive cross-sectional study. Can. J. Infect. Dis. Med. Microbiol. 2024(1), 5517662. Saitou, N. and Nei, M. 1987. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 4(4), 406–425. Satyaprakash, K., Pesingi, P.K., Das, A., Vineeth, M.R., Malik, S.V.S., Barbuddhe, S.B. and Rawool, D.B. 2024. Occurrence of multidrug-resistant (MDR) extended-spectrum beta-lactamase (ESBL)-producing Escherichia coli in wastewater and natural water sources from the Eastern part of Uttar Pradesh, India. Water Air Soil Pollut. 235(2), 125. Scott, L.C., Wilson, M.J., Esser, S.M., Lee, N.L., Wheeler, M.E., Aubee, A. and Aw, T.G. 2021. Assessing visitor use impact on antibiotic resistant bacteria and antibiotic resistance genes in soil and water environments of Rocky Mountain National Park. Sci. Total Environ. 785, 147122. Sinuraya, R.K., Wulandari, C., Amalia, R. and Puspitasari, I.M. 2023. Understanding public knowledge and behavior regarding antibiotic use in Indonesia. Infect. Drug Resist. 16(1), 6833–6842. Sornsenee, P., Chimplee, S., Arbubaker, A., Kongchai, S., Madimong, H. and Romyasamit, C. (2021) Occurrence, antimicrobial resistance profile, and characterization of extended-spectrum β-lactamase-producing Escherichia coli isolated from miced meat at local markets in Thailand. Foodborne Pathog. Dis. 19(3), 232–240. Ssebuganda, R., Lusiba, G. and Akampurira, E. 2023. Co-resistance between Flourquinolones, SXT), and aminoglycosides with extended spectrum β-lactamases (ESBLs) in Escherichia coli isolated from blood, pus and urine specimens at Makerere university clinical microbiology laboratory. [Dissertation]. Stastny, K., Hodkovicova, N., Jerabek, M., Petren, M., Viskova, M., Papouskova, A., Bartejsova, I., Putecova-Tosnerova, K., Charvatova, M., Zouharova, M., Matiaskova, K. and Nedbalcova, K. 2024. Dosage optimisation of trimrthroprim and sulfamethoxazole for treatment of avian pathogenic strain of Escherichia coli in broiler chickens. Antibiotics 13(1), 11. Tagkoskul, T., Thamthaweechok, N, Seenama, C. and Thamlikitkul, V. 2021. Antibiotic-resistant bacteria and antibiotic residue contamination in fresh raw food sold at Wholesale market in Thailand. J. Med. Assoc. Thailand 104(4), 654–622. Trott, D.J., Turnidge, J., Kovac, J.H., Simjee, S., Wilson, D. and Watts, J. 2021. Comparative macrolide use in humans and animals: should macrolides be moved off the World Health Organisation’s critically important antimicrobial list? J. Antimicrob. Chemother. 76(8), 1955–1961. Tseng, C.H., Liu, C.W. and Liu, P.Y. 2023. Extended-spectrum β-lactamases (ESBL) producing bacteria in animals. Antibiotics 12(4), 661. Tyasningsih, W., Ramandinianto, S.C., Ansharieta, R., Witaningrum, A.M., Permatasari, D.A., Wardhana, D.K., Effendi, M.H. and Ugbo, E.N. 2022. Prevalence and antibiotic resistance of Staphylococcus aureus and Escherichia coli isolated from raw milk in East Java, Indonesia. Vet. World 15(8), 2021–2028. Ugbo, E.N., Jacob, J.I., Effendi, M.H., Witaningrum, A.M., Agumah, B.N., Ugbo, A.I. and Moses, B.I. 2023. Poultry slaughterhouse wastewater as reservoirs for spreading extended-spectrum beta-lactamase-producing Escherichia coli in Abakaliki, Nigeria. Biodiversitas 24(9), 4960–4966. Vanstokstraeten, R., Pierard, D., Crombe, F., De Geyter, D., Wybo, I., Muyldermans, A., Seyler, L., Caljon, B., Janssen, T. and Demuyser, T. 2023. Genotypic resistance determined by whole genome sequencing versus phenotypic resistance in 234 Escherichia coli isolates. Sci. Rep. 13(1), 449. Wartiani, W., Maulida, A., Puspitasari, R. L. and Jumianto, S. 2023. Analysis of Escherichia coli extended-spectrum beta-lactamase ESBL bacteria in chicken farm waste in Cipondoh, Tangerang City. RSF Conf. Ser.: Bus. Manag. Soc. Sci. 3(2), 37–41. Wibisono, F.J., Sumiarto, B., Effendi, M.H., Permatasari, D.A. and Witaningrum, A.M. 2021. Molecular identification of ctxgene of extended spectrum beta-lactamases (ESBL) producing Escherichia coli on layer chicken in Blitar, Indonesia. J. Anim. Plant Sci. 31(4), 954–959. Widodo, A., Lamid, M., Effendi, M.H., Khairullah, A.R., Riwu, K.H.P., Yunistinasari, L.R., Kurniawan, S.C., Ansori, A.N.M., Silaen, O.S.M. and Dameanti, F.N.A.E.P. 2022 Antibiotic sensitivity profile of multidrug-resistant (MDR) Escherichia coli isolated from dairy cow’s milk in Probolinggo, Indonesia. Biodiversitas 23(10), 4971–4976. Widodo, A., Lamid, M., Effendi, M.H., Tyasningsih, W., Raharjo, D., Khairullah, A.R., Kurniawan, S.C., Yustinasari, L.R., Riwu, K.H.P. and Silaen, O.S.M. 2023. Molecular identification of bla TEM and bla CTX-M genes in multidrug-resistant Escherichia coli found in milk samples from dairy farms in Tulungagung, Indonesia. J. Vet. Res. 67(3), 381–388. Wulandari, L.P.L., Khan, M., Liverani, M., Ferdiana, A., Mahuri, Y.A., Probandari, A., Wibawa, T., Batura, N., Schierhout, G., Kaldor, J., Guy, R., Law, M., Day, R., Hanefeld, J., Parathon, H., Jan, S., Yeung, S. and Wiseman, V. 2021. Prevalence and determinants of inappropriate antibiotic dispensing at private drug retail outlets in urban and rural areas of Indonesia: a mixed methods study. BMJ Glob. Health 6(8), e004993. Yanestria, S.M., Dameanti, F.N.A.E.P., Musayannah, B.G., Pratama, J.W.A., Witaningrum, A.M., Effendi, M.H. and Ugbo, E.N. 2022. Antibiotic resistance pattern of Extended-Spectrum β-Lactamase (ESBL) producing Escherichia coli isolated from broiler farm environment in Pasuruan district, Indonesia. Biodiversitas 23(9), 4460–4465. Yu, Z., Liu, Z., Sun, L., Dong, C., Jin, Y., Hu, B. and Cheng, D. 2024. Mobile genetic elements mediate the cross-media transmissions of antibiotics resistance genes from pig farms and their risks. Sci. Total Environ. 926(1), 172115. Zalewska, M., Blazejewska, A., Czapko, A. and Popowska, M. 2023. Pig manure treatment strategies for mitigating the spread of antibiotic resistance. Sci. Rep. 13(1), 11999. Zurita, J., Yanez, F., Sevillano G., Orgeta-Paredes, D. and Mino, A.P.Y. 2020. Ready-to-eat street food: a potential source for dissemination of multidrug-resistant Escherichia coli epidemic clones in Quito, Equador. Lett. Appl. Microbiol. 70(3), 203–209. | ||
| How to Cite this Article |
| Pubmed Style Agustin ALD, Effendi MH, Tyasningsih W, Plumeriastuti H, Khairullah AR, Ekawasti F, Moses IB, Kinasih KN, Kusala MKJ, Mustika YR, Fauziah I, Wibowo S. Phylogenetic analysis blaTEM gene of Escherichia coli isolated from cave bats in West Nusa Tenggara Province, Indonesia. Open Vet. J.. 2024; 14(12): 3460-3473. doi:10.5455/OVJ.2024.v14.i12.30 Web Style Agustin ALD, Effendi MH, Tyasningsih W, Plumeriastuti H, Khairullah AR, Ekawasti F, Moses IB, Kinasih KN, Kusala MKJ, Mustika YR, Fauziah I, Wibowo S. Phylogenetic analysis blaTEM gene of Escherichia coli isolated from cave bats in West Nusa Tenggara Province, Indonesia. https://www.openveterinaryjournal.com/?mno=219520 [Access: November 23, 2025]. doi:10.5455/OVJ.2024.v14.i12.30 AMA (American Medical Association) Style Agustin ALD, Effendi MH, Tyasningsih W, Plumeriastuti H, Khairullah AR, Ekawasti F, Moses IB, Kinasih KN, Kusala MKJ, Mustika YR, Fauziah I, Wibowo S. Phylogenetic analysis blaTEM gene of Escherichia coli isolated from cave bats in West Nusa Tenggara Province, Indonesia. Open Vet. J.. 2024; 14(12): 3460-3473. doi:10.5455/OVJ.2024.v14.i12.30 Vancouver/ICMJE Style Agustin ALD, Effendi MH, Tyasningsih W, Plumeriastuti H, Khairullah AR, Ekawasti F, Moses IB, Kinasih KN, Kusala MKJ, Mustika YR, Fauziah I, Wibowo S. Phylogenetic analysis blaTEM gene of Escherichia coli isolated from cave bats in West Nusa Tenggara Province, Indonesia. Open Vet. J.. (2024), [cited November 23, 2025]; 14(12): 3460-3473. doi:10.5455/OVJ.2024.v14.i12.30 Harvard Style Agustin, A. L. D., Effendi, . M. H., Tyasningsih, . W., Plumeriastuti, . H., Khairullah, . A. R., Ekawasti, . F., Moses, . I. B., Kinasih, . K. N., Kusala, . M. K. J., Mustika, . Y. R., Fauziah, . I. & Wibowo, . S. (2024) Phylogenetic analysis blaTEM gene of Escherichia coli isolated from cave bats in West Nusa Tenggara Province, Indonesia. Open Vet. J., 14 (12), 3460-3473. doi:10.5455/OVJ.2024.v14.i12.30 Turabian Style Agustin, Alfiana Laili Dwi, Mustofa Helmi Effendi, Wiwiek Tyasningsih, Hani Plumeriastuti, Aswin Rafif Khairullah, Fitrine Ekawasti, Ikechukwu Benjamin Moses, Kurnia Nisa Kinasih, Muhammad Khaliim Jati Kusala, Yolla Rona Mustika, Ima Fauziah, and Syahputra Wibowo. 2024. Phylogenetic analysis blaTEM gene of Escherichia coli isolated from cave bats in West Nusa Tenggara Province, Indonesia. Open Veterinary Journal, 14 (12), 3460-3473. doi:10.5455/OVJ.2024.v14.i12.30 Chicago Style Agustin, Alfiana Laili Dwi, Mustofa Helmi Effendi, Wiwiek Tyasningsih, Hani Plumeriastuti, Aswin Rafif Khairullah, Fitrine Ekawasti, Ikechukwu Benjamin Moses, Kurnia Nisa Kinasih, Muhammad Khaliim Jati Kusala, Yolla Rona Mustika, Ima Fauziah, and Syahputra Wibowo. "Phylogenetic analysis blaTEM gene of Escherichia coli isolated from cave bats in West Nusa Tenggara Province, Indonesia." Open Veterinary Journal 14 (2024), 3460-3473. doi:10.5455/OVJ.2024.v14.i12.30 MLA (The Modern Language Association) Style Agustin, Alfiana Laili Dwi, Mustofa Helmi Effendi, Wiwiek Tyasningsih, Hani Plumeriastuti, Aswin Rafif Khairullah, Fitrine Ekawasti, Ikechukwu Benjamin Moses, Kurnia Nisa Kinasih, Muhammad Khaliim Jati Kusala, Yolla Rona Mustika, Ima Fauziah, and Syahputra Wibowo. "Phylogenetic analysis blaTEM gene of Escherichia coli isolated from cave bats in West Nusa Tenggara Province, Indonesia." Open Veterinary Journal 14.12 (2024), 3460-3473. Print. doi:10.5455/OVJ.2024.v14.i12.30 APA (American Psychological Association) Style Agustin, A. L. D., Effendi, . M. H., Tyasningsih, . W., Plumeriastuti, . H., Khairullah, . A. R., Ekawasti, . F., Moses, . I. B., Kinasih, . K. N., Kusala, . M. K. J., Mustika, . Y. R., Fauziah, . I. & Wibowo, . S. (2024) Phylogenetic analysis blaTEM gene of Escherichia coli isolated from cave bats in West Nusa Tenggara Province, Indonesia. Open Veterinary Journal, 14 (12), 3460-3473. doi:10.5455/OVJ.2024.v14.i12.30 |