| Research Article | ||
Open Vet. J.. 2025; 15(1): 289-299 Open Veterinary Journal, (2024), Vol. 15(1): 289-299 Research Article Genotypic characteristics and phylogenetic tree analysis of Staphylococcus aureus isolates from sheep subclinical mastitis milkAlaa Shamil Alalaf1*, Ayman Hani Taha2 and Omar Hashim Sheet21Department of Animal Production, College of Agriculture and Forestry, University of Mosul, Mosul, Iraq 2Department of Veterinary Public Health, College of Veterinary Medicine, University of Mosul, Mosul, Iraq *Corresponding Author: Alaa Shamil Alalaf. Department of Animal Production, College of Agriculture and Forestry, University of Mosul, Mosul, Iraq. Email: Alaa.shamil [at] uomosul.edu.iq Submitted: 06/10/2024 Accepted: 02/12/2024 Published: 31/01/2025 © 2025 Open Veterinary Journal
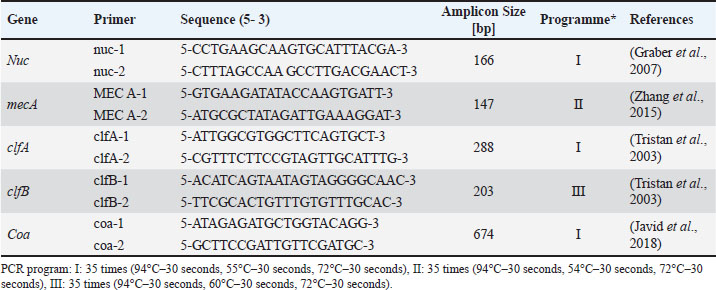
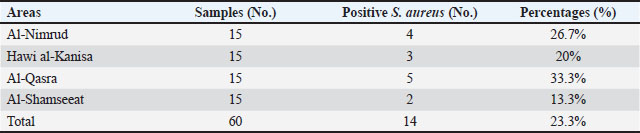
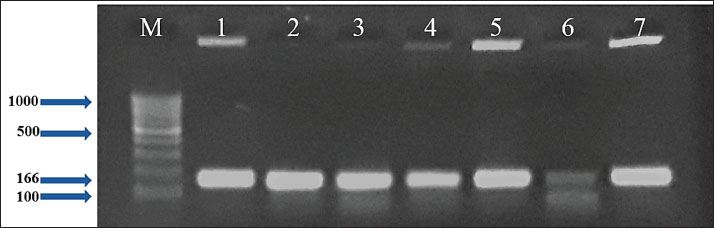
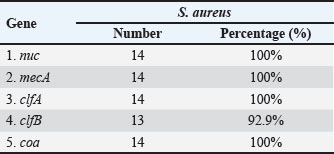
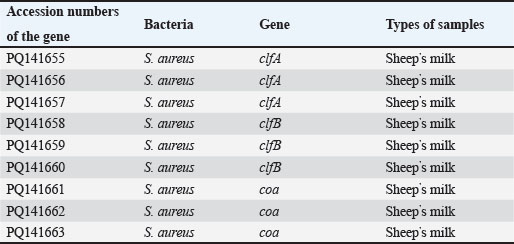
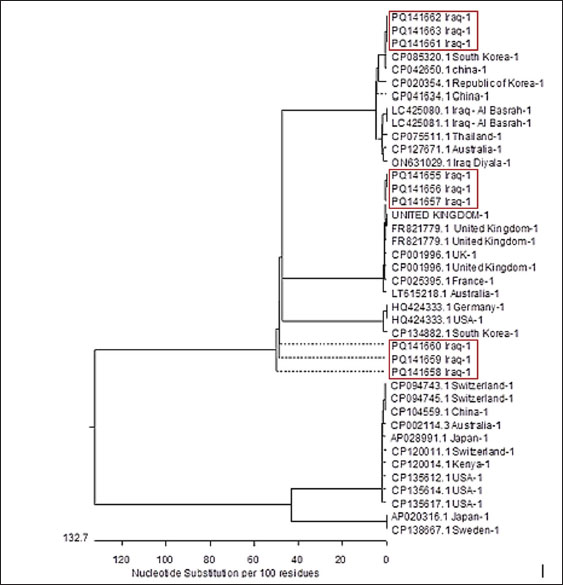
AbstractBackground: Staphylococcus aureus describes the greatest common cause of mastitis in sheep; however, little information was available concerning methicillin-resistant S. aureus (MRSA) in the milk of mastitis sheep. Aim: The current study aimed to find exactly the incidence of S. aureus and MRSA in milk samples from sheep farmhouses in Mosul city, in addition to discovering specific genes that encode virulence factors. Methods: Samples of 60 sheep’s subclinical milk were gathered between October 2023 and February 2024 from various places around the Nineveh governorate. Standard methods that include selective media and biochemical testing were utilized in the current study. Furthermore, the nuc, mecA, clfA, clfB, and coa genes in S. aureus were determined utilizing a polymerase chain reaction technique. Results: According to our findings, S. aureus was found in sheep subclinical mastitis at a rate of 23.3% (14/60) in sheep milk. Furthermore, all S. aureus isolates 100% appeared to carrying of the nuc, mecA, clfA, and coa genes, while 92.9% of isolates were positive for the clfB gene. The gene profiles of S. aureus isolates were divided into two primary categories. Based on the clfA, clfB, and coa genes, nine unique strains of S. aureus sequences were submitted to the NCBI GenBank. The phylogenetic tree analysis of all study isolates demonstrated its identity to various strains in the NCBI-BLAST. Conclusion: The incidence of S. aureus infections emphasized the presence of an issue in husbandry practices, insufficient adherence to basic hygiene protocols, and the random use of antibiotics. Keywords: Phylogenetic analysis tree, S. aureus, Sheep milk, Virulence factors. IntroductionDairy ruminants are frequently infected with intra-mammary infections (IMI), which represent a widespread disease that leads to significant financial losses (Kossaibati and Esslemont, 1997). The expenses associated with mastitis directly relate to veterinarian services and raise labor needs (Lescourret and Coulon, 1994). Poor milk quality and yield as a consequence of mastitis is the primary economic loss linked to all forms of mastitis (Seegers et al., 2003). In herds of sheep, the yearly prevalence of the clinical IMI was typically less of 5%. However, in a limited number of large groups of sheep, the frequency of a disease may surpass 30%–50% of the ruminant, resulting in a death rate (gangrenous mastitis) or killing the infected animals up to 70% of the herd (Bergonier et al., 2003). Furthermore, subclinical or clinical Staphylococcus aureus IMI cases have an effect on 3%–37% of groups of sheep and are significant of the legal, sanitary, and financial reasons. These situations additionally affect consumers of dairy products (Bergonier et al., 2003). Although over 100 distinct kinds of microorganisms are identified in the udder of ruminants, just a small percentage of these microbes can lead to mastitis (Owens and Watts, 1988). The microorganisms causing inflammation of mammary glands can be classified into two groups based on their reservoir, source, and route of transmission: contagious bacteria and environmental bacteria (Ruegg, 2012). Staphylococcus aureus is regarded as a major pathogen found in sheep, causing both preclinical and symptomatic intra-mammary infections; some of these isolates may have a broad geographical distribution (Smith et al., 2005). According to a number of previous studies, S. aureus isolated from cows’ mastitis (Sheet, 2022) and camel milk (Sheet et al., 2021). Many researchers showed that S. aureus is found in various types of food such as milk and its products (Vyletělová et al., 2011), meat (Sheet et al., 2023), and fish (Taha et al., 2024). Numerous virulence factors are found in S. aureus, and the various combinations of these factors, along with the infection place and the host immune response’s heterogeneity, result in the vast range of outcomes linked to these infections (Lowy, 1998). The degree and indicators of S. aureus infections correspond with virulence factors (Xing et al., 2014). Among them, foodborne poisoning is typically caused by staphylococcal enterotoxins (SEs). Food poisoning resulting from S. aureus harboring SEs may occur quickly, progress swiftly, and present a serious risk to human health (Basanisi et al., 2017). Furthermore, some S. aureus strains produce a variety of toxins, including exfoliative toxins A and B, toxic shock syndrome toxin-1 (tsst), SEs, and toxic shock syndrome toxin-like (SEls). It is widely recognized that food-related illnesses are mainly caused by SEs or SEls (Argudín et al., 2010). According to reports, these five classic enterotoxins account for 95% of staphylococcal food poisoning cases, with recently identified SEs responsible for the other 5% of infections (Papadopoulos et al., 2019). Staphylococcus aureus isolates can be detected using a variety of techniques. The conventional techniques depend on conventional biochemical testing and the structural characteristics of S. aureus colonies. Molecular techniques are more precise and quicker, yielding findings in 3–5 hours for the recognition of S. aureus (Ahmed et al., 2020). There are various molecular recognition techniques, such as loop-mediated isothermal amplification, real-time polymerase chain reaction, and polymerase chain reaction (PCR). Moreover, all genes encoding virulence factors have been identified by molecular techniques. Due to the significance of pathogenic S. aureus in subclinical mastitis in sheep, the objective of our study is to recognize the pathogen S. aureus in the milk of sheep infected via subclinical mastitis in the governorate of Nineveh, to determine which S. aureus strain is resistant to methicillin and which genes in S. aureus isolates encode virulence factors. Materials and MethodsSamples collectionBased on the California mastitis test, 60 sheep subclinical milk samples were obtained from different locations across the governorate between October 2023 and February 2024. These regions included Al-Nimrud, Hawi al-Kanisa, Al-Qasra, and Al-Shamseeat. The samples of milk were gathered in clean bottles and transferred directly to the health laboratory at the Faculty of Veterinary Medicine, Mosul University. After that, all of the peptone water containers were put in an incubator and permitted to go for 18–24 hours pre-enrichment process at 37°C. All samples streaked on media (blood and 7.5% mannitol salt agar), followed by they were incubated for a full day at 37°C. Staphylococcus aureus isolation and characterizationPhenotypic evaluation, coagulase and catalase activity tests, and gram staining were used for the investigation to identify characteristic S. aureus colonies (Quinn et al., 2011). Isolation of DNAThe positive isolates were grown overnight at 37°C on the mannitol salt medium with the purpose of getting the S. aureus genomic DNA. The DNA was isolated utilizing the DNeasy Blood and Tissue Kit (Qiagen, Germany), depending on the manufacturer’s instructions specific to Gram-positive bacteria. The isolated DNA was preserved at −20°C after being measured via Nanodrop (Biodrop, UK). Reaction of PCRThe nuc, mecA, clfA, clfB, and coa genes of S. aureus were detected by the PCR technique. The molecular weight of the nuc gene was 166 bp (Graber et al., 2007), mecA was 147 bp (Zhang et al., 2015), clfA was 288 bp (Tristan et al., 2003), clfB was 203 bp (Tristan et al., 2003), and coa was 674 bp (Javid et al., 2018). The PCR reaction required 30 μl in total volume, and the mixture was performed in a 200-μl tube (Biozym, Oldenhorf, Germany). The reaction mixture comprised 9 μl of DNeasy-free water from Promega Corporation (USA) and 4 μl of the S. aureus DNA template, 15 μl of the GoTaq Green Mix Master (2×) from Promega Corporation, 1 μl of each primer 1 and 2. The PCR product applied through a gel electrophoresis evaluation on a 2% agarose gel (Peqlab, Erlangen, Germany), utilizing a reference of a 100 bp ladder. DNA sequencingThe isolated DNA was sent to Macrogen Company, a commercial sequencing firm based in South Korea, for the purpose of purifying and sequencing nine PCR amplicons obtained from isolates of milk sheep that had all already been confirmed the S. aureus isolates were positive using conversation PCR. The clfA, clfB, and coa genes were intended to be the target genes for sequencing. The obtained sequences for the clfA, clfB, and coa genes were then contrasted using the NCBI BLASTn software, available at http://www.ncbi.nlm.nih.gov, to previously described S. aureus sequences that are accessible in GenBank. Additional examination of the alignment and comparability of these sequences was performed using GenomeNet’s online multiple sequence alignment tool CLUSTALW (http://www.genome.jp/tools/clustalw/). Phylogenetic trees were created using the neighbor-joining program and the same genome net tool CLUSTALW. To improve robustness, 500 duplicate sequences of the S. aureus clfA, clfB, and coa genes were included as an outgroup while creating the phylogenetic tree. Through purification, sequencing, and subsequent bioinformatics analysis, this all-encompassing technique sought to shed light on the genetic relationships among the S. aureus isolates and enhance comprehension of the phylogenetic context of the isolates from milk sheep. Ethical approvalAll samples were obtained with the owners’ agreement and used according to the ethical criteria issued by the Institutional Animal Care and Use Committee at Mosul University’s College of Veterinary Medicine, using the approved ID of UM. Vet. 2024.028. ResultsThe S. aureus colonies that provided positive results appeared on mannitol salt agar with a golden-yellowish color. Furthermore, specific biochemical tests that confirmed the existence of S. aureus, such as the coagulase and catalase assays, produced positive results. Our study found the prevalence rate of the S. aureus isolates in this investigation was 23.3% (14/60). The greatest percentage of S. aureus isolated in sheep was 33.3% (5/15) in the Al-Qasra district. Following, the prevalence rate of S. aureus in Al-Nimrud and Hawi al-Kanisa was 26.7% (4/15) and 20% (3/15). The lowest ratio of S. aureus isolated was in Al-Shamseeat 13.3 (2/15) as shown in Table 2. The PCR assay results corroborated the findings obtained from conventional methods Table 1, demonstrating that 100% (14/14) of S. aureus carried the nuc gene (Fig. 1). According to Table 3, all S. aureus isolated were methicillin-resistant S. aureus (MRSA), which possessed the mecA gene 100% (14/14) (Fig. 2). Furthermore, this study appeared that all S. aureus isolates have the clfA gene and coa gene 100% (14/14) (Fig. 3) (Fig. 5). Additionally, 92.9% (13/14) of the S. aureus had the clfB gene (Fig. 4). Moreover, the current study’s results showed that S. aureus was split into two distinct gene profiles according to the presence of distinct genes in each isolate (Table 4). The S. aureus isolates showed that the most frequent genes profile I (nuc + mecA + clfA + clfB + coa) was 13 (92.9%) and genes profile II (nuc + mecA + clfA + coa) was 1 (7.1%). The individual sequencing analysis (BLASTn) was conducted on nine novel gene sequences (three novel clfA gene sequences, three novel clfB gene sequences, and three novel coa gene sequences) that were derived from the subclinical mastitis milk of sheep, corresponding to the sequencing results of this research. Table 4 reveals that the NCBI Genbank has S. aureus sequences that are indexed under the following accession numbers: PQ141655, PQ141656, PQ141657, PQ141658, PQ141659, PQ141660, PQ141661, PQ141662, and PQ141663. Furthermore, a maximum likelihood phylogenetic tree analysis using the MegAlign program found that local variation gene sequences varied significantly from earlier identified ones that were listed in the GenBank database. Moreover, there is a strong 100% relationship among the S. aureus sequence types depending on the clfA gene (PQ141655, PQ141656, and PQ141657) and the CP001996.1 and CP025395.1 sequence types from the United Kingdom and France. Nevertheless, the sequence types (PQ141655, PQ141656, and PQ141657) in the current research are comparable to FR821779.1 from the United Kingdom and LT615218.1 from Australia by 99.6%–98.3%, respectively. In addition, the sequence types PQ141658, PQ141659, and PQ141660 of S. aureus depending on the clfB gene show 99.4% similarities with CP134882.1 from South Korea, and 98.8% similarities with each of HQ424333.1 from Germany and HQ424333.1 from the United States. The S. aureus sequence types identified via clfA and clfB failed to show any resemblance to the S. aureus sequence types isolated from Iraq. Additionally, an important relationship was seen between the sequence types and the S. aureus sequence types dependent on the coa gene (PQ141661, PQ141662, and PQ141663) and CP042650.1 Chinese and CP085320.1 South Korea acquired a perfect score was 100%. Further, there was a 95% resemblance between the sequence types (PQ141661, PQ141662, and PQ141663) and LC425080.1 and LC425081.1 Al Basra, Iraq. Furthermore, the sequence types of S. aureus (PQ141661, PQ141662, and PQ141663) had similarities with CP041634.1 Thailand and CP075511.1 China by 95% and 97.2%, respectively (Figs. 6 and 7). Table 1. Various primers used in PCR programs for detecting the genes of S. aureus.
Table 2. The number of samples and percentage of positive S. aureus isolated from sheep milk.
Fig. 1. Two percent agarose gel electrophoresis displaying the amplification of the nuc gene with molecular weight (166 bp) in S. aureus isolates. Table 3. The number and percentage of the genes found in S. aureus isolates.
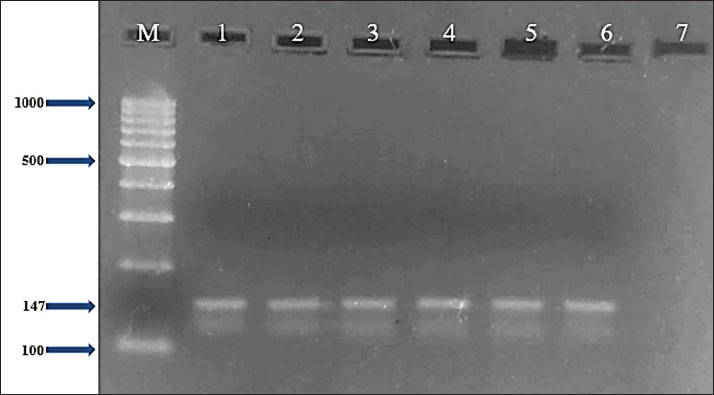
Fig. 2. Two percent agarose gel electrophoresis displaying the amplification of the mecA gene with molecular weight (147 bp) in S. aureus isolates.
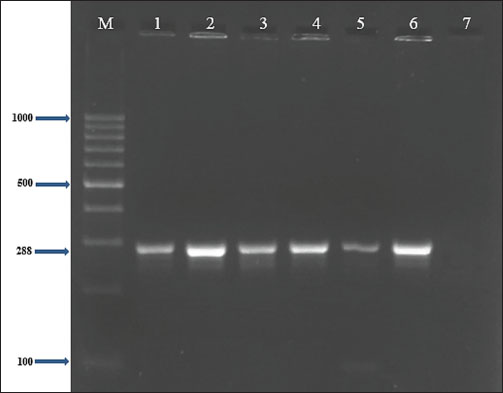
Fig. 3. Two percent agarose gel electrophoresis displaying the amplification of the clfA gene with molecular weight (288 bp) in S. aureus isolates.
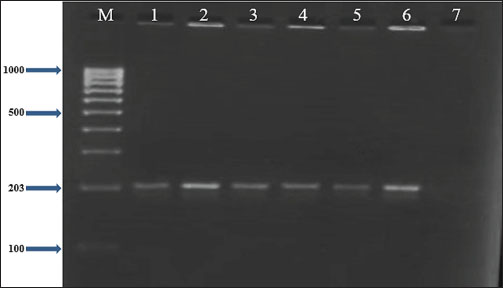
Fig. 4. Two percent agarose gel electrophoresis displaying the amplification of the clfB gene with molecular weight (203 bp) in S. aureus isolates.
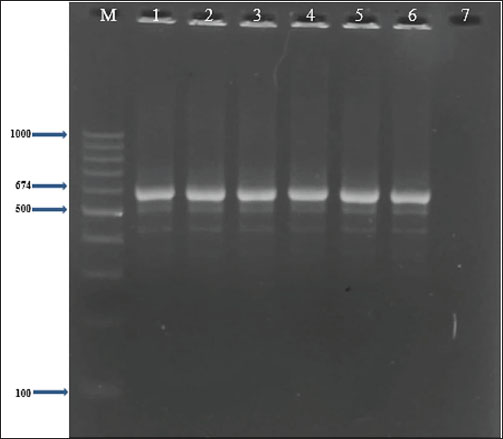
Fig. 5. Two percent agarose gel electrophoresis displaying the amplification of the coa gene with molecular weight (674 bp) in S. aureus isolates. Table 4. Types of the gene profiles of the S. aureus isolates (n=14) from sheep milk.
Table 5. The NCBI GenBank accession numbers for the clfA, clfB, and coa genes of S. aureus sequences in sheep’ s milk.
Fig. 6. Clustering analysis of S. aureus isolates’ gene sequences recovered from NCBI. DiscussionThe study intended to reveal the incidence of pathogenic S. aureus in sheep milk of subclinical mastitis-diagnosed cases in Nineveh Governorate, to identify the methicillin-resistant strain of Staphylococcus aureus and recognize the genes encoded virulence factors of S. aureus. Similar findings have been reported in Iraq, where S. aureus was isolated from subclinical mastitis in camels (Sheet, 2022), cattle (Sheet et al., 2021). In addition, this study found that the prevalence of S. aureus isolates from subclinical mastitis was 23.3% (14/60), with differences in prevalence ratios across different districts in Nineveh governorate. Globally, the prevalence of S. aureus in sheep and small ruminants is higher, ranging from 50% to 75% in other countries such as the USA, China, Slovakia, and Italy (De Centorbi et al.,1992; Moroni et al., 2005, Kotzamanidis et al., 2021; Király et al., 2024). The percentage in the examined sample from sheep milk with subclinical mastitis was 20% in Greece through the milking period (Vasileiou et al., 2019) and 27.38% in Algeria (Azzi et al., 2020), which is close to the ratio observed in our study. The variations in S. aureus prevalence rates come from the geographic distributions, hygienic practices in milk production facilities in farms, and the type of technique, which used for detecting the S. aureus in the samples. Recent investigations revealed that S. aureus was found in udder skin, workers’ hands, bedding, insects, and dust, significantly facilitating the transition of S. aureus among animals and their milk (Piccinini et al., 2012). Moreover, the detection of S. aureus in different cow body parts such as the vagina, muzzle, and skin injuries leads to the transmission of S. aureus to the surrounding environments, as well as from dairy herds to their offspring, either through ventilation or by feeding (Mørk et al., 2012). Insufficient cleanliness of milking tools, inadequate udder preparation, unhygienic farm conditions, and improper milk transportation without adhering to cold chain requirements are important factors in the distribution of S. aureus bacterium (Regasa et al., 2019; Altaey, 2025). In addition, the current study appeared that S. aureus isolated from sheep milk samples had the virulence genes (nuc, mecA, clfA, and coa) with a percentage of 100% (14/14) and clfB in 92.9% (13/14), the scientific literature correlated the presence of these genes with animal and human health concerns. Xing et al. (2016) mentioned that nuc gene was commonly present in the isolates of coagulase-positive S. aureus; other studies focus on mecA gene, the responsible for methicillin-resistant properties. The earlier studies showed that the percentage ratio of S. aureus isolates possess the mecA gene was 41.37% (24/58) in sheep milk flocks at some farms in Iran (Rahbarnia et al., 2023). Akgul et al. (2021) declared that the lower ratio of the mecA gene found in the S. aureus isolates in a group of sheep farms in Turkey was 17.2%; in addition, they found the percentage ratio of S. aureus isolated from cheese possessed the mecA gene was 46.7%. Furthermore, the percentage ratio of S. aureus isolated from sheep mastitis milk was 47.5%, and the ratio of the mecA gene detected in S. aureus isolates was 41.4 (Akgul et al., 2021). Moreover, a study in Spain showed that 99.5% of S. aureus isolated from 36 sheep farms were resistant to methicillin, and they possessed the mecA gene (Barrero-Domínguez et al., 2019).
Fig. 7. DNASTAR’s calculation of each pair’s gene sequence difference and similarity for S. aureus GenBank, and the NCBI accession number is indicated by the designation in parenthesis. Adhesion-encoding genes such as clfA and clfB genes were characteristic in S. aureus isolates from milk samples in the present study; these genes are important in the mastitis-associated udder lesions, the global investigations reported the detection of high percentages of the virulence genes such as clfA and clfB. Acosta et al. (2018) found the percentage ratio of clfA and clfB detected in S. aureus isolated from cow and goat milk in Brazil were 76%–76.67%. In Iran, the majority of S. aureus isolates from cow’s milk contained the clfA gene of 84%–65.3% of S. aureus isolates had the clfB genes (Momtaz et al., 2010). Studies appeared that the S. aureus isolated from cow mastitis infections have the clfA gene was 19%–100% of cases, while the clfB gene was detected in 91.8%–92.9% of S. aureus isolates (Momtaz et al., 2010; Klein et al., 2012). Our study showed that the coa gene was detected in all S. aureus isolates 100% of bacterial isolation, the following gene plays an important role in the pathogenicity of this bacterium, particularly in sheep mastitis cases. In India, Javid et al. (2018) found that 64.1% of coagulase—positive S. aureus isolated from sheep milk samples carried the coa gene with different genotypes, including 514 bp, 595 bp, 757 bp, and 802 bp, while another investigation revealed that all S. aureus isolates had the coa gene with a molecular weight of 580 bp (Stephan et al., 2001; Altaey, 2024). ConclusionStaphylococcus aureus and MRSA drive a significant global challenge to animal and human health and present an impediment to producing milk and its products. The elevated incidence of S. aureus infections can be attributed to inadequate husbandry practices, insufficient adherence to basic hygiene protocols, and the random use of antibiotics. Additionally, there is genetic variability in the virulence factors of S. aureus isolated from sheep milk based on the different geographical regions worldwide. Continued research and surveillance are essential to monitor and evaluate MRSA strains and to develop effective strategies for managing and attenuating their impact on both public health and dairy manufacturing. AcknowledgmentWe want to convey our appreciation to the College of Veterinary Medicine at the University of Mosul for furnishing the essential resources needed to carry out this research. Conflict of interestThe author declares no conflicts of interest in the preparation or analysis of this manuscript. FundingThe research is fully self-funded by the authors. Authors’ contributionOmar HS and Alaa SA conceived the idea for this project. Ayman HT performed DNA isolation and PCR reactions. All authors contributed to writing the initial draft and collaboratively revised and organized the final version of the research. Data availabilityThe data generated in this study are included in the article. ReferencesAcosta, A.C., Oliveira, P.R.F., Albuquerque, L., Silva, I.F., Medeiros, E.S., Costa, M.M., Pinheiro Junior, J.W. and Mota, R.A., 2018. Frequency of Staphylococcus aureus virulence genes in milk of cows and goats with mastitis. Pesq. Vet. Brasil. 38, 2029–2036. https://doi.org/10.1590/1678-5150-PVB-5786 Ahmed, I.M., Al-Sanjary, R.A. and Alkazaly, H.H., 2020. Detection of Mycobacterium paratuberculosis in raw cow’s milk using polymerase chain reaction (PCR) technique. Iraqi J. Vet. Sci. 34(1), 83–86. Available via https://www.iasj.net/iasj/download/b1ec70a657f79f08 Akgul, O., Bora, G. and GUDUCUOĞLU, H., 2021. Investigation of the gene carriage rates for Staphylococcus aureus, mecA, vanA and nuc genes in the nasal and milk specimens from the sheep caretakers with sheep. Large Anim. Rev. 27(5), 259–268. Altaey, O.Y., Hasan, A.A. and Alhaaik, A.G. 2025. Early-life development of spleen in white rabbit (Oryctolagus cuniculus): a morphometric and histochemical analysis. Vet. Integr. Sci. 23(1), 1–13. https://doi.org/10.12982/VIS.2025.012 Argudín, M.Á., Mendoza, M.C. and Rodicio, M.R., 2010. Food poisoning and Staphylococcus aureus enterotoxins. Toxins, 2(7), 1751–1773. http://dx.doi.org/10.1016/j.fm.2016.10.02010.3390/toxins2071751 Azzi, O., Lai, F., Tennah, S., Menoueri, M.N., Achek, R., Azara, E. and Tola, S., 2020. Spa-typing and antimicrobial susceptibility of Staphylococcus aureus isolated from clinical sheep mastitis in Médéa province, Algeria. Small Ruminant Res. 192, 106168. http://dx.doi.org/10.1016/j.smallrumres.2020.106168 Barrero-Domínguez, B., Luque, I., Galán-Relaño, Á., Vega-Pla, J.L., Huerta, B., Román, F. and Astorga, R.J., 2019. Antimicrobial resistance and distribution of Staphylococcus spp. pulsotypes isolated from goat and sheep bulk tank milk in Southern Spain. Foodborne Pathogens Dis. 16(10), 723–730. https://doi.org/10.1089/fpd.2018.2593 Basanisi, M.G., La Bella, G., Nobili, G., Franconieri, I. and La Salandra, G., 2017. Genotyping of methicillin-resistant Staphylococcus aureus (MRSA) isolated from milk and dairy products in South Italy. Food Microbiol. 62, 141–146. http://dx.doi.org/10.1016/j.fm.2016.10.020 Bergonier, D., De Crémoux, R., Rupp, R., Lagriffoul, G. and Berthelot, X., 2003. Mastitis of dairy small ruminants. Vet. Res. 34(5), 689–716. https://doi.org/10.1051/vetres:2003030 De Centorbi, O.P., de Cuadrado, A.M., Alcaraz, L.E., Laciar, A.L. and de Milán, M.C. 1992. Prevalence of Staphylococcus aureus isolated from subclinical bovine mastitis in dairies of the city of San Luis. Revis. Argentin. Microbiol. 24(2), 73–80. Graber, H.U., Casey, M.G., Naskova, J., Steiner, A. and Schaeren, W. 2007. Development of a highly sensitive and specific assay to detect Staphylococcus aureus in bovine mastitic milk. J. Dairy. Sci, 90(10), 4661–4669. http://dx.doi.org/10.3168/jds.2006-902 Javid, F., Taku, A., Bhat, M.A., Badroo, G.A., Mudasir, M. and Sofi, T.A., 2018. Molecular typing of Staphylococcus aureus based on coagulase gene. Vet. World. 11(4), 423. http://dx.doi.org/10.14202/vetworld.2018.423-430 Király, J., Hajdučková, V., Gregová, G., Szabóová, T. and Pilipčinec, E. 2024. Resistant S. aureus Isolates capable of producing biofilm from the milk of dairy cows with subclinical mastitis in Slovakia. Agriculture, 14(4), 571. http://dx.doi.org/10.3390/agriculture14040571 Klein, R.C., Fabres-Klein, M.H., Brito, M.A.V.P., Fietto, L.G. and Ribon, A.D.O.B. 2012. Staphylococcus aureus of bovine origin: genetic diversity, prevalence and the expression of adhesin-encoding genes. Vet. Microbiol. 160(1-2), 183–188. https://doi.org/10.1016/j.vetmic.2012.05.025 Kossaibati, M.A. and Esslemont, R.J. 1997. The costs of production diseases in dairy herds in England. Vet. J. 154(1), 41–51. https://doi.org/10.1016/s1090-0233(05)80004-8 Kotzamanidis C, Vafeas G, Giantzi V, Anastasiadou S, Mygdalias S, Malousi A, Loukia E, Daniel S and Zdragas A., 2021. Staphylococcus aureus isolated from ruminants with mastitis in northern Greece dairy herds: genetic relatedness and phenotypic and genotypic characterization. Toxins. 13(3), 176. http://dx.doi.org/10.3390/toxins13030176 Lescourret, F. and Coulon, J.B. 1994. Modeling the impact of mastitis on milk production by dairy cows. J. Dairy. Sci. 77(8), 2289–2301. https://doi.org/10.3168/jds.S0022-0302(94)77172-1 Lowy, F.D. 1998. Staphylococcus aureus infections. N. Engl. J. Med. 339(8), 520–532. http://dx.doi.org/10.1056/NEJM199808203390806 Momtaz, H., Rahimi, E. and Tajbakhsh, E., 2010. Detection of some virulence factors in Staphylococcus aureus isolated from clinical and subclinical bovine mastitis in Iran. African J. Biotechnol. 9(25), 3753–3758. Moroni, P., Pisoni, G., Vimercati, C., Rinaldi, M., Castiglioni, B., Cremonesi, P. and Boettcher, P., 2005. Characterization of Staphylococcus aureus isolated from chronically infected dairy goats. J. Dairy Sci. 88(10), 3500–3509. http://dx.doi.org/10.3168/jds.S0022-0302(05)73035-6 Mørk, T., Kvitle, B. and Jørgensen, H.J. 2012. Reservoirs of Staphylococcus aureus in meat sheep and dairy cattle. Vet. Microbiol. 155(1), 81–87. https://doi.org/10.1016/j.vetmic.2011.08.010 Owens, W.E. and Watts, J.L. 1988. Antimicrobial susceptibility and β-lactamase testing of staphylococci isolated from dairy herds. J. Dairy. Sci. 71(7), 1934–1939. https://doi.org/10.3168/jds.S0022-0302(88)79763-5 Papadopoulos, P., Angelidis, A.S., Papadopoulos, T., Kotzamanidis, C., Zdragas, A., Papa, A., Filioussis, G. and Sergelidis, D., 2019. Staphylococcus aureus and methicillin-resistant S. aureus (MRSA) in bulk tank milk, livestock and dairy-farm personnel in north-central and north-eastern Greece: prevalence, characterization and genetic relatedness. Food Microbiol. 84, 103249. http://dx.doi.org/10.1016/j.fm.2019.103249 Piccinini, R., Tassi, R., Daprà, V., Pilla, R., Fenner, J., Carter, B. and Anjum, M.F. 2012. Study of Staphylococcus aureus collected at slaughter from dairy cows with chronic mastitis. J. Dairy. Res. 79(2), 249–255. http://dx.doi.org/10.1017/S002202991200009X Quinn, P.J., Markey, B.K., Leonard, F.C., Hartigan, P., Fanning, S. and Fitzpatrick, E., 2011. Veterinary microbiology and microbial disease. Hoboken, NJ: John Wiley & Sons. Rahbarnia, L., Rad, R.K., Dehnad, A.R. and Naghili, B., 2023. The examination of some virulence factors in S. aureus isolates obtained from the healthy human population, sheep mastitis, and cheese. Iran. J. Vet. Res. 24(2), 110. https://doi.org/10.22099%2FIJVR.2023.43730.6410 Regasa, S., Mengistu, S. and Abraha, A., 2019. Milk safety assessment, isolation, and antimicrobial susceptibility profile of Staphylococcus aureus in selected dairy farms of Mukaturi and Sululta town, Oromia region, Ethiopia. Vet. Med. Int. 2019(1), 3063185. https://doi.org/10.1155/2019/3063185 Ruegg, P.L. 2012. Managing cows, milking and the environment to minimize mastitis. Adv. Dairy. Technol. 24, 351–359. https://doi.org/10.4190/jjlac.5.210 Seegers, H., Fourichon, C. and Beaudeau, F. 2003. Production effects related to mastitis and mastitis economics in dairy cattle herds. Vet. Res. 34(5), 475–491. https://doi.org/10.1051/vetres:2003027. Sheet, O.H. 2022. Molecular detection of mecA gene in methicillin-resistant Staphylococcus aureus isolated from dairy mastitis in Nineveh governorate, Iraq. Iraqi J. Vet. Sci. 36(4), 939–943. https://doi.org/10.33899/ijvs.2022.132643.2115 Sheet, O.H., Al-Mahmood, O.A., Othamn, S.M., Al-Sanjary, R.A., Alsabawi, A.H. and Abdulhak, A.A., 2023. Detection of positive mecA Staphylococcus aureus isolated from meat and butchers’ shops by using PCR technique in Mosul city. Iraqi J. Vet. Sci 37(4), 865–870. http://dx.doi.org/10.33899/ijvs.2023.136964.2632 Sheet, O.H., Jwher, D.M., Al-Sanjary, R.A. and Alajami, A.D. 2021. Direct detection of Staphylococcus aureus in camel milk in the Nineveh governorate by using the PCR technique. Iraqi J. Vet. Sci. 35(4), 669–672. https://doi.org/10.33899/ijvs.2020.127725.1524 Smith, E.M., Green, L.E., Medley, G.F., Bird, H.E., Fox, L.K., Schukken, Y.H., Kruze, J.V., Bradley, A.J., Zadoks, R.N. and Dowson, C.G. 2005. Multilocus sequence typing of intercontinental bovine Staphylococcus aureus isolates. J. Clin. Microbiol. 43(9), 4737–4743. https://doi.org/10.1128/JCM.43.9.4737-4743.2005 Stephan, R., Annemüller, C., Hassan, A.A. and Lämmler, C. 2001. Characterization of enterotoxigenic Staphylococcus aureus strains isolated from bovine mastitis in north-east Switzerland. Vet. Microbiol. 78(4), 373–382. https://doi.org/10.1016/S0378-1135(00)00341-2 Taha, A.H., Al-Mahmood, O.A., Sheet, O.H., Hamed, A.A., Al-Sanjary, R.A. and Abdulmawjood, A.A. 2024. Molecular detection of methicillin resistant Staphylococcus aureus isolated from local fish in Mosul city. Iraqi J. Vet. Sci. 38(2), 437–441. http://dx.doi.org/10.33899/ijvs.2023.142707.3191 Tristan, A., Ying, L., Bes, M., Etienne, J., Vandenesch, F. and Lina, G. 2003. Use of multiplex PCR to identify Staphylococcus aureus adhesins involved in human hematogenous infections. J. Clin. Microbiol. 41(9), 4465–4467. http://dx.doi.org/10.1128/jcm.41.9.4465-4467.2003 Vasileiou, N.G., Chatzopoulos, D.C., Sarrou, S., Fragkou, I.A., Katsafadou, A.I., Mavrogianni, V.S., Petinaki, E. and Fthenakis, G.C. 2019. Role of staphylococci in mastitis in sheep. J Dairy Res. 86(3), 254–266. Vyletělová, M., Hanuš, O., Karpíšková, R. and Šťástková, Z., 2011. Occurrence and antimicrobial sensitivity in staphylococci isolated from goat, sheep and cow’s milk. Acta Univ Agric Silvic Mendel Brun, 59, pp. 209–214. Xing, X., Zhang, Y., Wu, Q., Wang, X., Ge, W. and Wu, C., 2016. Prevalence and characterization of Staphylococcus aureus isolated from goat milk powder processing plants. Food Control, 59, pp.644–650. http://dx.doi.org/10.1016/j.foodcont.2015.06.042 Zhang, K., McClure, J.A., Elsayed, S., Louie, T. and Conly, J.M., 2005. Novel multiplex PCR assay for characterization and concomitant subtyping of staphylococcal cassette chromosome mec types I to V in methicillin-resistant Staphylococcus aureus. J. clinic. Microbiol. 43(10), pp.5026–5033. http://dx.doi.org/10.1128/JCM.43.10.5026-5033.2005 | ||
| How to Cite this Article |
| Pubmed Style Alalaf AS, Taha AH, Sheet OH. Genotypic characteristics and phylogenetic tree analysis of Staphylococcus aureus isolates from sheep subclinical mastitis milk. Open Vet. J.. 2025; 15(1): 289-299. doi:10.5455/OVJ.2025.v15.i1.27 Web Style Alalaf AS, Taha AH, Sheet OH. Genotypic characteristics and phylogenetic tree analysis of Staphylococcus aureus isolates from sheep subclinical mastitis milk. https://www.openveterinaryjournal.com/?mno=223447 [Access: January 15, 2026]. doi:10.5455/OVJ.2025.v15.i1.27 AMA (American Medical Association) Style Alalaf AS, Taha AH, Sheet OH. Genotypic characteristics and phylogenetic tree analysis of Staphylococcus aureus isolates from sheep subclinical mastitis milk. Open Vet. J.. 2025; 15(1): 289-299. doi:10.5455/OVJ.2025.v15.i1.27 Vancouver/ICMJE Style Alalaf AS, Taha AH, Sheet OH. Genotypic characteristics and phylogenetic tree analysis of Staphylococcus aureus isolates from sheep subclinical mastitis milk. Open Vet. J.. (2025), [cited January 15, 2026]; 15(1): 289-299. doi:10.5455/OVJ.2025.v15.i1.27 Harvard Style Alalaf, A. S., Taha, . A. H. & Sheet, . O. H. (2025) Genotypic characteristics and phylogenetic tree analysis of Staphylococcus aureus isolates from sheep subclinical mastitis milk. Open Vet. J., 15 (1), 289-299. doi:10.5455/OVJ.2025.v15.i1.27 Turabian Style Alalaf, Alaa Shamil, Ayman Hani Taha, and Omar Hashim Sheet. 2025. Genotypic characteristics and phylogenetic tree analysis of Staphylococcus aureus isolates from sheep subclinical mastitis milk. Open Veterinary Journal, 15 (1), 289-299. doi:10.5455/OVJ.2025.v15.i1.27 Chicago Style Alalaf, Alaa Shamil, Ayman Hani Taha, and Omar Hashim Sheet. "Genotypic characteristics and phylogenetic tree analysis of Staphylococcus aureus isolates from sheep subclinical mastitis milk." Open Veterinary Journal 15 (2025), 289-299. doi:10.5455/OVJ.2025.v15.i1.27 MLA (The Modern Language Association) Style Alalaf, Alaa Shamil, Ayman Hani Taha, and Omar Hashim Sheet. "Genotypic characteristics and phylogenetic tree analysis of Staphylococcus aureus isolates from sheep subclinical mastitis milk." Open Veterinary Journal 15.1 (2025), 289-299. Print. doi:10.5455/OVJ.2025.v15.i1.27 APA (American Psychological Association) Style Alalaf, A. S., Taha, . A. H. & Sheet, . O. H. (2025) Genotypic characteristics and phylogenetic tree analysis of Staphylococcus aureus isolates from sheep subclinical mastitis milk. Open Veterinary Journal, 15 (1), 289-299. doi:10.5455/OVJ.2025.v15.i1.27 |