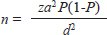| Research Article | ||
Open Vet. J.. 2025; 15(1): 437-445 Open Veterinary Journal, (2025), Vol. 15(1): 437-445 Research Article Molecular and serological detection of Leptospira interrogans among wild rats in flood-prone residential areas of IndonesiaDewi Masyithah Darlan1, Evita Mayasari2*, Sunna Vyatra Hutagalung1, Ahadi Kurniawan3, Ledy Afrida Sinaga3, Alemina Pinem3 and Bernike Ambarita31Department of Parasitology, Faculty of Medicine, Universitas Sumatera Utara, Medan, Indonesia 2Department of Microbiology, Faculty of Medicine, Universitas Sumatera Utara, Medan, Indonesia 3Medan Public Health Laboratory Center, Ministry of Health Republic Indonesia, Medan, Indonesia *Corresponding Author: Evita Mayasari. Department of Microbiology, Faculty of Medicine, Universitas Sumatera Utara, Medan, Indonesia. Email: evita [at] usu.ac.id Submitted: 18/10/2024 Accepted: 19/12/2024 Published: 31/01/2025 © 2025 Open Veterinary Journal
AbstractBackground: Indonesia is a tropical country with heavy rainfall, mostly in low-lying areas. Floods are one of the most common natural disasters in Indonesia, with climate change causing continuous flooding in some regions. The spread of human pathogens as a severe consequence of flooding, such as Leptospira, which may cause Weil’s disease, is a concern for public health. Aim: In this cross-sectional study, we compared DNA from the rat kidney to serum samples to identify pathogenic Leptospira using polymerase chain reaction (PCR) amplification to promote a less invasive method of collecting samples from the rat vectors. Methods: Fifty-nine rodents inhabiting highly populated, flood-prone suburban regions were captured inside and outside houses. Following DNA extractions, we analyzed the quantity and quality of DNA concentration from the kidney and serum specimens using a nanophotometer. The lipL32 gene was amplified to detect the pathogenic Leptospira. Results: The mean value of kidney DNA was 151.67 ng/µl with an average A260/A280 value of 1.836, whereas the mean value of serum DNA was 22.08 ng/µl with an average A260/A280 value of 1.233. Twenty (33.9%) kidney DNA and 10 (16.9%) serum DNA samples showed the target DNA (lipl32). The multiple sequence alignment analysis revealed the lipL32 sequences homology to Leptospira interrogans ser. Copenhageni. Conclusion: Rat kidneys exhibited higher DNA amount and purity than the serum. Moreover, the PCR detection of lipl32 revealed higher positive results in kidney DNA than serum DNA samples, with high similarity to L. interrogans lipl32 sequences. Therefore, the kidney remains a better DNA source than serum for the molecular analysis of Leptospira in rats. Keywords: Kidney, Leptospira interrogans, lipL32, Rat, Serum. IntroductionLeptospirosis, or Weil’s disease, is ubiquitous yet tends to occur in tropical countries with heavy rainfall (World Health Organization, 2020). The causative agent, mainly Leptospira interrogans, is maintained in the environment by reservoir animals. Rats are among the most common sources of human infection, as they carry the pathogen without symptoms, and both live alongside each other in the environment (Boey et al., 2019). Leptospira survives in the renal tubules of the infected rats, which subsequently shed the pathogen in their urine (Boey et al., 2019). Heavy rainfall that causes inundation in areas contaminated by the pathogen-contained urine facilitates the disease being transmitted to humans. People living in flood-prone areas face a high risk of contracting the disease because soil and floodwaters often contain Leptospira from animal urine, particularly from rodents, which are a common reservoir for bacteria. Some people infected by the pathogenic Leptospira may suffer from severe symptoms that consequently cause kidney or liver failure or meningitis that can be fatal (Brito Monteiro et al., 2021). In Indonesia, leptospirosis is one of the re-emerging bacterial zoonoses with an annual morbidity of ~39.2 per 100,000 persons in the population (World Health Organization, 2020). Data on leptospirosis in Indonesia revealed the occurrences from Java, Maluku, Sulawesi, Bali, Sumatra, and Kalimantan islands (Gasem et al., 2020; World Health Organization, 2020; Sunaryo and Priyanto, 2022). About 920 cases of leptospirosis were related to 122 deaths all around Indonesia in 2019 (World Health Organization, 2020). However, accessible data on leptospirosis occurrence in North Sumatra is rare. Pathogenic Leptospira significantly impacts kidney health and serum parameters in infected individuals (Sayyadi et al., 2022). The presence of Leptospira in kidney tissue and serum is well-documented (Pedersen et al., 2016; Sayyadi et al., 2022). The choice between using the rat kidney and rat serum for DNA analysis depends on the specific objectives of a study because each sample type offers distinct advantages and limitations. However, collecting serum samples from animals allows for easier and less invasive collection than kidney tissue. As part of public health surveillance activity for Leptospira in the human environment, this study reported the identification of Leptospira in the kidney and serum of rats captured from flood-prone suburban areas of North Sumatra. Materials and MethodsSample collectionWe collected rats from residential areas of the densely suburban population in the North Sumatra province of Indonesia from September 2022 to January 2023. Three regencies were chosen based on a preliminary survey: Asahan, Serdang Bedagai, and Batubara. We chose areas mostly affected by floods, especially after heavy rain. The sampling locations are in the residential areas of each regency. Cage traps, each with fish bait, were placed inside and outside houses. Rats were identified based on morphological traits according to the literature (Aplin et al., 2003). Whole blood was collected from rats using a disposable syringe for an immediate transfer to a 1.5-ml polypropylene microtube, followed by microcentrifugation at 6,000 rpm to separate the clot. The supernatant (serum) was immediately transferred into a clean microtube and kept in a cold storage box at ~4°C. Following the rat dissection mentioned earlier, we collected the kidneys and stored them individually in a glass tube containing ethanol. Samples were collected aseptically, stored in a cold storage box, and transported as soon as possible to the laboratory for further investigation. The sample size was calculated using a formula for the single population proportion:
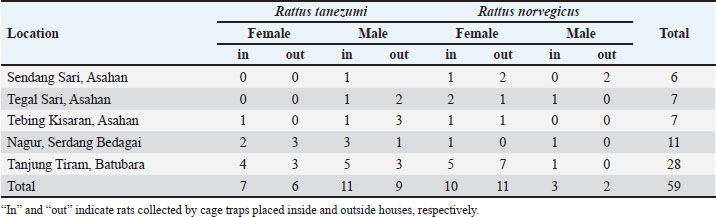
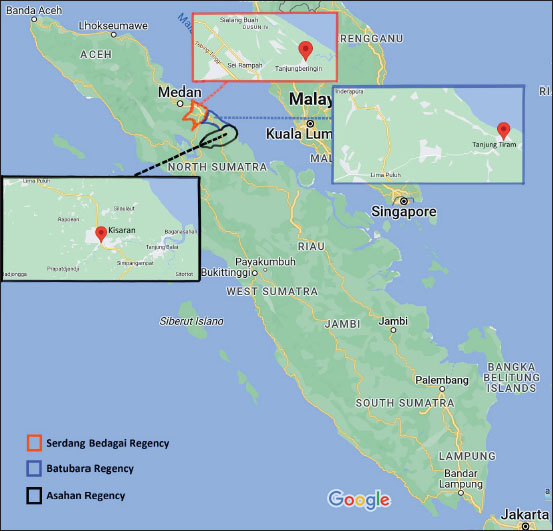
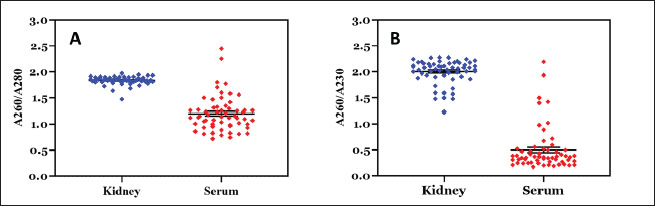
n=sample size za=95% confidence level (1.96) P=prevalence (1.5%) d=margin of error (8%) Thus, the minimum sample size=25 DNA extraction and spectrophotometric analysisThe rat kidney was weighed in a microtube and then crushed aseptically using a disposable pellet pestle. After microcentrifugation at 12,000 × g for 10 minutes at room temperature, the rat serum was transferred to another 1.5 ml sterile microtube. All samples were processed individually for genomic DNA extraction according to the DNeasy Blood & Tissue kit (Qiagen, Germany) protocol. The kit was designed for the rapid purification of total DNA in a sample. The purity and yield of genomic DNA were analyzed using 1 µl of each sample and the IMPLEN NanoPhotometer N50 (Implen GmbH, Germany) at 260, 280, and 230 nm absorbances. Detection of Leptospira by polymerase chain reaction (PCR)Leptospira interrogans genomic DNA was obtained from the Jakarta Center for Environmental Health and Disease Control Engineering and used as a positive control. The genomic DNA (10–100 ng) was mixed with 12.5 µl KAPA2G Fast ReadyMix (Kapa Biosystems, Inc., Roche, South Africa), 0.5 μM of each forward and reverse primer, and PCR-grade water was added to reach a total volume of 25 μl for standard PCR amplification by following the kit’s protocol. PCR using the Veriti thermal cycler (Applied Biosystems, Brazil) was performed with a primer set (forward: 5′-AAG CAT TAC CGC TTG TGG TG-3′ and reverse: 5′-GAA CTC CCA TTT CAG CGA TT-3′) to amplify lipL32 of pathogenic Leptospira (Stoddard et al., 2009). The PCR condition consists of initial denaturation at 95°C for 3 minutes, 35 cycles of 95°C for 15 seconds, 60°C for 15 seconds, and 72°C for 4 seconds, followed by a final extension at 72°C for 15 seconds. GelRed nucleic acid stain (EMD Millipore Corporation, USA) was added to 2% Omnipur agarose (Merck, Germany) in 1x TAE buffer (Merck, Germany) before gel agarose electrophoresis. Along with a DNA marker (Perfect DNA 100 bp Ladder, Novagen, Germany), the PCR products were loaded individually into the appropriate wells of the agarose gel in the Mini-Sub Cell GT (Bio-Rad, USA) containing 1x TAE buffer. The gel tray was then exposed to an electromotive force at 110 volts for 1 hour. The DNA bands were then visualized by exposure to UV light from a gel documentation system (Uvitec, France). Sequencing of PCR products and data analysisOne milliliter of PCR product of each sample was subjected to DNA sequencing of the lipL32 sequence by following the protocol of the Sanger sequencing kit using the ABI PRISM 3730xl Genetic Analyzer (Applied Biosystems, USA). The signal generated from the genetic analyzer was further monitored using sequencing analysis software v5.3 with KB Basecaller v1.4 (Applied Biosystems, USA) to determine the results. Descriptive statistics were done using Microsoft Office Excel 16 and GraphPad Prism 10. The McNemar test method was chosen to analyze the paired nominal data using SPSS version 23. Ethics approvalRodent trapping and investigation were approved by the Ethics Committee of the Universitas Sumatera Utara in agreement with the Declaration of Helsinki for animals, as registered in letter number 986/KEPK/USU/2022. Euthanasia, blood collection, dissection, and disposition of rats were performed by trained personnel of the Indonesian Ministry of Health by following the available guidelines (American Veterinary Medical Association, 2007; Committee for the Update of the Guide for the Care and Use of Laboratory Animals, 2011). ResultsRat collection from flood-prone areasA total of 59 rats were collected from the highly populated residential areas of three regencies in the North Sumatra province (Table 1), which is often affected by floods. Twenty rats were collected from Asahan Regency at around GPS coordinates of 2.982111 N 99.612722 E, 2.979190 N 99.629414 E, and 2.982695 N 99.627196 E (Fig. 1). Eleven rats from Serdang Bedagai Regency at Nagur village, Tanjungberingin subdistrict, around GPS coordinates of 3.499888 N 99.199556 E. Twenty-eight rats from Batubara Regency at around GPS coordinates of 3.217128 N 99.580925 E and 3.216558 N 99.589907 E. We morphologically identified 33 Rattus tanezumi and 26 Rattus norvegicus. Leptospira lipL32 was detected from the kidney samples of 9 R. tanezumi and 11 R. norvegicus. Rattus tanezumi was the dominant species (81.8%) found in the target location at Serdang Bedagai Regency, and the dominant species (85.7%) carried pathogenic Leptospira in the area. However, both rat species were found equally in the target locations at Asahan and Batubara regencies. Spectrophotometry analysis of rat kidney and serum DNAQualified (high-purity, high-yield) DNA samples most likely produce reliable results in many molecular biology applications. After DNA extraction, it is crucial to analyze the nucleic acid quality for further experiments. We used at least 1 µl of the sample for each UV spectrometry analysis with the low-salt DNA elution buffer as the blank. First, we measured the DNA absorption at 260 nm (A260) and analyzed the typical DNA absorbance spectrum of the samples individually. Then, we obtained the absorbance ratio at 260 and 280 nm (A260/A280) to analyze the relative purity of each DNA sample (Sambrook and Russell, 2001). As the absorption wavelength A230 may detect not only protein (Liu et al., 2009) but also chemical contaminants such as salts, EDTA, and other organic compounds that were used in DNA extraction (Sambrook and Russell, 2001; Lucena-Aguilar et al., 2016), we further examined the absorbance ratio at A260/A230. Table 1. Characteristics of samples collected from rats.
Fig. 1. Locations of rat sample collection in North Sumatra.
Fig. 2. Nucleic acid absorption of rat kidney and serum DNA samples by UV spectrometry analysis. A. The DNA absorbance ratio at 260 and 280 nm (A260/A280). The concentrated blue dots indicate the relative purity of kidney DNA samples with a mean value of 1.836. The scattered red dots show most of the serum DNA below the A260/A280 ratio of 1.8 (mean value=1.223). B. The DNA absorbance ratio at A260/A230. The relative purity of serum DNA (red dots) was lower (mean value=1.577) than that of kidney DNA (blue dots, mean value=2.002). The black line in the plotted graphs reveals the standard error of the mean. DNA samples with an A260/A280 ratio of 1.8 to 2.0 and an A260/A230 ratio of 2.0 to 2.2 are considered “pure” in a UV spectrometry analysis (Sambrook an Russell, 2001; Lucena-Aguilar et al., 2016). The mean value of all A260/A280 ratios from the kidney DNA samples was 1.836 (Fig. 2A). Most of the rat kidney DNA was at a relative purity with 49 of 59 (~83%) samples showing an A260/A280 ratio between a range of 1.804 and 1.972. However, only 39 (~66.1%) of the kidney DNA samples showed an A260/A230 ratio between 2.001 and 2.275 (the mean value from total samples was 2.002) (Fig. 2B). Contrary to the kidney DNA, most of the serum DNA showed low purity with the mean values of 1.233 and 1.577 for the A260/A280 and A260/A230 ratios, respectively. UV spectrophotometric reading at A260 is suitable to calculate the nucleic acid in a sample as described in Beer–Lambert law (Sambrook and Russell, 2001), with A260 between 0.1 and 1.0 considered a reliable value (Lucena-Aguilar et al., 2016). Thus, we quantified the DNA amount of each sample using this method and obtained the mean values of 151.67 and 22.08 ng/µl for the A260 readings of the kidney and serum DNA samples, respectively. PCR detection of pathogenic LeptospiraThe primers used in this study were designed following the alignment of lipL32 sequences of several pathogenic Leptospira to obtain a highly consistent consensus sequence (Stoddard et al., 2009). Molecular assays using the primers were guaranteed to be 100% sensitive and specific for lipL32 detection of Leptospira (Stoddard et al., 2009). After standard PCR and gel agarose electrophoresis, we detected ~242 bp fragments of lipL32 from 33.9% (20/59) samples of rat kidney DNA. However, only 16.9% (10/59) of serum DNA samples showed similar amplicons. Figure 3A presents amplicons from the kidney DNA that were also detected from the serum DNA. We further analyzed the PCR results data in Figure 3B statistically to check whether the rat kidney and serum DNA samples are equally effective as a DNA source for PCR amplification of the target sequences. The McNemar test is an appropriate method to analyze the paired nominal data (Adedokun and Burgess, 2012), and the results showed a significant p-value=0.021 (alpha=0.05, confidence interval=95%) and implied differences between the two DNA sources. Therefore, the rat kidney could be more effective as a DNA source for PCR analysis than rat serum.
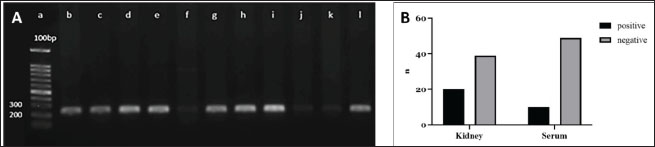
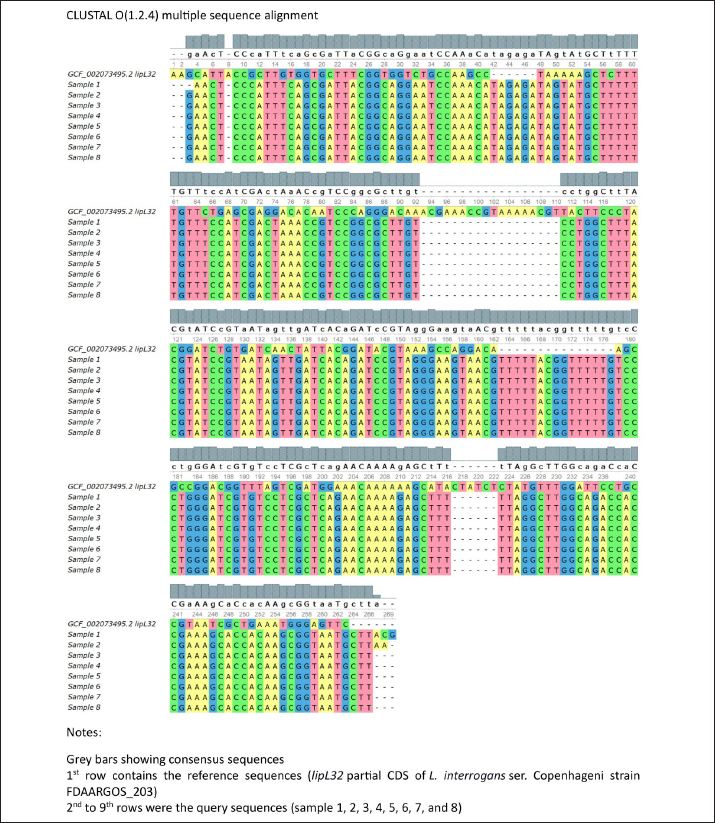
Fig. 3. Leptospira lipL32 PCR amplification from the rat kidney and serum DNA. A. Agarose gel electrophoresis of the PCR products. The primers amplified fragments of lipL32 sequences in the positive control DNA (lane l) and kidney DNA samples (lanes b-k), as shown by the ~242 bp bands. A 100 bp DNA ladder was used as the molecular weight marker (lane a). B. A diagram showing lipL32 detection results from the kidney and serum DNA. Filled bars represent samples with ~242 bp bands (positive). Gray bars represent samples without the target DNA bands (negative). Leptospira lipL32 sequence analysislipL32 is a protein-coding gene for an outer membrane surface lipoprotein of the Spirochaetales order (Paysan-Lafosse et al., 2022). LipL32 is the most abundant surface protein in pathogenic Leptospira (Vivian et al., 2009; Paysan-Lafosse et al., 2022). Oligonucleotides that were constructed from a consensus sequence of pathogenic Leptospira lipL32 provided a highly sensitive detection in the PCR assay (Stoddard et al., 2009). Eight out of 10 samples of PCR products in Figure 3A were successfully sequenced by Sanger methods. We analyzed the chromatograms of the sequences individually using FinchTV 1.4.0 (Geospiza, Inc.; Seattle, WA; http://www.geospiza.com). The individual sequence alignments of the eight samples using the BLAST (Altschul et al., 1990) showed similar hits of homologous sequences. The first hit (97.71% identity) for sample #6 (Fig. 3A, lane h) was L. interrogans strain B43 lipL32 partial CDS (GenBank accession number OM830322.1), whereas the uncultured Leptospira sp. clone C1 lipL32 partial CDS (GenBank accession number MG831575.1) appeared as the first hit (100% identity) for samples #1, 2, 3, 4, 5, 7, 8 (Fig. 3A, lane b, c, d, e, g, i, k). Moreover, the BLAST identified lipL32 partial CDS of L. interrogans isolate RnGZ52-2019 (GenBank accession number OK560918.1) as homologous sequences (100% identity) for all samples. Further, we performed multiple sequence alignment (MSA) of the sample sequences and lipL32 partial CDS of L. interrogans ser. Copenhageni strain FDAARGOS_203 (GCF_002073495.2) as the reference sequences using Clustal Omega 1.2.4 (Sievers et al., 2011). The matrix results indicated 51.87% identity between sample 1 and the reference sequences. Alignment between the rest of the samples (#2, 3, 4, 5, 6, 7, 8) and the reference sequences indicated 52.09% identity. The MSA result was visualized and provided in Additional File 1. DiscussionRodents are the major reservoir of pathogenic Leptospira and carry this infectious agent without any sign of illness (Boey et al., 2019). Various species of rodents exist in different regions of Indonesia (Hadi et al., 2021; Sunaryo and Priyanto, 2022), yet there is no accessible seroprevalence data or molecular analysis to confirm the species (Boey et al., 2019; Hadi et al., 2021). We identified R. tanezumi and R. norvegicus as the most prevalent carriers of pathogenic Leptospira in suburban residences. Rattus norvegicus has been frequently reported as a dominant species in human habitations (Koizumi et al., 2022; Sun et al., 2024). Although both species are currently equally dominant in the study area, the R. tanezumi population can significantly surpass R. norvegicus in correlation with climate change (Jing et al., 2022). A population genomics approach would be suitable to analyze further the rodents’ population, origin, and migration histories. Efficient and effective nucleic acid extraction methods are crucial to obtaining a qualified DNA sample for molecular analysis. DNA extraction methods using a proteolytic enzyme (proteinase K) effectively lyse the rat kidney and serum components (Read, 2001). Moreover, proteinase K prevents DNA degradation by digesting nucleases and removing contamination in a sample (Nakajima et al., 1994), providing relatively clean DNA compared to another method (Peñafiel et al., 2019). The spin column with a silica matrix adsorbed DNA from the rat kidney and serum samples, allowing quick DNA purification (QIAGEN, 2020). As a result, this nucleic acid extraction method produced relatively pure DNA as shown by the rat kidney samples (Fig. 2) and a relatively clean image of the corresponding PCR amplicons (Fig. 3A). In contrast to the kidney DNA, the overall rat serum DNA showed lower A260/A280 and A260/A230, indicating sample impurity. Contaminants such as proteins could be the reason for the serum DNA impurity. Despite its practicability as a source for Leptospira DNA detection (Budihal and Perwez, 2014), the serum has lower sensitivity compared to other blood fractions (whole blood and plasma) (Stoddard et al., 2009; Bourhy et al., 2011). The persistence of Leptospira in the blood clot and the subsequent clot removal may reduce its amount in the serum (Stoddard et al., 2009). A previous study demonstrated a dramatic decrease in DNA quantity referred to the lysis of whole blood cells (WBC) during storage (Huang et al., 2017). Moreover, there was evidence of nucleic acid inhibition in serum specimens (Bourhy et al., 2011) that presumably related to the immunoglobulin G effect in lowering DNA polymerase activity (Sidstedt et al., 2018). The low sensitivity of DNA detection from serum samples in this study could be due to WBC lysis upon storage and the small concentration of Leptospira in serum. The latter was confirmed by measurement using a nanophotometer that showed lower serum DNA yield compared to kidney specimens. Given the fact that rats are the main carriers of Leptospira, there is a possibility that some rats captured in this study were in the immune phase, thus the blood-borne pathogens were eliminated by anti-Leptospira immunoglobulin (mainly IgM) (Vernel-Pauillac et al., 2021; Rajapakse, 2022). Consequently, lipl32 detection from rat serum DNA was significantly lower than the results shown by similar methods using the rat kidney DNA (Fig. 3B). Leptospira lipL32 is found exclusively in the pathogenic strain and demonstrated highly conserved sequences (Haake et al., 2000; Vivian et al., 2009; Fernandes et al., 2022; Paysan-Lafosse et al., 2022). Thus, lipL32 is likely one of the most specific and common targets for PCR detection of the pathogenic Leptospira. The product, LipL32, is an outer membrane protein that mediates Leptospira interaction with a broad range of the host’s extracellular matrix proteins (Fernandes et al., 2022). We detected 33.9% and 16.9% lipL32 amplicons from the rat kidney and serum specimens, respectively. A previous study reported lipL32 detection in environmental samples (water and soil) from the flood-prone regions of Jakarta, which implies the water contamination from the vectors’ excreta, including the rodents’ urine (Widiyanti et al., 2019). The amplicons that were positive from both specimens were sequenced and analyzed for homology with the lipL32 sequence available in the National Institutes of Health database. The amplicons’ DNA sequence homology to L. interrogans ser. Copenhageni strain FDAARGOS_203 was confirmed by MSA with an overall 52% identity. However, the partial lipL32 sequence of our samples hindered the analysis of a relevant protein sequence. Further investigation of the lipL32 complete sequence of the rat DNA samples may predict a functional protein sequence to conduct a more sensitive MSA for homology analysis of the pathogenic strains. ConclusionThe amount and purity of the rat kidney DNA were higher than the serum DNA. We detected lipl32 in 33.9% of rat kidney DNA with high similarity to L. interrogans lipl32 sequences, yet only 16.9% of serum DNA samples were positive for lipl32. Therefore, the kidney remains an ideal source for DNA analysis of Leptospira in rats. Despite the limitations in design and methods, this study revealed the detection of pathogenic Leptospira in rats from flood-prone residential areas, which will be crucial for relevant epidemiological studies in the future. Future field-based studies are required to assess the prevalence and transmission of Leptospira in humans, particularly in urban and rural settings, and to understand the interaction between Leptospira and animal reservoirs in various ecosystems for leptospirosis outbreak prediction. AcknowledgmentsWe thank the head and secretary of Lembaga Penelitian Universitas Sumatera Utara. We express our gratitude to the Head of Dinas Kesehatan (Public Health Office), Mr. Harmein Harahap, MKM (Administrator of Communicable Diseases Prevention and Control of the Public Health Office) of Asahan Regency, and the team. Many thanks to the Heads of Sendang Sari, Tegal Sari, and Tebing Kisaran at Asahan Regency. We appreciate the Head of the Public Health Office and the Head of Nagur at Serdang Bedagai Regency. We extend courtesy to the Head of the Public Health Office of Batubara Regency. Our thanks as well to the Head of Tanjung Tiram Subdistrict (Sukamaju) of Batubara Regency. Conflicts of interestAll the authors declare that they have no conflicts of interest in terms of institution, research, and funding. FundingThe funding source of this study was the TALENTA research grant of Universitas Sumatera Utara from the USU non-PNBP fund. Authors’ contributionsEM conceptualized the research design, was involved in the fieldwork, collected and analyzed the data, wrote the original draft, and reviewed and edited the manuscript draft. DMD was involved in the research grant application and review of the original draft. SVH conceptualized the methodology and was involved in the fieldwork and data collection. AK was involved in the fieldwork and sample collection. LAS was involved in the fieldwork, sample collection, and data collection. AP and BA supervised the fieldwork and sample collection process and verified the results. All authors have read and approved the manuscript. Data availabilityThe data and material obtained during this study are available from the corresponding author upon reasonable request. ReferencesAdedokun, O.A. and Burgess, W.D. 2012. Analysis of paired dichotomous data: a gentle introduction to the McNemar test in SPSS. J. Multidiscip. Eval. 8(17), 125–131; doi:10.56645/jmde.v8i17.336 Altschul, S.F., Gish, W., Miller, W., Myers, E.W. and Lipman, D.J. 1990. Basic local alignment search tool. J. Mol. Biol. 215(3), 403–410; doi:10.1016/S0022-2836(05)80360-2 American Veterinary Medical Association. 2007. AVMA guidelines on euthanasia, 2007 update. Schaumburg, IL, USA. Available via http://www.avma.org/issues/animal_welfare/euthanasia.pdf (Accessed 7 July 2023) Aplin, K.A., Brown, P.R., Jacob, J., Krebs, C.J. and Singleton, G.R. 2003. Field methods for rodent studies in Asia and the Indo Pacific. Aust Centre Int Agric Res Monogr. pp: 33–41. Available via https://www.aciar.gov.au/sites/default/files/legacy/node/528/mn100field_methods_for_rodent_studies_in_asia_and__19800.pdf (Accessed 11 October 2024) Boey, K., Shiokawa, K. and Rajeev, S. 2019. Leptospira infection in rats: a literature review of global prevalence and distribution. PLoS Negl. Trop. Dis. 13(8), e0007499; doi:10.1371/journal.pntd.0007499 Bourhy, P., Bremont, S., Zinini, F., Giry, C. and Picardeau, M. 2011. Comparison of real-time PCR assays for detection of pathogenic Leptospira spp. in blood and identification of variations in target sequences. J. Clin. Microbiol. 49(6), 2154–2160; doi:10.1128/jcm.02452-10 Brito Monteiro, M., Egidio de Sousa, I., Piteira, M., Coelho, S. and Freitas, P. 2021. Leptospirosis, a re-emerging threat. Cureus 13(4), e14295; doi:10.7759/cureus.14295 Budihal, S. and Perwez, K. 2014. Leptospirosis diagnosis: competancy of various laboratory tests. J. Clin. Diagn. Res. 8(1), 199–202; doi:10.7860/JCDR/2014/6593.3950 Committee for the Update of the Guide for the Care and Use of Laboratory Animals. 2011. Guide for the care and use of laboratory animals, 8th ed. Washington, DC: National Academies Press. Available via https://grants.nih.gov/grants/olaw/guide-for-the-care-and-use-of-laboratory-animals.pdf (Accessed 11 October 2024) Gasem, M.H., Hadi, U., Alisjahbana, B., Tjitra, E., Hapsari, M.M.D.E.A.H., Lestari, E.S., Aman, A.T., Lokida, D., Salim, G., Kosasih, H. and Merati, K.T.P. 2020. Leptospirosis in Indonesia: diagnostic challenges associated with atypical clinical manifestations and limited laboratory capacity. BMC. Infect. Dis. 20(1), 179; doi:10.1186/s12879-020-4903-5 Fernandes, L.G.V., Putz, E.J., Stasko, J., Lippolis, J.D., Nascimento, A.L.T.O. and Nally, J.E. 2022. Evaluation of LipL32 and LigA/LigB knockdown mutants in Leptospira interrogans serovar copenhageni: impacts to proteome and virulence. Front. Microbiol. 12, 799012; doi:10.3389/fmicb.2021.799012 Haake, D.A., Chao, G., Zuerner, R.L., Barnett, J.K., Barnett, D., Mazel, M., Matsunaga, J., Levett, P.N. and Bolin, C.A. 2000. The leptospiral major outer membrane protein LipL32 is a lipoprotein expressed during mammalian infection. Infect. Immun. 68(4), 2276–2285; doi:10.1128/IAI.68.4.2276-2285.2000 Hadi, I., Zamroni, Y., Tresnani, G., Afrizal, Y.M. and Suana, I.W. 2021. Rat and mice species (sub family: murinae) diversity from east lombok indonesia. IOP Conf Series: Earth. Environ. Sci. 913, 012045; doi:10.1088/1755-1315/913/1/012045 Huang, L.H., Lin, P.H., Tsai, K.W., Wang, L.J., Huang, Y.H., Kuo, H.C. and Li, S.C. 2017. The effects of storage temperature and duration of blood samples on DNA and RNA qualities. PLOS One 12(9), e0184692; doi:10.1371/journal.pone.0184692 Jing, M., Chen, Y., Yao, K., Wang, Y. and Huang, L. 2022. Comparative phylogeography of two commensal rat species (Rattus tanezumi and Rattus norvegicus) in China: insights from mitochondrial DNA, microsatellite, and 2b-RAD data. Ecol. Evol. 12(10), e9409; doi:10.1002/ece3.9409 Koizumi, N., Morita, M., Nuradji, H., Noor, S.M., Dharmayanti, N.I., Randusari, P., Mu, J.J., Solante, R.M., Saito, N., Ariyoshi, K. and Ha, H.T.T. 2022. Comparative genomic analysis of Leptospira spp. isolated from Rattus norvegicus in Indonesia. Infect. Genet. Evol. 102, 1–6; doi:10.1016/j.meegid.2022.105306 Liu, P.F., Avramova, L.V. and Park, C. 2009. Revisiting absorbance at 230nm as a protein unfolding probe. Anal. Biochem. 389(2), 165–170; doi:10.1016/j.ab.2009.03.028 Lucena-Aguilar, G., Sánchez-López, A.M., Barberán-Aceituno, C., Carrillo-Ávila, J.A., López-Guerrero, J.A. and Aguilar-Quesada, R. 2016. DNA source selection for downstream applications based on DNA quality indicators analysis. Biopreserv. Biobank. 4(4), 264–270; doi:10.1089/bio.2015.0064 Nakajima, H., Itoh, K., Arakawa, E., Inoue, M., Mori, T. and Watanabe, H. 1994. Degradation of a polymerase chain reaction (PCR) product by heat-stable deoxyribonuclease (DNase) produced from Yersinia enterocolitica. Microbiol. Immunol. 38(2), 153–156; doi:10.1111/j.1348-0421.1994.tb01757.x Paysan-Lafosse, T., Blum, M., Chuguransky, S., Grego, T., Pinto, B.L., Salazar, G.A., Bileschi, M.L., Bork, P., Bridge, A., Colwell, L. and Gough, J. 2022. InterPro in 2022. Nucleic Acids Res. 51(D1), D418–D427; doi:10.1093/nar/gkac993 Pedersen, K.K., Anderson, T.D., Bevins, S.N., Pabilonia, K.N., Whitley, P.N., Virchow, D.R. and Gidlewski, T. 2016. Evidence of leptospirosis in the kidneys and serum of feral swine (Sus scrofa) in the United States. Epidemiol. Infect. 145, 87–94; doi: 10.1017%2FS0950268816002247 Peñafiel, N., Flores, D.M., de Aguilar, J.R., Guayasamin, J.M. and Bonaccorso, E. 2019. A cost-effective protocol for total DNA isolation from animal tissue. Neotrop. Biodivers. 5(1), 69–74; doi: 10.1080/23766808.2019.1706387 QIAGEN. 2020. DNeasy blood & tissue handbook. Available via https://www.qiagen.com/us/resources/download.aspx?id=68f29296-5a9f-40fa-8b3d-1c148d0b3030&lang=en (Accessed 2 January 2024) Rajapakse, S. 2022. Leptospirosis: clinical aspects. ClinMed 22(1), 14–17; doi: 10.7861/clinmed.2021-0784 Read, S.J. 2001. Recovery efficiences on nucleic acid extraction kits as measured by quantitative LightCycler PCR. Mol. Pathol. 54(2), 86–90; doi: 10.1136/mp.54.2.86 Sambrook, J. and Russell, DW. 2001. The condensed protocols from molecular cloning: a laboratory manual, 3rd edition. New York, NY: Cold Spring Harbor Laboratory Press, p: A6.1 Sayyadi, M., Hosseinzadeh, S., Abdollahpour, G., Shekarforoush, S.S., Samiei, A. and Hosseinzadeh, M. 2022. Identification of Leptospira in patients with renal failure using serological, molecular and pathological based techniques, in Shiraz, Iran. Res. Sq. [Preprint]; doi: 10.21203/rs.3.rs-1149322/v1 Sidstedt, M., Hedman, J., Romsos, E.L., Waitara, L., Wadsö, L., Steffen, C.R., Vallone, P.M. and Rådström, P. 2018. Inhibition mechanisms of hemoglobin, immunoglobulin G, and whole blood in digital and real-time PCR. Anal. Bioanal. Chem. 410, 2569–2583; doi: 10.1007/s00216-018-0931-z Sievers, F., Wilm, A., Dineen, D., Gibson, T.J., Karplus, K., Li, W., Lopez, R., McWilliam, H., Remmert, M., Söding, J. and Thompson, J.D. 2011. Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Mol. Syst. Biol. 7, 539; doi: 10.1038/msb.2011.75 Stoddard, R.A., Gee, J.E., Wilkins, P.P., McCaustland, K. and Hoffmaster, A.R. 2009. Detection of pathogenic Leptospira spp. through TaqMan polymerase chain reaction targeting the LipL32 gene. Diagn. Microbiol. Infect. Dis. 64(3), 247–255; doi: 10.1016/j.diagmicrobio.2009.03.014 Sun, Q., Liu, Y., Han, Y., Liu, W., Cao, X., Li, B. and Wang, X. 2024. Rodent ecology and etiological investigation in China: results from vector biology surveillance, Shandong Province, China, 2012–2022. China CDC wkly. 6(36), 911–917; Available via https://weekly.chinacdc.cn/en/article/doi/10.46234/ccdcw2024.193 Sunaryo, S. and Priyanto, D. 2022. Leptospirosis in rats and livestock in Bantul and Gunungkidul district, Yogyakarta, Indonesia. Vet. World. 15(6), 1449–1455; doi: 10.14202/vetworld.2022.1449-1455 Vernel-Pauillac, F., Murray, G.L., Adler, B., Boneca, I.G. and Werts, C. 2021. Anti-Leptospira immunoglobulin profiling in mice reveals strain specific IgG and persistent IgM responses associated with virulence and renal colonization. PLoS Negl. Trop. Dis. 15(3), e0008970; doi: 10.1371/journal.pntd.0008970 Vivian, J.P., Beddoe, T., McAlister, A.D., Wilce, M.C., Zaker-Tabrizi, L., Troy, S., Byres, E., Hoke, D.E., Cullen, P.A., Lo, M. and Murray, G.L. 2009. Crystal structure of LipL32, the most abundant surface protein of pathogenic Leptospira spp. J. Mol. Biol. 387(5), 1229–1238; doi: 10.1016/j.jmb.2009.02.038 Widiyanti, D., Djannatun, T., Astuti, I.I.P. and Maharsi, E.D. 2019. Leptospira detection in flood-prone environment of Jakarta, Indonesia. Zoonoses Public Health. 66(6), 597–602; doi: 10.1111/zph.12610 World Health Organization. 2020. Leptospirosis prevention and control in Indonesia. Jakarta, Indonesia: WHO Indonesia. Available via https://www.who.int/indonesia/news/detail/24-08-2020-leptospirosis-prevention-and-control-in-indonesia (Accessed 24 August 2023) Additional File 1
Additional File 1. Multiple sequence alignment (MSA) of the sample sequences and lipL32 partial CDS of L. interrogans ser. Copenhageni (RefSeq: GCF_002073495.2). | ||
| How to Cite this Article |
| Pubmed Style Darlan DM, Mayasari E, Hutagalung SV, Kurniawan A, Sinaga LA, Pinem A, Ambarita B. Molecular and serological detection of Leptospira interrogans among wild rats in flood-prone residential areas of Indonesia. Open Vet. J.. 2025; 15(1): 437-445. doi:10.5455/OVJ.2025.v15.i1.39 Web Style Darlan DM, Mayasari E, Hutagalung SV, Kurniawan A, Sinaga LA, Pinem A, Ambarita B. Molecular and serological detection of Leptospira interrogans among wild rats in flood-prone residential areas of Indonesia. https://www.openveterinaryjournal.com/?mno=225182 [Access: January 15, 2026]. doi:10.5455/OVJ.2025.v15.i1.39 AMA (American Medical Association) Style Darlan DM, Mayasari E, Hutagalung SV, Kurniawan A, Sinaga LA, Pinem A, Ambarita B. Molecular and serological detection of Leptospira interrogans among wild rats in flood-prone residential areas of Indonesia. Open Vet. J.. 2025; 15(1): 437-445. doi:10.5455/OVJ.2025.v15.i1.39 Vancouver/ICMJE Style Darlan DM, Mayasari E, Hutagalung SV, Kurniawan A, Sinaga LA, Pinem A, Ambarita B. Molecular and serological detection of Leptospira interrogans among wild rats in flood-prone residential areas of Indonesia. Open Vet. J.. (2025), [cited January 15, 2026]; 15(1): 437-445. doi:10.5455/OVJ.2025.v15.i1.39 Harvard Style Darlan, D. M., Mayasari, . E., Hutagalung, . S. V., Kurniawan, . A., Sinaga, . L. A., Pinem, . A. & Ambarita, . B. (2025) Molecular and serological detection of Leptospira interrogans among wild rats in flood-prone residential areas of Indonesia. Open Vet. J., 15 (1), 437-445. doi:10.5455/OVJ.2025.v15.i1.39 Turabian Style Darlan, Dewi Masyithah, Evita Mayasari, Sunna Vyatra Hutagalung, Ahadi Kurniawan, Ledy Afrida Sinaga, Alemina Pinem, and Bernike Ambarita. 2025. Molecular and serological detection of Leptospira interrogans among wild rats in flood-prone residential areas of Indonesia. Open Veterinary Journal, 15 (1), 437-445. doi:10.5455/OVJ.2025.v15.i1.39 Chicago Style Darlan, Dewi Masyithah, Evita Mayasari, Sunna Vyatra Hutagalung, Ahadi Kurniawan, Ledy Afrida Sinaga, Alemina Pinem, and Bernike Ambarita. "Molecular and serological detection of Leptospira interrogans among wild rats in flood-prone residential areas of Indonesia." Open Veterinary Journal 15 (2025), 437-445. doi:10.5455/OVJ.2025.v15.i1.39 MLA (The Modern Language Association) Style Darlan, Dewi Masyithah, Evita Mayasari, Sunna Vyatra Hutagalung, Ahadi Kurniawan, Ledy Afrida Sinaga, Alemina Pinem, and Bernike Ambarita. "Molecular and serological detection of Leptospira interrogans among wild rats in flood-prone residential areas of Indonesia." Open Veterinary Journal 15.1 (2025), 437-445. Print. doi:10.5455/OVJ.2025.v15.i1.39 APA (American Psychological Association) Style Darlan, D. M., Mayasari, . E., Hutagalung, . S. V., Kurniawan, . A., Sinaga, . L. A., Pinem, . A. & Ambarita, . B. (2025) Molecular and serological detection of Leptospira interrogans among wild rats in flood-prone residential areas of Indonesia. Open Veterinary Journal, 15 (1), 437-445. doi:10.5455/OVJ.2025.v15.i1.39 |