| Research Article | ||
Open Vet. J.. 2025; 15(4): 1848-1857 Open Veterinary Journal, (2025), Vol. 15(4): 1848-1857 Research Article Newcastle disease virus in Egyptian domestic poultry, 2019–2021: Molecular characterization, phylogenetic analysis, and coinfection with avian influenza A virusAhmed El Taweel1, Mohamed El Sayes1, Asmaa Maatouq2, Mokhtar Gomaa1, Yassmin Moatasim1, Omnia Kutkat1, Pamela P. McKenzie3, Ahmed Kandeil1,3, Richard J. Webby3, Mohamed Ahmed Ali1, Ghazi Kayali4 and Rabeh El-Shesheny1*1Center of Scientific Excellence for Influenza Virus, National Research Centre, Giza, Egypt 2Poultry Diseases Department, National Research Centre, Giza, Egypt 3Department of Infectious Diseases, St. Jude Children’s Research Hospital, Memphis, TN, USA 4Human Link, Dubai, United Arab Emirates *Corresponding Author: Rabeh El-Shesheny. Center of Scientific Excellence for Influenza Virus, National Research Centre, Giza, Egypt. Email:rabeh.elshesheny [at] human-link.org Submitted: 20/12/2024 Accepted: 19/03/2025 Published: 30/04/2025 © 2025 Open Veterinary Journal
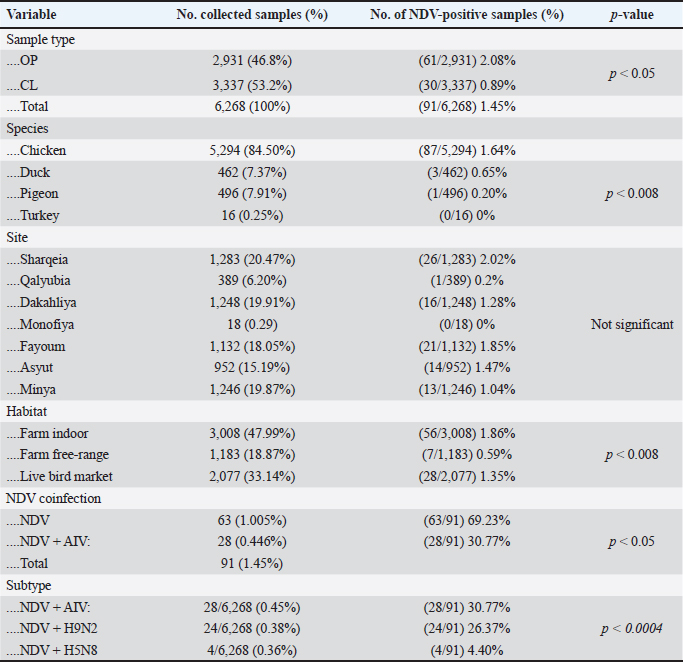
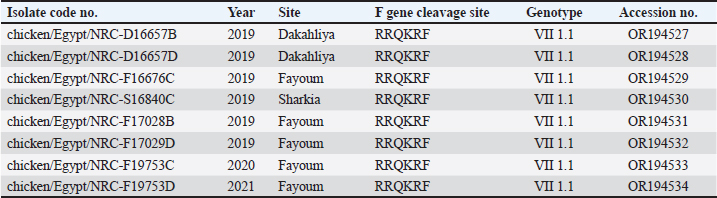
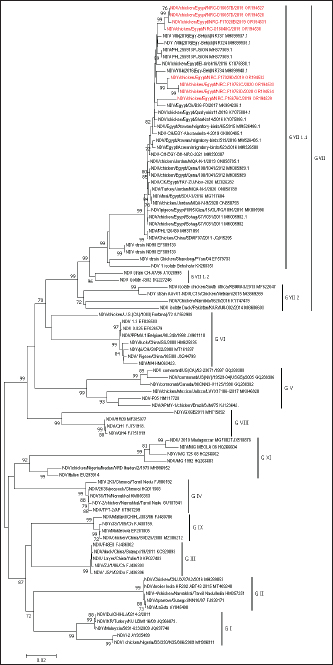
AbstractBackground: Newcastle disease virus (NDV) is a major viral disease of poultry that causes outbreaks in chickens in Egypt. Aim: The aim of this study was to determine the prevalence and molecular characteristics of NDV and its cocirculation with avian influenza virus (AIV) in Egyptian domestic poultry. Methods: Samples collected from January 2019 to February 2021 from different flock types of vaccinated and nonvaccinated domestic birds in seven governorates in Egypt were tested for NDV. A total of 6,268 swab samples from healthy, sick, and recently dead domestic poultry that showed typical clinical signs of NDV or AIV were collected. NDV was successfully isolated from swabs via inoculation of specific pathogen-free embryonated chicken eggs. Hemagglutination assay (HA) was performed followed by RT-PCR of positive HA samples. Genotyping of these isolates was performed by sequencing the full fusion (F) gene. Results: NDV was detected by RT-PCR in 1.45% (91/6268) of samples. This percentage differed significantly according to sample type, site, species, and habitat. Sixty-three (69.23%) samples were positive for NDV and 28 (30.77%) were coinfected with AIVs. This percentage differed significantly based on the AIV subtype, with 26.37% (24/91) being H9N2-positive samples and 4.40% (4/91) being H5N8-positive samples. Results indicated that NDV isolates exhibited the characteristic cleavage site motif (112RRQKRF117) of the velogenic strains of NDV. Phylogenetic analysis of the full F gene clustered with isolates in Group I of genotype VII from Egyptian and Jordanian strains. Conclusion: This study highlights the need for continued surveillance of NDV genotype VII to investigate the evolution of NDV in Egypt. Keywords: Avian influenza viruses, Egypt, Newcastle diseases virus, poultry. IntroductionPoultry production is an important economic sector with backyard flocks making up the majority of these activities, especially in developing countries (Moharam et al., 2019). Low biosecurity standards and high risk of infectious diseases, such as Newcastle disease and highly pathogenic avian influenza (HPAI), are common in backyard production (Conan et al., 2012). The Newcastle disease virus (NDV) and avian influenza virus (AIV) are the two main viral diseases that significantly impact the poultry industry due to morbidity, high mortality, and a drop in egg production (Samy and Naguib, 2018). NDV, also known as avian paramyxovirus-1, is a species within the genus orthoavulavirus and belongs to the Paramyxoviridae family (Rima et al., 2019). NDV is a negative-sense, single-stranded RNA virus with a genome composed of 15 kb encoding seven proteins: nucleocapsid protein, V protein, phosphoprotein, matrix protein (M ), fusion protein (F), large RNA-dependent polymerase protein, and hemagglutinin-neuraminidase protein (HN) (Steward et al., 1993). The envelope of NDV has two surface glycoproteins; the HN protein, which enables the virus to be attached to the host cell, and the F protein, which is associated with the fusion between the virus envelope and the host cell membrane (Mahon et al., 2011). NDV is categorized based on its pathogenicity into three groups (Saif et al., 2003): Lentogenic NDV, known to be used as a live-virus vaccine as it lacks the ability to cause disease in adult birds; mesogenic NDV, known as a non-fatal virus causing respiratory signs such as gasping, coughing, sneezing, and rales in young birds, decrease in egg production which returns to normal in a few weeks, and neurologic signs in protracted cases (Hanson and Brandly, 1955); and velogenic NDV, which is a highly virulent form of NDV isolates causing lethargy, inappetence, respiratory distress, and clear mucus discharge from the mouth prostration (Saif et al., 2003; Miller, 2014). The pathogenicity of NDV relies on the amino acid sequence of the F protein protease cleavage site (Nagai et al., 1976; Glickman et al., 1988) as well as the cleavability of the F protein of different NDV pathotypes by specific cellular proteases (Gotoh et al., 1992; Ogasawara et al., 1992). The systemic spread of velogenic NDV is associated with the presence of dibasic amino acids in the F protein sequence, whereas lentogenic NDV can only replicate in the mucosal surfaces of the host (Ogasawara et al., 1992). NDV strains are classified into two classes based on F gene sequencing (class I and II). Class I is a single genotype that includes nonvirulent NDV strains that are often asymptomatic in aquatic wild birds. Class II included all virulent NDV strains isolated from domestic birds with at least 21 genotypes from I to XXI (Dimitrov et al., 2019). In Egypt, the first detection of NDV was in 1948 (Daubney and Mansy, 1948). NDV genotypes II, VI, and VII are the most prevalent genotypes in Egypt and other North African countries (Megahed et al., 2020). In 2011, NDV genotype VII was first detected in Egypt (Radwan et al., 2013). Moreover, the vaccination programs in the commercial poultry farm sectors were not enough to curb the virus and reduce its prevalence, which is clearly shown by the new cases that continue to be detected (Naguib et al., 2020). Additionally, the NDV 2. II and NDV 2. VII genotypes have been detected in recent years in Egypt (Radwan et al., 2013). On the other hand, co-infections of NDV with either AIVs or infectious bronchitis viruses in poultry have been reported (Naguib et al., 2017; Ahmed and Naguib, 2018; Moharam et al., 2019), causing morbidity, high mortality rates, and decreased egg production. Each year, millions of birds are affected by AIVs (Alexander, 2000), causing tremendous losses to the poultry industry and posing a threat to human health. Both HPAI subtypes H5N1, H5N8, and H5N2 and low pathogenic avian influenza (LPAI) virus subtype H9N2 have been detected in Egypt (Kayali et al., 2016; El-Shesheny et al., 2022; Kandeil et al., 2022). Co-infection of NDV and AIV was detected in migratory wild birds in most wetlands during three successive migration seasons from September to March (2015–2018) in Southern Egypt (Sultan et al., 2022). The aim of this study was to determine the prevalence and molecular characteristics of NDV and its cocirculation with AIV in Egyptian domestic poultry. Materials and MethodsSample collectionSamples were collected from domestic birds between January 2019 and February 2021. A total of 6,268 swab samples, including 3,337 cloacal (CL) and 2,931 oropharyngeal (OP) samples, were collected from birds from 39 commercial poultry farms, 22 backyard flocks, two abattoirs, and 22 live bird markets in Egypt. Sampling was performed in four governorates from the Nile Delta (Dakahliya, Monofiya, Qalyobiya, and Sharqeia), one governorate from Middle Egypt (Fayoum), and two governorates from Southern Egypt (Asyut and Minya). Samples were collected from healthy, sick, and dead birds, including chickens (n=5,294), ducks (n=462), pigeons (n=496), and turkey (n=16). Virus isolation, subtyping, and genomic analysisAll collected samples were individually inoculated in the allantoic cavities of 10-day-old specific pathogen-free embryonated hen eggs, incubated for 2–5 days, and checked daily for embryo death. The eggs were then chilled to 4°C. Then, 100 μl of each allantoic fluid was tested for hemagglutination activity using 0.5% chicken red blood cells (Huang et al., 2004). AIV and NDV molecular detection All positive culture samples by hemagglutination activity assay were subjected to viral RNA extraction using a QIAamp viral mini kit (Qiagen, Hilden, Germany) according to the manufacturer’s protocol. NDV molecular detection by RT-PCR amplification was performed using primers targeting a partial region of the F gene of NDV, forward NDV-F primer: 5′-GGAGGATGTTGGCAGCATT-3′ and reverse NDV-F primer: 5′-GTCAACATATACACCTCATC-3′ (Pang et al., 2002). RT-PCR reactions were carried out using one-step RT-PCR according to the manufacturer’s instructions (Qiagen, Hilden, Germany). The final 320-bp PCR product was gel-purified using a PCR purification kit (Qiagen, Hilden, Germany). RT-PCR amplification to detect AIV in NDV-positive samples was performed using primers targeting the partial Matrix (M). Samples positive for the M gene underwent AIV subtyping using primers H5, H9, N1, N2, and N8, as previously reported (El Sayes et al., 2022; Kandeil et al., 2022). Full fusion fene in NDV sequencing Positive samples of the partial F gene of NDV were subjected to RT-PCR for full F gene detection. RT-PCR amplification was performed using a one-step RT-PCR kit according to the manufacturer’s instructions (Qiagen, Hilden, Germany). For the amplification of the full-fusion gene at 1,662 bp using forward primer F_Fusion_Nhe1-His 5-ACAAATGGCTCAAAACCTTCCAGGA-3 and reverse primer R_Fusion_-Xho1 5-GATTTACTCGAGTCATGCTCTCGTGGTGGCTCTC-3. The electrophoresis of PCR products was performed on 1.5% agarose gels, and the amplified products were visualized using the Image capture gel documentation system (Biometra, Göttingen, Germany). The concentrations of the amplified products were measured using a Qubit fluorometer, and eight samples with the highest DNA concentrations underwent sequencing. DNA libraries were prepared using Nextera XT DNA kits (Illumina, San Diego, CA) according to the manufacturer’s instructions. The libraries were sequenced using an Illumina MiSeq personal genome sequencer with 150-bp paired-end reads. Total reads were used for assembly based on a reference genome sequence from GenBank using the Merge Overlapping Pairs, Trim Reads, Map Reads to Reference, and Extract Consensus Sequence tools using CLC Genomics Workbench v21 (CLC Bio, Qiagen). The eight full fusion gene nucleotide sequences were submitted to GenBank. Genetic analysis of full F gene and phylogenetic analysis Complete coding sequences of the full F gene were used for comparative genetic analyses. To determine the genotypes of NDV viruses, we used previously published NDV reference strains, and a neighbor-joining algorithm phylogenetic tree of the full F segment was constructed using the nucleotide sequences. Alignment and phylogenic analyses were performed using MEGA 7 (Molecular Evaluation Genetic Analysis, version 7) (Kumar et al., 2016). Bootstrap support values were generated using 1,000 rapid bootstrap replicates. Statistical analysisThe chi-square test was used for statistical analysis. A p value of < 0.05 was considered to indicate statistical significance. The analysis was performed using GraphPad Prism v.8. Ethical approvalEthical approval was obtained from the Ethics Committee of the National Research Centre, Egypt. ResultsPrevalence of NDV and AIV in domestic birdsThe overall percentage of NDV detected was 1.45% (91/6,268) with 2.08% (61/2,931) positives in OP samples and 0.89% (30/3,337) positive in CL samples. This percentage differed significantly according to sample type, species, and habitat. The detection percentage by species was 1.64% (87/5,294) in chickens, 0.65% (3/462) in ducks, and 0.20% (1/496) in pigeons. NDV was not detected in turkeys. The detection percentage of NDV by sites was the highest in Sharqeia at 2.02% (26/1,283), followed by 1.85% (21/1,132), 1.28% (16/1,248), 1.47% (14/952), 1.04% (13/1,246), and 0.2% (1/389) in Fayoum, Dakahliya, Asyut, Minya, and Qalyubiam, respectively, while NDV was not detected in Monofiya. The detection percentage by habitat ranged from 1.86% (56/3,008) in indoor farms to 1.35% (28/2,077) in live bird markets, and the lowest was detected in free-range farms at 0.59% (7/1,183) (Table 1). Of the 91 NDV-positive samples, 63 (69.23%) samples were positive for NDV only and 28 (30.77%) samples were coinfected with AIV. This percentage differed significantly based on the AIV subtype, where 26.37% (24/91) were co-infected with H9N2 and 4.40% (4/91) were co-infected with H5N8. H5N1 co-infection was not detected (Table 1). Molecular features of NDV isolatesFor full fusion gene sequencing, we selected eight samples with the highest DNA concentrations from full F gene amplification. The eight NDV isolates, compared with the La Sota reference strain and other reference NDV isolates, showed a distinctive cleavage site motif (112RRQKRF117) for the velogenic strains of NDV amino acids. The macular features were determined according to the date of sample collection, site of sample collection, and genotypes of NDV obtained by sequencing the full F-gene. These nucleotide sequences were submitted to GenBank (accession numbers are listed in Table 2). Sequence and phylogenetic analysis of the F geneWe studied the genetic relationships among the eight NDV isolates, other viruses isolated from Egypt, and other viruses available in GenBank. The F genes of NDV/chicken/Egypt/NRC-D16657B/2019, NDV/chicken/Egypt/NRC-D16657D/2019, NDV/chicken/Egypt/NRC-S16840C/2019, and NDV/chicken/Egypt/NRC-F17028B/2019 shared 99.10%–99.46% nucleotide identity with NDV/chicken/Egypt/Qualyobia26/2016. NDV/chicken/Egypt/NRC-F16676C/2019 shared 99.22% nucleotide identity with VII-b|2016|Egy-Beh|ck|NR726, whereas NDV/chicken/Egypt/NRC-F17029D/2019, NDV/chicken/Egypt/NRC-F19753C/2020, and NDV/chicken/Egypt/NRC-F19753D/2020 shared 99.04%–99.70% nucleotide identity with NM85/2019. The homology analysis of the eight sequences demonstrated that the F-gene shared a very high nucleotide sequence identity (97.2%–100%) (Table S1). All Egyptian isolates were related to the genotype VII subtype. They were very close to other Egyptian isolates with 99% similarity, and 94% similarity with other strains from China, Egypt, and Jordan (Fig. 1). Mutation analysis of the fusion gene of NDVThe deduced amino acid sequences at the fusion protein cleavage site revealed the presence of multiple basic amino acid residues in all studied NDV isolates with motif 112RRKKRF117 for the velogenic strains of NDV amino acids compared with the La Sota reference strain and other NDV isolates. The eight isolates had (56–65) amino acid mutations compared with the La Sota strain, as shown in Figure 2. Table 1. Prevalence of NDVs in domestic birds.
DiscussionIn Egypt, there have been continuous reports of NDV outbreaks among domestic poultry, which have affected the economy of the poultry industry (El-Zoghby, 2014; Hassan, 2020; Lebdah et al., 2022). The poultry sector plays a significant role in NDV spread in the environment and among susceptible hosts, leading to outbreaks and financial losses. Most resting areas in wetland habitats where migratory birds shed their viruses could potentially be transmitted to domesticated birds (Kim et al., 2007; Ruenphet et al., 2011; Habib et al., 2018). Birds traveling from Europe and Asia to Africa in pursuit of warm weather may have brought NDV and AIV along with them (Olsen et al., 2006). This study revealed that NDV was detected in domestic bird samples from chickens, ducks, pigeons, and turkeys coinfected with AIV. This outcome is consistent with previous studies reporting that some NDV strains are common in birds (Sakai et al., 2007; Ruenphet et al., 2011; Habib et al., 2018; Hasan Mohammed et al., 2020; Abd Elfatah et al., 2021). Our results showed an overall percentage of NDV detection of 1.45% (91/6,268), 63 samples (69.23%) were infected with NDV only, and 28 samples (30.77%) were co-infected with AIVs. This percentage differed significantly according to AIV subtype, with 26.37% (24/91) were coinfected with H9N2, and 4.40% (4/91) were coinfected with H5N8. H5N1 subtype was not detected. By host, NDV prevalence ranged from 1.64% in chicken to 0.65% in duck and 0.20% in pigeon. NDV was not detected in turkey, whereas AIV detection was higher in chicken (3.15%) than in ducks (2.16%) and pigeons (1.81%). H9N2 was most common in chickens in all regions of Egypt (2.34%), followed by H5N8 (0.622%). H5N1 was not detected. Table 2. Epidemiology and molecular features of NDV isolates.
Fig. 1. The phylogenetic tree of eight selected isolates of NDV genotype VII compared with other related reference strains of NDV genotypes. The phylogenetic tree was constructed using the neighbor-joining algorithm with the Kimura two-parameter model. The reliability of the phylogenetic inference at each branch node was estimated using the bootstrap method with 1,000 replications. Phylogenetic analyses were conducted using MEGA 7. The NDV isolates from this study are represented in red.
Fig. 2. F protein amino acid alignment between the eight studied strains, the lentogenic La Sota vaccine, and other reference strains. Several reports have been published on the genetic evolution and pathogenesis of velogenic NDV among domesticated birds (Radwan et al., 2013; Umali et al., 2014; Selim et al., 2018; Nagy et al., 2020; Tran et al., 2020) rather than in wild birds (Hasan Mohammed et al., 2020; Abd Elfatah et al., 2021). In this study, to characterize NDV virulence strains, eight positive samples with sufficient viral loads were selected for sequencing of the full F gene to detect the presence of a multi-basic amino acid sequence at the F protein cleave site. These viruses were isolated from chickens during 2018–2021 in Egypt. The results indicated that NDVs isolated from domestic birds are pathogenic, with velogenic NDV criteria related to the F gene cleavage site 112RRQKRF117, in agreement with previous studies (Zanaty et al., 2019; Meng et al., 2021). This result contrasts with previous studies (Sakai et al., 2007; Ramey et al., 2013; Hasan Mohammed et al., 2020) reporting that isolates from wild birds are lentogenic strains with 111GERQER/L117 cleavage sites. The eight isolates were phylogenetically classified as the VII 1.1 sub-genotype, the predominant sub-genotype circulating among poultry flocks in Egypt (Radwan et al., 2013; Orabi et al., 2017; Saad et al., 2017; El Naggar et al., 2018; Nagy et al., 2020; Tran et al., 2020) and worldwide (Wang et al., 2017). Other studies detected velogenic strains in different sites in Egypt that were different from our studied areas. The full F gene sequences of the 10 NDV isolates showed that they contained multibasic amino acid motifs (111E/GRRQKR/F117). They also phylogenetically clustered into genotype VII.1.1 groups, closely related to NDVs isolated from Egyptian chickens. These findings demonstrate the role of migrating birds in the ongoing spread of virulent NDV among farmed chickens in Southern Egypt (Sultan et al., 2022). These data imply that genotype VII is still widespread, endemic in various nations, and genetically evolving in several countries, including Egypt. Despite administering mass vaccination using live and inactivated vaccines of NDV under several programs to all commercial flocks, Egypt’s poultry sector is experiencing severe financial losses because of NDV (GVII) and AIV infections. Our study provided an epidemiological picture of NDV and AIV coinfection and spread among domestic birds in Egypt. ConclusionThe current study highlights the possibility of velogenic NDV, HPAI (H5N8), and LPAI (H9N2) coinfections among domesticated birds in Egypt. Additionally, the introduction of evolved strains can have a negative effect on vaccination programs and virus control efforts in general. The coinfection of birds with NDV and AIV highlights the necessity of precise and stringent investigation procedures to stop the spread of new viral variants and reduce health hazards to the general population. AcknowledgmentsThe authors acknowledge Human Link DMCC for providing administrative and technical support for this project. This publication is funded by Human Link DMCC. Conflict of interestThe authors declare no conflict of interest. FundingThis project was funded by the National Institute of Allergy and Infectious Diseases, National Institutes of Health, Department of Health and Human Services under contract number 5N93021C00016 and by the STDF for the research fund under the Center of Scientific Excellence upgrading Fund (46654). Authors’ contributionAhmed El Taweel: Methodology, Investigation, Formal analysis, and Writing – original draft. Mohamed El Sayes: Investigation, Formal analysis. Asmaa Maatouq: Investigation. Mokhtar Gomaa: Investigation. Yassmin Moatasim: Investigation. Omnia Kutkat: Investigation. Pamela P. McKenzie: Project administration. Ahmed Kandeil: Conceptualization, Data curation, Investigation, Writing – review and editing. Richard J. Webby: Funding acquisition. Mohamed Ahmed Ali: Conceptualization, Supervision, Funding acquisition, Writing – review and editing. Ghazi Kayali: Conceptualization, Project administration, Funding acquisition, and Writing – review and editing. Rabeh El-Shesheny: Conceptualization, Data curation, Investigation, Methodology, Supervision, Writing – review and editing. Data availabilityThe data supporting the findings of this study are not openly available due to sensitivity reasons. However, they are available from the corresponding author upon reasonable request. ReferencesAbd Elfatah, K.S., Elabasy, M.A., El-khyate, F., Elmahallawy, E.K., Mosad, S.M., El-gohary, F.A., Abdo, W., Al-brakati, A., Seadawy, M.G., Tahoon, A.E. and El-gohary, A.E. 2021. Molecular characterization of velogenic newcastle disease virus (sub-genotype VII.1.1) from wild birds, with assessment of its pathogenicity in susceptible chickens. Animals 11, 505. Ahmed, S. and Naguib, M.M. 2018. Avian respiratory coinfection and impact on avian influenza pathogenicity in domestic poultry: field and experimental findings. Vet. Sci. 5, 23. Alexander, D.J. 2000. A review of avian influenza in different bird species. Vet. Microbiol. 74, 3–13. Conan, A., Goutard, F.L., Sorn, S. and Vong, S. 2012. Biosecurity measures for backyard poultry in developing countries: a systematic review. BMC Vet. Res. 8, 240. Daubney, R. and Mansy, W. 1948. The occurrence of Newcastle disease in Egypt. J. Comp. Pathol. Ther. 58, 189–200. Dimitrov, K.M., Abolnik, C., Afonso, C.L., Albina, E., Bahl, J., Berg, M., Briand, F.X., Brown, I.H., Choi, K.S., Chvala, I., Diel, D.G., Durr, P.A., Ferreira, H.L., Fusaro, A., Gil, P., Goujgoulova, G.V., Grund, C., Hicks, J.T., Joannis, T.M., Torchetti, M.K., Kolosov, S., Lambrecht, B., Lewis, N.S., Liu, H., Liu, H., Mccullough, S., Miller, P.J., Monne, I., Muller, C.P., Munir, M., Reischak, D., Sabra, M., Samal, S.K., Servan De Almeida, R., Shittu, I., Snoeck, C.J., Suarez, D.L., Van borm, S., Wang, Z. and Wong, F.Y.K. 2019. Updated unified phylogenetic classification system and revised nomenclature for Newcastle disease virus. Infect. Genet. Evol. 74, 103917. El-shesheny, R., Moatasim, Y., Mahmoud, S.H., Song, Y., El Taweel, A., Gomaa, M., Kamel, M.N., Sayes, M.E., Kandeil, A., Lam, T.T.Y., Mckenzie, P.P., Webby, R.J., Kayali, G. and Ali, M.A. 2022. Highly pathogenic avian influenza A(H5N1) virus clade 2.3.4.4b in wild birds and live bird markets, Egypt. Pathogens 12, 36. El-zoghby, E.F. 2014. Epidemiological investigations and molecular characterization of avian influenza virus in poultry in Egypt. El naggar, R.F., Rohaim, M.A., Bazid, A.H., Ahmed, K.A., Hussein, H.A. and Munir, M. 2018. Biological characterization of wild-bird-origin avian avulavirus 1 and efficacy of currently applied vaccines against potential infection in commercial poultry. Arch. Virol. 163, 2743–2755. El Sayes, M., Kandeil, A., Moatasim, Y., El taweel, A., Rubrum, A., Kutkat, O., Kamel, M.N., Badra, R., Barakat, A.B. and Mckenzie, P.P. 2022. Insights into genetic characteristics and virological features of endemic avian influenza A (H9N2) viruses in Egypt from 2017–2021. Viruses 14, 1484. Glickman, R.L., Syddall, R.J., Iorio, R.M., Sheehan, J.P. and Bratt, M.A. 1988. Quantitative basic residue requirements in the cleavage-activation site of the fusion glycoprotein as a determinant of virulence for Newcastle disease virus. J. Virol. 62, 354–356. Gotoh, B., Ohnishi, Y., Inocencio, N., Esaki, E., Nakayama, K., Barr, P., Thomas, G. and Nagai, Y. 1992. Mammalian subtilisin-related proteinases in cleavage activation of the paramyxovirus fusion glycoprotein: superiority of furin/PACE to PC2 or PC1/PC3. J. Virol. 66, 6391–6397. Habib, M., Yaqub, T., Nazir, J., Shehzad, W., Aziz Ul, R., Sohail, T., Mukhtar, N., Mehboob, A., Munir, M. and Shabbir, M.Z. 2018. Genomic and biological characterization of Newcastle disease viruses isolated from migratory mallards (Anas platyrhynchos). Arch. Virol. 163, 2179–2188. Hanson, R. and Brandly, C. 1955. Identification of vaccine strains of Newcastle disease virus. Science 122, 156–157. Hasan Mohammed, M., Kandeil, A., Alkhazindar, M., Tarek Abdelsalam, E. and Ahmed Ali, M. 2020. Isolation of newcastle disease virus from wild migratory birds in Egypt. J. World Poult. Res. 10, 520–526. Hassan, K. 2020. Avian influenza infections in poultry farms in Egypt, a continuous challenge: current problems related to pathogenesis, epidemiology and diagnosis. Berlin, Germany: Freie Universitaet Berlin. Huang, Z., Panda, A., Elankumaran, S., Govindarajan, D., Rockemann, D.D. and Samal, S.K. 2004. The hemagglutinin-neuraminidase protein of Newcastle disease virus determines tropism and virulence. J. Virol. 78, 4176–4184. Kandeil, A., Moatasim, Y., El Taweel, A., El Sayes, M., Rubrum, A., Jeevan, T., Mckenzie, P.P., Webby, R.J., Ali, M.A. Kayali, G. and El-Shesheny, R. 2022. Genetic and antigenic characteristics of highly pathogenic avian influenza A (H5N8) viruses circulating in domestic poultry in Egypt, 2017–2021. Microorganisms 10, 595. Kayali, G., Kandeil, A., El-Shesheny, R., Kayed, A.S., Maatouq, A.M., Cai, Z., Mckenzie, P.P., Webby, R.J., El Refaey, S., Kandeel, A. and Ali, M.A. 2016. Avian influenza A(H5N1) virus in Egypt. Emerg. Infect. Dis. 22, 379–88. Kim, L.M., King, D.J., Curry, P.E., Suarez, D.L., Swayne, D.E., Stallknecht, D.E., Slemons, R.D., Pedersen, J.C., Senne, D.A. and Winker, K. 2007. Phylogenetic diversity among low-virulence Newcastle disease viruses from waterfowl and shorebirds and comparison of genotype distributions to those of poultry-origin isolates. J. Virol. 81, 12641–12653. Kumar, S., Stecher, G. and Tamura, K. 2016. MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 33, 1870–1874. Lebdah, M., El-Rahman, S., Attia, A., Karam, R., Awad, N. and El-Bagoury, M. 2022. Phylogenetic and histopathological characterization of newcastle disease virus (VII. 1.1) recently isolated from naturally infected quails in Egypt. Adv. Anim. Vet. Sci. 10, 1481–1491. Mahon, P.J., Mirza, A.M. and Iorio, R.M. 2011. Role of the two sialic acid binding sites on the newcastle disease virus HN protein in triggering the interaction with the F protein required for the promotion of fusion. J. Virol. 85, 12079–12082. Megahed, M., Mohamed, W. and Hassanin, O. 2020. A complex genetic diversity of newcastle disease virus (ndv) in africa continent: an updated review. J. Anim. Health Prod. 9, 97–109. Meng, L., Zhang, S., Guo, X., Akhtar, R.W., Shah, S.A.H., Zhao, K. and Yuan, W. 2021. Complete genome and molecular characterization of genotype VII velogenic Newcastle disease virus isolated in China. Acta Virol. 65, 149–159. Miller, P. 2014. Newcastle disease in poultry. U: avian pneumoencephalitis, exotic or velogenic Newcastle disease. Darmstadt, Germany: Merck Publishing Group. Moharam, I., Razik, A.A.E., Sultan, H., Ghezlan, M., Meseko, C., Franzke, K., Harder, T., Beer, M. and Grund, C. 2019. Investigation of suspected Newcastle disease (ND) outbreaks in Egypt uncovers a high virus velogenic ND virus burden in small-scale holdings and the presence of multiple pathogens. Avian Pathol. 48, 406–415. Nagai, Y., Klenk, H.-D. and Rott, R. 1976. Proteolytic cleavage of the viral glycoproteins and its significance for the virulence of Newcastle disease virus. Virology 72, 494–508. Naguib, M., Höper, D., El-Kady, M., Afify, M., Erfan, A., Abozeid, H., Hassan, W., Arafa, A.-S.A., Beer, M. and Harder, T. 2020. Genomic and antigenic properties of Newcastle disease virus genotypes 2. XX1 and 2. VII from Egypt do not point to antigenic drift as a driving force of spread. Authorea [Preprints] 1–16; doi: 10.22541/au.160637962.28915681/v1 Naguib, M.M., El-Kady, M.F., Lãschow, D., Hassan, K.E., Arafa, A.-S., El-Zanaty, A., Hassan, M.K., Hafez, H.M., Grund, C. and Harder, T.C. 2017. New real time and conventional RT-PCRs for updated molecular diagnosis of infectious bronchitis virus infection (IBV) in chickens in Egypt associated with frequent co-infections with avian influenza and Newcastle Disease viruses. J. Virol. Methods 245, 19–27. Nagy, A., Ali, A., Zain El-Abideen, M.A., Kilany, W. and Elsayed, M. 2020. Characterization and genetic analysis of recent and emergent virulent newcastle disease viruses in Egypt. Transbound. Emerg. Dis. 67, 2000–2012. Ogasawara, T., Gotoh, B., Suzuki, H., Asaka, J., Shimokata, K., Rott, R. and Nagai, Y. 1992. Expression of factor X and its significance for the determination of paramyxovirus tropism in the chick embryo. EMBO J. 11, 467–472. Olsen, B., Munster, V.J., Wallensten, A., Waldenstrom, J., Osterhaus, A.D. and Fouchier, R.A. 2006. Global patterns of influenza a virus in wild birds. Science 312, 384–388. Orabi, A., Hussein, A., Saleh, A.A., El-Magd, M.A. and Munir, M. 2017. Evolutionary insights into the fusion protein of Newcastle disease virus isolated from vaccinated chickens in 2016 in Egypt. Arch. Virol. 162, 3069–3079. Pang, Y., Wang, H., Girshick, T., Xie, Z. and Khan, M.I. 2002. Development and application of a multiplex polymerase chain reaction for avian respiratory agents. Avian Dis. 46, 691–699. Radwan, M., Darwish, S., El-Sabagh, I., El-Sanousi, A. and Shalaby, M. 2013. Isolation and molecular characterization of Newcastle disease virus genotypes II and VIId in Egypt between 2011 and 2012. Virus Genes 47, 311–316. Ramey, A.M., Reeves, A.B., Ogawa, H., Ip, H.S., Imai, K., Bui, V.N., Yamaguchi, E., Silko, N.Y. and Afonso, C.L. 2013. Genetic diversity and mutation of avian paramyxovirus serotype 1 (Newcastle disease virus) in wild birds and evidence for intercontinental spread. Arch. Virol. 158, 2495–2503. Rima, B., Balkema-Buschmann, A., Dundon, W.G., Duprex, P., Easton, A., Fouchier, R., Kurath, G., Lamb, R., Lee, B., Rota, P., Wang, L. and ICTV Report, C. 2019. ICTV virus taxonomy profile: paramyxoviridae. J. Gen. Virol. 100, 1593–1594. Ruenphet, S., Jahangir, A., Shoham, D., Morikawa, K., Miyoshi, Y., Hanawa, E., Okamura, M., Nakamura, M. and Takehara, K. 2011. Surveillance and characterization of Newcastle disease viruses isolated from northern pintail (Anas acuta) in Japan during 2006-09. Avian Dis. 55, 230–235. Saad, A., Samy, A., Soliman, M., Arafa, A., Zanaty, A., Hassan, M., Sultan, A., Bazid, A. and Hussein, A. 2017. Genotypic and pathogenic characterization of genotype VII Newcastle disease viruses isolated from commercial farms in Egypt and evaluation of heterologous antibody responses. Arch. Virol. 162, 1985–1994. Saif, Y., Barnes, H., Glisson, J., Fadlyam, M. and Swayne, D. 2003. Diseases of poultry, 11th ed. Ames, IA: Iowa State University Press, p: 56. Sakai, K., Sakabe, G., Tani, O., Watanabe, Y., Jahangir, A., Nakamura, M. and Takehara, K. 2007. Characterization of Newcastle disease virus isolated from northern pintail (Anas acuta) in Japan. J. Vet. Med. Sci. 69, 1307–1311. Samy, A. and Naguib, M.M. 2018. Avian respiratory coinfection and impact on avian influenza pathogenicity in domestic poultry: field and experimental findings. Vet. Sci. 5, 23. Selim, K.M., Selim, A., Arafa, A., Hussein, H.A. and Elsanousi, A.A. 2018. Molecular characterization of full fusion protein (F) of Newcastle disease virus genotype VIId isolated from Egypt during 2012-2016. Vet. World 11, 930–938. Steward, M., Vipond, I.B., Millar, N.S. and Emmerson, P.T. 1993. RNA editing in Newcastle disease virus. J. Gen. Virol. 74, 2539–2547. Sultan, S., Eldamarany, N.M.I., Abdelazeem, M.W. and Fahmy, H.A. 2022. Active surveillance and genetic characterization of prevalent velogenic Newcastle disease and highlypathogenic avian influenza H5N8 viruses among migratory wild birds in Southern Egypt during 2015-2018. Food Environ. Virol. 14, 280–294. Tran, G.T.H., Sultan, S., Osman, N., Hassan, M.I., Van Dong, H., Dao, T.D., Omatsu, T., Katayama, Y., Mizutani, T. and Takeda, Y. 2020. Molecular characterization of full genome sequences of Newcastle disease viruses circulating among vaccinated chickens in Egypt during 2011–2013. J. Vet. Med. Sci. 82, 809–816. Umali, D.V., Ito, H., Shirota, K., Katoh, H. and Ito, T. 2014. Characterization of complete genome sequence of genotype VI and VII velogenic Newcastle disease virus from Japan. Virus Genes 49, 89–99. Wang, Y., Yu, W., Huo, N., Wang, W., Guo, Y., Wei, Q., Wang, X., Zhang, S., Yang, Z. and Xiao, S. 2017. Comprehensive analysis of amino acid sequence diversity at the F protein cleavage site of Newcastle disease virus in fusogenic activity. PLoS One 12, e0183923. Zanaty, A.M., Hagag, N.M., Rabie, N., Saied, M., Selim, K., Mousa, S.A., Shalaby, A.G., Arafa, A.-S. and Hassan, M.K. 2019. Epidemiological, phylogenetic analysis and pathogenicity of newcastle disease virus circulating in poultry farms, Egypt during 2015-2018. Hosts Viruses 6, 50. Supplementary MaterialTable S1. Genetic similarity among the NDV isolates sequenced in this study.
| ||
| How to Cite this Article |
| Pubmed Style Taweel AE, Sayes ME, Maatouq A, Gomaa M, Moatasim Y, Kutkat O, Mckenzie PP, Kandeil A, Webby RJ, Ali MA, Kayali G, El-shesheny R. Newcastle disease virus in Egyptian domestic poultry, 2019–2021: Molecular characterization, phylogenetic analysis, and coinfection with avian influenza A virus. Open Vet. J.. 2025; 15(4): 1848-1857. doi:10.5455/OVJ.2025.v15.i4.37 Web Style Taweel AE, Sayes ME, Maatouq A, Gomaa M, Moatasim Y, Kutkat O, Mckenzie PP, Kandeil A, Webby RJ, Ali MA, Kayali G, El-shesheny R. Newcastle disease virus in Egyptian domestic poultry, 2019–2021: Molecular characterization, phylogenetic analysis, and coinfection with avian influenza A virus. https://www.openveterinaryjournal.com/?mno=233845 [Access: January 13, 2026]. doi:10.5455/OVJ.2025.v15.i4.37 AMA (American Medical Association) Style Taweel AE, Sayes ME, Maatouq A, Gomaa M, Moatasim Y, Kutkat O, Mckenzie PP, Kandeil A, Webby RJ, Ali MA, Kayali G, El-shesheny R. Newcastle disease virus in Egyptian domestic poultry, 2019–2021: Molecular characterization, phylogenetic analysis, and coinfection with avian influenza A virus. Open Vet. J.. 2025; 15(4): 1848-1857. doi:10.5455/OVJ.2025.v15.i4.37 Vancouver/ICMJE Style Taweel AE, Sayes ME, Maatouq A, Gomaa M, Moatasim Y, Kutkat O, Mckenzie PP, Kandeil A, Webby RJ, Ali MA, Kayali G, El-shesheny R. Newcastle disease virus in Egyptian domestic poultry, 2019–2021: Molecular characterization, phylogenetic analysis, and coinfection with avian influenza A virus. Open Vet. J.. (2025), [cited January 13, 2026]; 15(4): 1848-1857. doi:10.5455/OVJ.2025.v15.i4.37 Harvard Style Taweel, A. E., Sayes, . M. E., Maatouq, . A., Gomaa, . M., Moatasim, . Y., Kutkat, . O., Mckenzie, . P. P., Kandeil, . A., Webby, . R. J., Ali, . M. A., Kayali, . G. & El-shesheny, . R. (2025) Newcastle disease virus in Egyptian domestic poultry, 2019–2021: Molecular characterization, phylogenetic analysis, and coinfection with avian influenza A virus. Open Vet. J., 15 (4), 1848-1857. doi:10.5455/OVJ.2025.v15.i4.37 Turabian Style Taweel, Ahmed El, Mohamed El Sayes, Asmaa Maatouq, Mokhtar Gomaa, Yassmin Moatasim, Omnia Kutkat, Pamela P. Mckenzie, Ahmed Kandeil, Richard J. Webby, Mohamed Ahmed Ali, Ghazi Kayali, and Rabeh El-shesheny. 2025. Newcastle disease virus in Egyptian domestic poultry, 2019–2021: Molecular characterization, phylogenetic analysis, and coinfection with avian influenza A virus. Open Veterinary Journal, 15 (4), 1848-1857. doi:10.5455/OVJ.2025.v15.i4.37 Chicago Style Taweel, Ahmed El, Mohamed El Sayes, Asmaa Maatouq, Mokhtar Gomaa, Yassmin Moatasim, Omnia Kutkat, Pamela P. Mckenzie, Ahmed Kandeil, Richard J. Webby, Mohamed Ahmed Ali, Ghazi Kayali, and Rabeh El-shesheny. "Newcastle disease virus in Egyptian domestic poultry, 2019–2021: Molecular characterization, phylogenetic analysis, and coinfection with avian influenza A virus." Open Veterinary Journal 15 (2025), 1848-1857. doi:10.5455/OVJ.2025.v15.i4.37 MLA (The Modern Language Association) Style Taweel, Ahmed El, Mohamed El Sayes, Asmaa Maatouq, Mokhtar Gomaa, Yassmin Moatasim, Omnia Kutkat, Pamela P. Mckenzie, Ahmed Kandeil, Richard J. Webby, Mohamed Ahmed Ali, Ghazi Kayali, and Rabeh El-shesheny. "Newcastle disease virus in Egyptian domestic poultry, 2019–2021: Molecular characterization, phylogenetic analysis, and coinfection with avian influenza A virus." Open Veterinary Journal 15.4 (2025), 1848-1857. Print. doi:10.5455/OVJ.2025.v15.i4.37 APA (American Psychological Association) Style Taweel, A. E., Sayes, . M. E., Maatouq, . A., Gomaa, . M., Moatasim, . Y., Kutkat, . O., Mckenzie, . P. P., Kandeil, . A., Webby, . R. J., Ali, . M. A., Kayali, . G. & El-shesheny, . R. (2025) Newcastle disease virus in Egyptian domestic poultry, 2019–2021: Molecular characterization, phylogenetic analysis, and coinfection with avian influenza A virus. Open Veterinary Journal, 15 (4), 1848-1857. doi:10.5455/OVJ.2025.v15.i4.37 |