| Research Article | ||
Open Vet. J.. 2025; 15(6): 2551-2561 Open Veterinary Journal, (2025), Vol. 15(6): 2551-2561 Research Article Detection of biofilm genes in MDR Staphylococcus aureus isolated from human and cattle urinary tract infections in Babylon Governorate, IraqSumod Abdul Kadhem Salman1 and Balsam Miri Mizher Al Muhana1*1Department of Microbiology, College of Veterinary Medicine, University of Al-Qadisiyah, Al-Diwaniyah, Iraq * Correspondence to: Balsam Miri Mizher Al Muhana. Department of Microbiology, College of Veterinary Medicine, University of Al-Qadisiyah, Al-Diwaniyah, Iraq. Email: Balsam.mizher [at] qu.edu.iq Submitted: 10/02/2025 Revised: 26/04/2025 Accepted: 12/05/2025 Published: 30/06/2025 © 2025 Open Veterinary Journal
AbstractBackground: Staphylococcus aureus (S. aureus) is a major pathogenic bacterium in veterinary medicine and human health. It is one of the most important bacterial agents causing urinary tract infection (UTI). The increasing incidence of multidrug-resistant and biofilm-forming S. aureus is a great problem today. The aim of the study was to investigate the phenotypic and genotypic aspects of biofilm formation in multiple drug-resistant (MDR) S. aureus isolated from UTI infection in human and cattle in Babylon, Iraq. Methodology: A total of 168 and 172 urine samples were collected from UTI infection in humans and cattle, respectively, during the period from November 2023 to February 2024. Morphological and biochemical identification was used to diagnose S. aureus. Additional confirmation was performed by the automated Vitek 2 compact system. Results: Among the 168 human isolates, 24 (14.2%) and 172 cattle isolates, 16 (9.3%) isolates were diagnosed as S. aureus. Genotypically identification of the isolates was performed using the 16s RNA gene. Twenty-two (91.6%) from 24 human source S. aureus isolates and 8 (50%) from 16 animal source S. aureus isolates were found to be MDR according to the Vitek 2 compact system. Results revealed that 19 (86.3%) and 7(87.5%) MDR isolates were phenotypically positive for biofilm production in concern to human and animal source isolates, respectively. Genotypically, three polysaccharide intracellular adhesion genes and one MSCRAMMs (fib) were screened in MDR bacteria using specific primers. The prevalence rate of genes was icaA (100%), icaB (100%), and icaC (100%) in all 30 MDR isolates. fib gene were present in 63.6% of human isolates and 75% of cattle source isolates. Conclusion: The study has shown that the biofilm-forming S. aureus that causes UTI were resistant to multiple antibiotic agents. These findings underscore the necessity of development effective treatment approaches to control UTI infections in humans and animals. Keywords: Staphylococcus aureus, Vitek II, Biofilm genes, UTI infections. IntroductionUrinary tract infection (UTI) is the most common bacterial infection in hospital- and community-acquired infections (Tekin et al., 2012). Bacteria are considered the primary cause of UTI, followed by fungi and viruses which are sometimes considered as an uncommon phenomenon. UTI is not dangerous, but patients have many symptoms as the stage progresses may lead to death in severe cases (Parveen et al., 2011; Demilie et al., 2012). Staphylococcus aureus is supposed as one of the main pathogens in humans and animals (Sheet et al., 2021). It is one of the most important bacterial agents causing UTI because of the high distribution of this species in the hospital environment, high antibiotic resistance, and high resistance against extreme conditions (Tong et al., 2015). UTI sources in neonatal animals may be associated with ascending infection from the urachus, involving the kidney, bladder, and ureter. In bovine species, an additional possible pathway for this disease is through the vulva, which may play a main role as an entry site for numerous urinary pathogens. Furthermore, in cattle bacteremia, septic catheterization, and post-calving illnesses may be predisposing factors for UTI (Baxter, 1989). Worldwide, antimicrobial resistance is a growing and emerging public health problem (Olopade et al., 2023). Methicillin-resistant S. aureus (MRSA) can harm public health as they are resistant to multiple antibiotics (Sheet et al., 2023). The unselective antibiotic treatment increased the number of multiple drug-resistant (MDR) strains, which is considered a great challenge in UTI treatment (Farajzadah et al., 2019). In MDR bacteria, the pathogen is resistant to several chemotherapeutic agents and has acquired non-susceptibility to at least one agent in three or more (≤3) antimicrobial categories. This phenomenon increases the mortality and morbidity rates (Nikaido, 2009). Although methicillin has been used to eliminate resistant S. aureus, the development of MRSA isolates has restricted their efficiency (Sheet et al., 2023). The resistance to β-lactam antibiotics occurring in MRSA strains could be associated with the presence of transferable genomic islands, called SCCmec, in the bacterial genome, where the mecA gene determines resistance to methicillin. Within the different types of SCCmec, resistance genes to other classes of antibiotics like macrolides, lincosamide, aminoglycosides, tetracyclines (T), and streptogramin B (Mlynarczyk-Bonikowska et al., 2022). Moreover, the capability of S. aureus to produce biofilms assistance to the bacteria in the urinary tract for growth and persistence by providing a nutrient-rich environment and protecting it from host defense and antimicrobial agents (Kot et al., 2016). The important component of S. aureus biofilm is polysaccharides of intercellular adhesion (PIA), which is synthesis by icaADBC operon. The characterization of bacteria will enhance our understanding of their pathogenesis and aid in the development of effective therapeutic approaches for controlling UTI infections. Thus, our objective was to investigate the phenotypic and genotypic aspects of biofilm formation in MDR S. aureus isolated from UTI infection in human and cattle in Babylon, Iraq. Materials and MethodsSamples CollectionHumanUrine samples were collected in a clean container from 168 patients with UTI, representing both sexes and different age groups during the period from November 2023 to February 2024. A survey was made with patients to yield the full history of general information, such as age, clinical cases associated, and type of antibiotic administration. CattleThis study was carried out on 172 cattle of different ages and sexes from different regions in Babylon Governorate through a period from November 2023 to February 2024. The animals appeared clinically to be suffering from UTI based on the following symptoms: signs of pain, increased urination frequency with discomfort during urination, presence of blood in urine, urine color, and odor changes may be found, loss of appetite, rise of body temperature, and general weakness. Urine samples were collected under an aseptic condition using a urinary catheter, and the first urine drops from living animals were excluded in sterile bottles (Twafik, 2023). Isolation and identification of S. aureus isolatesHuman and animal urine samples were cultured in brain heart infusion broth and then incubated at 37 C for 24 hours, the growing bacteria were inoculated on blood agar, mannitol salt agar and then incubated overnight at 37 C. Biochemical tests, staining, and microscopic examination were performed (MacFaddin, 2000). All bacterial isolates were identified using an automated Vitek 2 compact system (Biomerieux, America). Furthermore, the 16S rRNA gene, the marker of S. aureus, was screened in all identified isolates by using a specific primer was forward 5’-TATAGATGGATCCGCGCTGC, reverse 5’-GGGTCCCCGTCAATTCCTTT with a product size of 711 bp. Detection of MDR S. aureus using a Vitek 2 compact systemStaphylococcus aureus isolates were tested against 18 antibiotics belonging to different classes using a Vitek 2 compact system (Biomerieux, America). The antibiotics were penicillin G (PG), Pipercillin/ tazobactam (PT), oxacillin (OX), gentamicin (GM), tobramycin (TOB), levofloxacin (LEV), moxifloxacin (MXF), erythromycin (E), clindamycin (CD), linezolid (LZD), teicoplanin (TE), vancomycin (V), T, tigecycline (TG), nitrofurantoin (NIT), fusic acid (FD), rifampicin (RA), and trimethoprim-sulfamethoxazole (TS). Any strains resistant to at least one agent in three or more (≤3) antimicrobial categories were considered MDR. Results were interpreted according to the Clinical and Laboratory Standards Institute breakpoints (CLSI, 2023). Biofilm assayPhenotypic detection of biofilmThe isolates were screened for biofilm formation using the Congo red agar (CRA) assay. Briefly, CRA plates were prepared using the following components: Congo red stain (0.8 gm/l), agar (10 gm/l), brain heart infusion broth (37 gm/l), and sucrose (5 gm/l). The stain was prepared independently as an aqueous solution and then sterilized in an autoclave at 1210C for 15 minutes. This solution was then mixed with sterilized brain heart infusion agar at 550C. The sucrose was added to the other components of the medium. Plates were cultured with S. aureus isolates and incubated at 370C for 24 hours. The red color colonies interpreted as a negative result for biofilm production, whereas the black colonies with dry crystalline were interpreted as a positive result (Melo et al., 2013). The assay was performed in triplicate. Genotypic detection of biofilmMDR S. aureus isolates were tested by PCR technique for the detection of four biofilm genes (icaA, icaB, icaC, fib) by using a specific primer as shown in Table 1. Genomic DNA was extracted by using (Presto™ Mini gDNA Bacteria Kit) and done according to company instructions. The extracted total DNA was assessed using a Nanodrop (Thermo Scientific NanoDrop Lite UV Visible Spectrophotometer, USA). DNA concentration is measured as ng/µl and the purity was checked at absorbance (260/280 nm). Table 1. Table 1. Primers used in detection of biofilm genes.
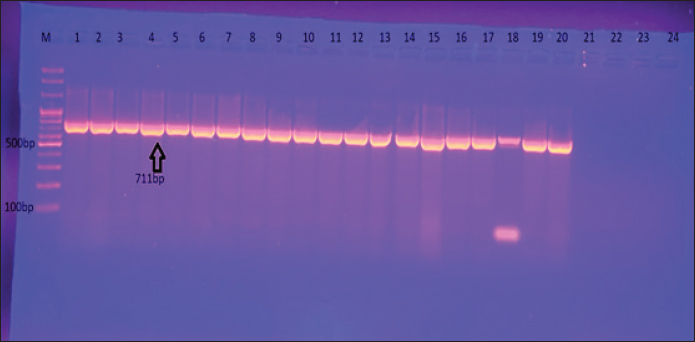
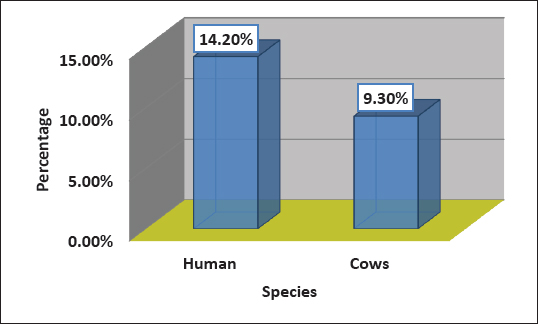
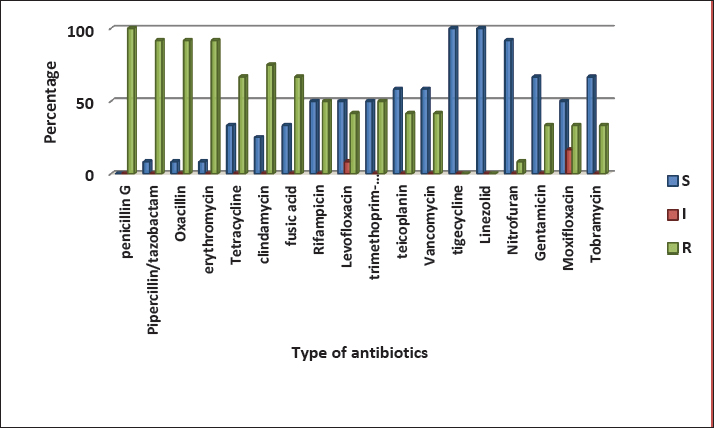
Fig 1. Gel electrophoresis of the 16S rRNA amplicon of S. aureus on (1%) agarose at 100 volt for1 hour M: DNA Marker 2,000 bp/ show the positive results for this gene with amplicon size 711 bp. Ethical ApprovalAll procedures in this study were approved by the Animal Ethics Commission of the School of Veterinary Medicine (certificate number 2326) on 29/5/2024. ResultsIsolation and Identification of S. aureusAll bacterial isolates in the current study were identified using morphological and biochemical tests, as described by Brooks et al. (2010). Genotypical identification of isolates was performed using the 16s rRNA gene (the marker of S. aureus ), which was detected in all identified isolates as shown in Figure 1. A total of 168 human urine samples and 172 cow urine samples were used in our study. Human urine samples were collected from hospitalized patients. Out of the 168 samples, 24 (14.2%) were diagnosed as S. aureus-positive strains, as shown in Figure 2. From 172 cows’ urine samples, 16 (9.3%) were positive for S. aureus (Fig 2). Antibiotic susceptibility patternOut of 24 tested isolates, the following susceptibilities were detected: with regard to human source isolates, complete susceptibility (100%) was observed for TG and LZD, and 91.7% for nitrofuran. The most resistance rates of the isolates were PG 100%, PT, and OX 91.6%, and E 83 %. On the other hand, different levels of resistance were observed for other antibiotics, such as T (66.6%), CD (60%), FD 58.3 %, RA, LEV, and TS 50%), TE and V 41.6 %, GM, MXF, and TOB 33% (Fig 3).
Fig. 2. Percentage of S. aureus isolates in human and cattle urine samples.
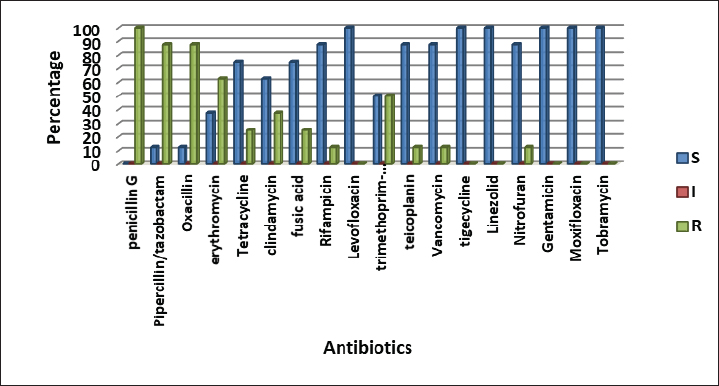
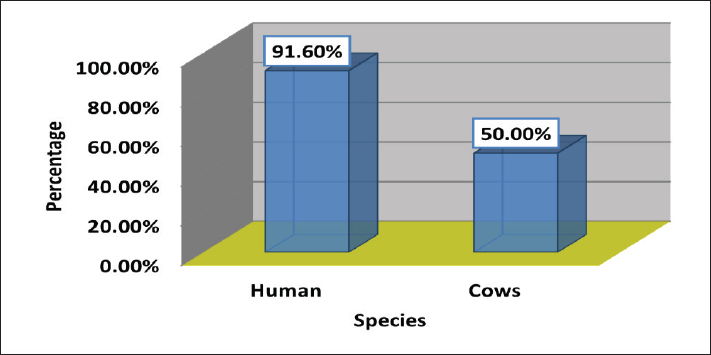
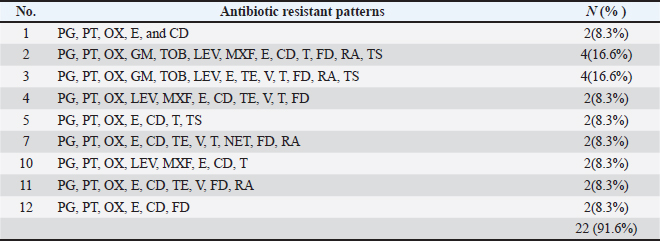
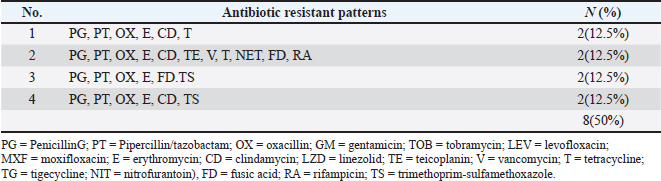
Fig. 3. Antimicrobials susceptibility test of S. aureus isolated from a human with urinary tract infection. Cattle isolates showed different levels of sensitivity; complete susceptibility (100%) was observed for TG, LZD, GM, TOB, LEV, and MXF. In addition, 87.5 % of susceptibility was shown for TE, V, nitrofuran, and RA. The complete resistance rates (100%) of the isolates were detected for PG. Resistance rate to other antibiotics was observed for PT and OX 87.5%, 62.5% for E, 37.5% for CD and TS, and 25% for T and FD (Fig 4). Detection of MDR S. aureusMDR strains of S. aureus were investigated using the Vitek 2 compact system, 22(91.6%) from 24 human source S. aureus isolates and 8 (50%) from 16 animal source S. aureus isolates were found to be MDR (Fig 5) because it has resistance to ≤3 classes of antibiotics (Table 2 and Table 3).
Fig. 4. Antimicrobials susceptibility test of S. aureus isolated from cattle with urinary tract infection.
Fig. 5. Prevalence of MDR S. aureus in human and cattle urine samples. Biofilm AssayPhenotypic detection of biofilmThe results revealed that 19 (86.3%) and 7(87.5%) MDR isolates were positive for biofilm production in concern to human and animal sources, respectively. Table 2. Table 2. MDR patterns of S. aureus isolated from human with UTI.
Table 3. Table 3. MDR Patterns of S. aureus isolated from cattle with UTI.
Table 4. Percentage of biofilm genes of MDR S. aureus isolated from human and cattle urine samples according to PCR technique.
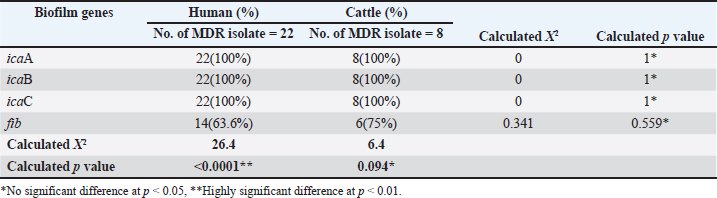
Genotypic detection of biofilmThree PIA genes and one MSCRAMMs (fib gene) were screened in 30 MDR isolates using specific primers. Genotypic assay revealed that these isolates harbored all tested genes (Table 4). The incidence rates of biofilm genes were icaA (100%), icaB (100%), and icaC (100%) in human and cattle isolates, as shown in Fig 6, Fig 7, Fig 8–Fig 9. DiscussionIsolation and identification of S. aureusGenotypical identification of isolates was done by 16s rRNA gene (the marker of S. aureus), and this gene was detected in all identified isolates. Our study appeared that the results of classical methods and the PCR technique were similar. Other studies also showed no difference between classical and molecular methods to identify S. aureus (Sheet, 2022; Sheet et al., 2022). The high frequency of this bacteria in human UTI (14.2%) was similar to other study in Baghdad governorate found that the percentage of S. aureus in urine is 13.8% (Khalaf et al., 2022). The high frequency of this bacteria in UTI found in other studies in Al-Mosul governorate reached 40.4% (Abdul Razzaq, 2013). Regarding to cows’ urine samples, a previous study in Baghdad found that the rate of S. aureus in sheep and cows urine infected with UTI is 8 % (Abdul-Ratha et al., 2013). Al-Yassaree (2005) found that the percentage of S. aureus in urine of cows is 40%.
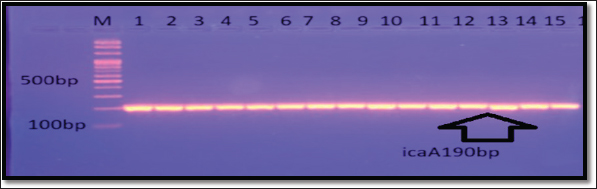
Fig. 6. Gel electrophoresis of icaA amplicon of S. aureus on (1%) agarose at 100 volt for 1 hour/M: DNA Marker 2,000 bp/ show the positive results for this gene with amplicon size 190 bp.
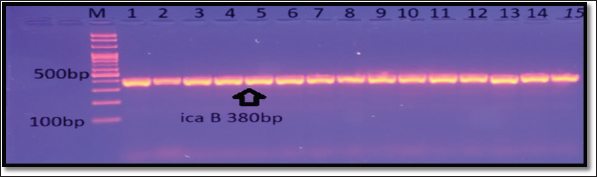
Fig. 7. Gel electrophoresis of icaB amplicon of S. aureus on (1%) agarose at 100 volt for 1 hour/M: DNA Marker 2,000 bp/ show the positive results for this gene with amplicon size 380 bp.
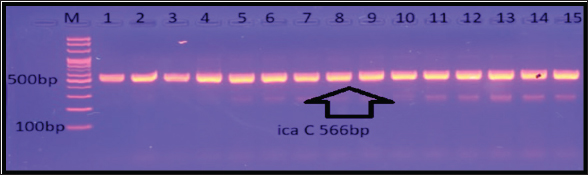
Fig. 8. Gel electrophoresis of the icaC amplicon of S. aureus on (1%) agarose at 100 volt for 1 hour/M: DNA Marker 2,000 bp/show the positive results for this gene with amplicon size 566 bp.
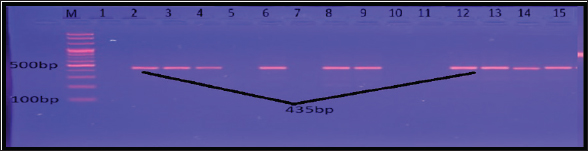
Fig. 9. Gel electrophoresis of fib amplicon of S. aureus on (1%) agarose at 100 volt for 1 hour /M: DNA Marker 2,000 bp/ show some positive results for this gene with amplicon size 435 bp. Antibiotic susceptibility patternAntibiotics have the ability to disrupt the peri-urethral flora, permitting uropathogens to thrive and enter the urinary system. Furthermore, this state gives bacteria the chance to exchange their genetic material containing antibiotic-resistant genes via horizontal gene transfer (Ouno et al., 2013). Microorganisms undergo mutations in either their chromosomal DNA or RNA to survive, which may lead to the emergence of resistance to several antimicrobial agents. The bacterial cell wall plays a critical role as a barrier, but due to genetic mutations in chromosomal DNA, the structures of the cell wall or the cell membrane changes and this changing increase the resistance phenomenon. Drug Efflux Pumps are consider one of the important ways for the MDR mechanism (Mukherjee et al., 2021). Detection of MDR S. aureusThe rate of MDR S. aureus was higher in this study compared with a previous study by Khalaf et al. (2022), who found that the percentage of MDR S. aureus in urine was 41.1%. MDR is an unavoidable phenomenon in which pathogens are resistant to multiple antibiotic group (Nikaido, 2009). This phenomenon increases the rates of mortality and morbidity, therefore it called “Superbugs.” The increasing rates of this state is belonged to several causes, such as the use of undefined antimicrobial agents, the overuse of antibiotics, and unhygienic conditions. Horizontal gene transfer between different species of bacteria and mutation are responsible for Antibiotic resistance (Von Wintersdorff et al., 2016). There are other reasons for the higher prevalence of MDR in the current study, which may be attributed to the random use of antibiotics for human and animal treatment, which is common in our country. Biofilm AssayPhenotypic Detection of BiofilmBiofilms can be defined as aggregates of monospecies or multispecies bacterial communities enclosed in a protective extracellular matrix. This matrix is composed of secreted exopolysaccharide (EPS), cellular proteins, environmental DNA, lipids, and water (Tan et al., 2015). The greatly incidence of bacterial biofilm in this study may reflect the virulence determinants of bacteria in the urinary tract. A local study by Al-Mtory (2016) showed that 100% of S. aureus clinical isolates were biofilm production. Biofilm is an important feature of S. aureus infection and is composed of numerous layers of bacteria enclosed within an EPS glycocalyx. This material prevents the bacteria from the host immune defenses and prevents the delivery of drugs. The black color formed by biofilm-producer bacteria on Congo red media indicates the capacity of dye to stain the polysaccharide layer formed during biofilm formation (Bose et al., 2009). Genotypic Detection of BiofilmMDR and biofilm formation are known virulent factors responsible for S. aureus chronic infections that can threaten public health (Manandhar et al., 2018). Persisted cells in the biofilm matrix resisted antimicrobial agents and host defense. It has been suggested that the intracellular adhesion (ica) operon plays a vital role in the control of biofilm production. The ica locus (icaADBC) encodes important proteins mediate the synthesis of PIA molecules. (Motallebi et al., 2016). The present results are similar to those of Atshan et al. (2012), who found that icaADBC locus were existing in all sixty MRSA and MSSA isolates. Parastan et al. (2020) and Chen et al. (2020) found high incidence rates (100%) of icaADBC genes in human and food S. aureus source isolates, respectively. Rodhe et al. found that all of the 80 S. aureus isolates had these genes using the PCR method. In this study, the rate of these genes prevalence was significantly more than other previous studies like Goudarzi et al., who reported that the incidence of icaADBC genes were 76%, 77.3%, 57.3%, and 50.7%, respectively, in 75 S. aureus isolated from patients with UTIs (Bimanand et al., 2018; Goudarzi et al., 2019). Antibiotic resistance among bacteria exist in biofilms layer may rise up to 1000 times (Neupane et al., 2016). The reasons for this resistance may be due to difficulty in antibiotic penetration in biofilm layers, the slow growth rate of bacteria, and antibiotic degradation. Furthermore, biofilm production provides a stand for horizontal gene transfer among bacterial species, causing an exchange of drug-resistance genes and other virulence factors (Madsen et al., 2012). Fibrinogen is a glycoprotein found in blood plasma. Fibrinogen have a critical role in the blood coagulation, fibrinogen is converted to fibrin (insoluble form). The fibrinogen-binding protein (fib) is a main virulence factor in S. aureus infections and can interfere with platelet aggregation and the complement cascade within the host (Shannon and Flock, 2004). In our study, the fib gene was found in 63.6% of human isolates and 75% of cattle source isolates. Previous studies by Drożdż et al. found the fib gene in 79% of S. aureus isolates, whereas a local study on S. aureus isolated from different regions of cattle found that 69.7% of isolates harbored this gene (Mohammed and Al-Iedani, 2020). Differences in the occurrence of MSCRAMMs and icaADBC genes among different researches may be attributed to epidemiological variations, periods of isolate collection, or methodological differences (Rodhe et al., 2001). In general, antimicrobial resistance and biofilm formation become escalating and intractable problems for health in humans and animals. ConclusionsThe current study demonstrated that the rise of antibiotic resistance in S. aureus, which causes UTI, limits the therapeutic use of antimicrobials. Additionally, MDR isolates had the ability to produce biofilm, which made their treatment complicated, confirming the critical need to seek a new antibacterial agent that treats the infection associated with the development of biofilm. Conflict of interestThe authors declare no conflict of interest. Author contributionsSD: conceptualization, data collection, and writing of the original manuscript draft. SD: sharing in writing the manuscript draft and editing. BA: review and editing. All authors have approved the final manuscript for publication. FundingThis research received no specific grant. Data availabilityAll data supporting the findings of this study are available in the manuscript, and no additional data sources are required. ReferencesAbdul-Ratha, H.A. and Mohammad, A.J. 2013. The occurrence of urinary tract infection by bacteria in humans and animals in Baghdad city and its susceptibility to antibiotics. J. Genet. Environ. Resour. Conserv. 1(3), 204–208. AbdulRazzaq, M.G. 2013. Pattern of antibiotic sensitivity and resistance of uropathogens among pediatric patients with urinary tract infection. Iraqi J. Pharm. 13(1), 64–76. Al-Mtory, H.K. 2016. Bacteriological and molecular study of Staphylococcus spp. isolated from clinical specimens. MSc thesis. College of Medicine, Babylon University, Iraq. Al-Yassaree, A.S.S. 2005. Investigation of the role of Staphylococcus saprophyticus in urinary tract infections in humans and some animals. MSc Thesis, College of Veterinary Medicine, Baghdad University, Iraq. Artini, M., Papa, R., Scoarughi, G.L., Galano, E., Barbato, G., Pucci P. and Selan, L. 2013. Comparison of the action of different proteases on virulence properties related to the staphylococcal surface. J. Appl. Microbiol. 114(1), 266–277. Atshan, S.S., Nor Shamsudin, M., Sekawi, Z., Lung, L.T.T., Hamat, R.A., Karunanidhi, A. and Pei, C. 2012. Prevalence of adhesion and regulation of biofilm-related genes in different clones of Staphylococcus aureus. Biomed Res. Int. 2012(1), 976972. Baxter, G.M. 1989. Umbilical masses in calves: diagnosis, treatment, and complications. Compend. Contin. Educ. Pract. Vet. 11, 505–513. Bimanand, L, Taherikalani, M., Jalilian, F.A., Sadeghifard, N., Ghafourian, S., Mahdavi Z. and Pakzad, I. 2018. Association between biofilm production, adhesion genes, and drug resistance in different SCCmec types of methicillin-resistant Staphylococcus aureus strains isolated from several major hospitals of Iran. Iran. J. Basic Med. Sci. 21(4), 400. Bose, S., Khodke, M., Basak, S. and Mallick, S.K. 2009. Detection of biofilm-producing staphylococci: need of the hour. J. Clin. Diagn. Res. 3(6), 1915–1920. Brooks, G.F., Carroll, K.C., Butel, J.S., Morse, S.A. and Mietzner, T.A. 2010. Medical microbiology. Jawetz, Melnick and Adelbergs, 25th Edition, New York, NY: McGraw-Hill Companies, pp: 213–219. Chen, Q., Xie, S., Lou, X., Cheng, S., Liu, X., Zheng W. and Wang, H. 2020. Biofilm formation and prevalence of adhesion genes among Staphylococcus aureus isolates from different food sources. Microbiol. Open 9(1), e00946. Demilie, T., Beyene, G., Melaku S. and Tsegaye, W. 2012. Urinary bacterial profile and antibiotic susceptibility pattern among pregnant women in northwest Ethiopia. Ethiop. J. Health Sci. 22(2), 223–226. Drożdż, K., Ochońska, D., Ścibik, Ł., Gołda-Cępa, M., Biegun, K. and Brzychczy-Włoch, M. 2022. The frequency of occurrence of resistance and genes involved in the process of adhesion and accumulation of biofilm in Staphylococcus aureus strains isolated from tracheostomy tubes. Microorganisms 10(6), 1210. Farajzadah Sheikh, A., Goodarzi, H., Yadyad, M.J., Aslani, S., Amin, M., Jomehzadeh, N. and Hashemzadeh, M. 2019. Virulence-associated genes and drug susceptibility patterns of uropathogenic Escherichia coli isolated from patients with urinary tract infection. Infect. Drug Resist. 12, 2039–2047. Gajdács, M. 2019. The continuing threat of methicillin- resistant Staphylococcus aureus. Antibiotics 8(2), 52. Goudarzi, M., Mohammadi, A., Amirpour, A., Fazeli, M., Nasiri, M.J., Hashemi, A. and Goudarzi, H. 2019. Genetic diversity and biofilm formation analysis of Staphylococcus aureus causing urinary tract infections in Tehran, Iran. J. Infect. Dev. Ctries. 13(09), 777–785. Kot, B., Wicha, J., Gruzewska, A., Piechota, M., Wolska K. and Obrebska, M. 2016. Virulence factors, biofilm-forming ability, and antimicrobial resistance of urinary Escherichia coli strains isolated from hospitalized patients. Turk. J. Med. Sci. 46(6), 1908–1914. Lebeaux, D., Ghigo, J.M. and Beloin, C. 2014. Biofilm- related infections: bridging the gap between clinical management and fundamental aspects of recalcitrance toward antibiotics. Microbiol. Mol. Biol. Rev. 78(3), 510–543. MacFaddin, J.F. 2000. Biochemical tests for identification of medical Bacteria (3rd ed.). Baltimore, MA: Williams and Wilkins. Madsen, J.S., Burmølle, M., Hansen, L.H. and Sørensen, S.J. 2012. The interconnection between biofilm formation and horizontal gene transfer. FEMS Immunol. Med. Microbiol. 65(2), 183–195. Manandhar, S., Singh, A., Varma, A., Pandey, S. and Shrivastava, N. 2018. Biofilm-producing clinical Staphylococcus aureus isolates increased the prevalence of antibiotic-resistant cases in tertiary care hospitals of Nepal. Front. Microbiol. 9, 2749. Melo, P.D.C., Ferreira, L.M., Nader Filho, A., Zafalon, L.F., Vicente, H.I.G. and Souza, V.D. 2013. Comparison of methods for the detection of biofilm formation by Staphylococcus aureus isolated from bovine subclinical mastitis. Brazil. J. Microbiol. 44, 119–124. Mlynarczyk-Bonikowska, B., Kowalewski, C., Krolak- Ulinska, A. and Marusza, W. 2022. Molecular mechanisms of drug resistance in Staphylococcus aureus. Int. J. Mol. Sci. 23(15), 8088. Mohammed, A.L. and Al-Iedani, A.A. 2020. Molecular detection of ica gene and some surface proteins in biofilm producer of methicillin-resistant and methicillin-sensitive Staphylococcus aureus. Biochem. Cell. Arch. 20, 3623–3631. Motallebi, M., Jabalameli, F., Asadollahi, K., Taherikalani, M. and Emaneini, M. 2016. Spreading of genes encoding enterotoxins, haemolysins, adhesin and biofilm among methicillin-resistant Staphylococcus aureus strains with staphylococcal cassette chromosome mec type IIIA isolated from burn patients. Microb. Pathog. 97, 34–37. Mukherjee, R., Priyadarshini, A., Pati Pandey, R. and Samuel Raj, V. 2021. Antimicrobial resistance in Staphylococcus aureus. London: IntechOpen. doi: 10.5772/intechopen.96888. Neupane, S., Pant, N.D., Khatiwada, S., Chaudhary, R. and Banjara, M.R. 2016. Correlation between biofilm formation and resistance toward different commonly used antibiotics along with extended spectrum beta lactamase production in uropathogenic Escherichia coli isolated from the patients suspected of urinary tract infections visiting Shree Birendra Hospital, Chhauni, Kathmandu, Nepal. Antimicrob. Resist. Infect. Control 5, 1–5. Nikaido, H. 2009. Multidrug resistance in bacteria. Annu. Rev. Biochem. 78(1), 119–146. Olopade, A., Bitrus, A.A., Momoh-Zekeri, A.H. and Bamaiyi, P.H. 2022. Multi-drug resistant phenotypes of extended-spectrum β-lactamase (ESBL)-producing E. coli from layer chickens. Iraqi J. Vet. Sci. 36(4), 945–951. doi: 10.33899/ ijvs.2022.132655.2117. Ouno, G.A., Korir, S.C., Cheruiyot, J., Ratemo, D.O., Mabeya, M.B., Mauti, O.G. and Kiprono, S.J. 2013. Isolation, identification, and characterization of urinary tract infectious bacteria and the effect of different antibiotics. J. Nat. Sci. Res. 3(6), 2224– 3186. Parastan, R., Kargar, M., Solhjoo, K. and Kafilzadeh, F. 2020. A synergistic association between adhesion-related genes and multidrug resistance patterns of Staphylococcus aureus isolates from different patients and healthy individuals. J. Glob. Antimicrob. Resist. 22, 379–385. Parveen, K., Momen, A., Begum, A.A. and Begum, M. 2011. Prevalence of urinary tract infection during pregnancy. J. Dhaka Natl. Med. Coll. Hosp. 17(2), 8–12. Rohde, H., Knobloch, J.K., Horstkotte, M.A. and Mack, D. 2001. Correlation of Staphylococcus aureus icaADBC genotype and biofilm expression phenotype. J. Clin. Microbiol. 39(12), 4595–4596. Selim, S., Faried, O.A., Almuhayawi, M.S., Saleh, F.M., Sharaf, M., El Nahhas, N. and Warrad, M. 2022. Incidence of vancomycin-resistant Staphylococcus aureus strains among patients with urinary tract infections. Antibiotics 11(3), 408. Shannon, O. and Flock, J.I. 2004. Extracellular fibrinogen binding protein, Efb, from Staphylococcus aureus binds to platelets and inhibits platelet aggregation. Thromb. Haemost. 91(04), 779–789. Sheet, O.H. 2022. Molecular detection of mecA gene in methicillin-resistant Staphylococcus aureus isolated from dairy mastitis in Nineveh governorate, Iraq. Iraqi J. Vet. Sci. 36(4), 939–943. Sheet, O.H., Al-Mahmood, O.A., Othamn, S.M., Al- Sanjary, R.A., Alsabawi, A.H. and Abdulhak, A.A. 2023. Detection of positive mecA Staphylococcus aureus isolated from meat and butchers’ shops by using PCR technique in Mosul city. Iraqi J. Vet. Sci. 37(4), 865–870. Sheet, O.H., Hussein, S.A. and Al-Chalaby, A.Y. 2021. Detection of methicillin-resistant Staphylococcus aureus from broiler carcasses in Mosul city. Iraqi J. Vet. Sci. 35(3), 489–493. Sheet, O.H., Talat, R.A., Kanaan, I.I., Najem, A.A. and Alchalabi, A.S.S. 2022. Detection of the nuc gene in Staphylococcus aureus isolated from swamps and ponds in Mosul city by using PCR techniques. Iraqi J. Vet. Sci. 36(3), 821–824. Silver, L.L. 2011. Challenges of antibacterial discovery. Clin. Microbiol. Rev. 24(1), 71–109. Tan, X., Qin, N., Wu, C., Sheng, J., Yang, R., Zheng, B. and Jia, A. 2015. Transcriptome analysis of the biofilm formed by methicillin-susceptible Staphylococcus aureus. Sci. Rep. 5(1), 11997. Tekin, A., Dal T.D.Ö., Tekin, R., Bozdağ, H. and Özekinci, T. 2012. In-vitro efficacy of nitrofurantoin and some antibiotics in Escherichia coli strains isolated from urine cultures. New J. Med. 29(2), 88–91. Tong, S.Y., Davis, J.S., Eichenberger, E., Holland, T.L. and Fowler, Jr. V.G. 2015. Staphylococcus aureus infections: epidemiology, pathophysiology, clinical manifestations, and management. Clin. Microbiol. Rev. 28(3), 603–661. Twafik, J.H. 2023. Studies on renal bacterial affections in sheep in Matrouh Governorate. Assiut Vet. Med. J. 69(179), 160–171. Vivas, R., Barbosa, A.A.T., Dolabela, S.S. and Jain, S. 2019. Multidrug-resistant bacteria and alternative methods to control them: an overview. Microb. Drug Resist. 25(6), 890–908. Von Wintersdorff, C.J., Penders, J., Van Niekerk, J.M., Mills, N.D., Majumder, S., Van Alphen, L.B. and Wolffs, P.F. 2016. Dissemination of antimicrobial resistance in microbial ecosystems through horizontal gene transfer. Front. Microbiol. 7, 173. | ||
| How to Cite this Article |
| Pubmed Style Salman SAK, Muhana BMMA. Detection of biofilm genes in MDR Staphylococcus aureus isolated from human and cattle urinary tract infections in Babylon Governorate, Iraq. Open Vet. J.. 2025; 15(6): 2551-2561. doi:10.5455/OVJ.2025.v15.i6.27 Web Style Salman SAK, Muhana BMMA. Detection of biofilm genes in MDR Staphylococcus aureus isolated from human and cattle urinary tract infections in Babylon Governorate, Iraq. https://www.openveterinaryjournal.com/?mno=242150 [Access: January 25, 2026]. doi:10.5455/OVJ.2025.v15.i6.27 AMA (American Medical Association) Style Salman SAK, Muhana BMMA. Detection of biofilm genes in MDR Staphylococcus aureus isolated from human and cattle urinary tract infections in Babylon Governorate, Iraq. Open Vet. J.. 2025; 15(6): 2551-2561. doi:10.5455/OVJ.2025.v15.i6.27 Vancouver/ICMJE Style Salman SAK, Muhana BMMA. Detection of biofilm genes in MDR Staphylococcus aureus isolated from human and cattle urinary tract infections in Babylon Governorate, Iraq. Open Vet. J.. (2025), [cited January 25, 2026]; 15(6): 2551-2561. doi:10.5455/OVJ.2025.v15.i6.27 Harvard Style Salman, S. A. K. & Muhana, . B. M. M. A. (2025) Detection of biofilm genes in MDR Staphylococcus aureus isolated from human and cattle urinary tract infections in Babylon Governorate, Iraq. Open Vet. J., 15 (6), 2551-2561. doi:10.5455/OVJ.2025.v15.i6.27 Turabian Style Salman, Sumod Abdul Kadhem, and Balsam Miri Mizher Al Muhana. 2025. Detection of biofilm genes in MDR Staphylococcus aureus isolated from human and cattle urinary tract infections in Babylon Governorate, Iraq. Open Veterinary Journal, 15 (6), 2551-2561. doi:10.5455/OVJ.2025.v15.i6.27 Chicago Style Salman, Sumod Abdul Kadhem, and Balsam Miri Mizher Al Muhana. "Detection of biofilm genes in MDR Staphylococcus aureus isolated from human and cattle urinary tract infections in Babylon Governorate, Iraq." Open Veterinary Journal 15 (2025), 2551-2561. doi:10.5455/OVJ.2025.v15.i6.27 MLA (The Modern Language Association) Style Salman, Sumod Abdul Kadhem, and Balsam Miri Mizher Al Muhana. "Detection of biofilm genes in MDR Staphylococcus aureus isolated from human and cattle urinary tract infections in Babylon Governorate, Iraq." Open Veterinary Journal 15.6 (2025), 2551-2561. Print. doi:10.5455/OVJ.2025.v15.i6.27 APA (American Psychological Association) Style Salman, S. A. K. & Muhana, . B. M. M. A. (2025) Detection of biofilm genes in MDR Staphylococcus aureus isolated from human and cattle urinary tract infections in Babylon Governorate, Iraq. Open Veterinary Journal, 15 (6), 2551-2561. doi:10.5455/OVJ.2025.v15.i6.27 |