| Research Article | ||
Open Vet. J.. 2025; 15(7): 3185-3192 Open Veterinary Journal, (2025), Vol. 15(7): 3185-3192 Research Article Clinical xsKlebsiella pneumoniae isolates and their efflux pump mechanism for antibiotic resistance challengeAli A. Kadhum*, Mohammed H. Khudor and Hanaa K. IbraheimDepartment of Microbiology, College of Veterinary Medicine, University of Basrah, Basrah, Iraq *Corresponding Author: Ali A. Kadhum. Department of Microbiology, College of Veterinary Medicine, University of Basrah, Basrah, Iraq. Email: ali.abd [at] stu.edu.iq Submitted: 19/03/2025 Revised: 04/06/2025 Accepted: 11/06/2025 Published: 31/07/2025 © 2025 Open Veterinary Journal
ABSTRACTBackground: Klebsiella pneumoniae is a serious pathogen that causes many disorders in humans and animals. Klebsiella pneumoniae, which is one of the most important pathogens in hospitals, often causes many clinical manifestations, including pneumonia, urinary tract infections, and meningitis. Interest in this bacterium has increased due to the increasing incidence of infection caused by it, as well as its high resistance to antibiotics, especially broad-spectrum antibiotics. Aim: This study showed the efflux pump mechanism of clinical K. pneumoniae isolates and antibiotic resistance in samples collected from sheep and human respiratory tract infection in southern Iraq. Methods: Three hundred samples were collected, and the samples included: 150 nasal swabs from sheep and 150 sputum samples from humans. Through bacteriological and biochemical examinations. The isolates were identified K. pneumoniae isolates were also confirmed by 16S rRNA. Susceptibility testing of the antibiotics used in the study. To determine the phenotypic efflux pump activity, the agar ethidium bromide cartwheel method was used. Results: Of 150 sputum human specimens and 150 nasal swabs from sheep were tested, 25 and 17 K. pneumoniae species isolates from patients and sheep, respectively, for the resistance of the bacteria isolated from humans to antibiotics. The highest rate of resistance was to piperacillin (88%), and the lowest rate was to antibiotics (36%), imipenem. The highest of bacterial susceptibility to the antibiotic imipenem was (44%) and (36%) for levofloxacin, respectively. For the bacterial isolates from sheep, the highest percentage of resistance to rifampin was (82.3%), and the highest percentage of sensitivity was to imipenem and Levofloxacin antibiotics. The results showed that most of the 39 bacterial isolates (92.8%) possessed an efflux pump mechanism. The result of genotyping to identify the efflux pump genes tolC and acrAB revealed that all isolates carried the genes. Conclusion: All the isolates were resistant to antibiotics, and the bacterial isolates under study most possess the efflux pump mechanism. All bacteria also have efflux pump genes, and this gives the bacteria more resistance against many antibiotics. Keywords: Klebsiella pneumoniae, Efflux pump, Antibiotic resistance. IntroductionKlebsiella pneumoniae is the most important cause of nosocomial infections belonging to the Enterobacteriaceae Gram-negative family (Miftode et al., 2021; AlKhafaji et al., 2024). Bacteria are the cause of some serious diseases when the immune defenses are weak, especially acute pneumonia. K. pneumoniae is considered an opportunistic pathogen that causes pneumonia (Carvalho et al., 2021; Al-Kanany et al., 2023). Some animal products, particularly animal proteins and milk, have become important for global population growth. Therefore, it is necessary to prevent bacterial infections in animals and ruminants, and this is what veterinarians have sought to do (Al-Tememe and Abbas, 2022; Abdulla et al., 2024). There are many virulence factors shared between K. pneumoniae and other bacteria, which enable the transmission of K. pneumoniae bacteria, especially in hospitals. The most important of these factors is resistance to antibiotics of more than 80%. Immunocompromised patients can also be infected with enterobacteriaceae, most notably K. pneumoniae bacteria (Thamer and Shareef, 2022; Baqer et al., 2023). Klebsiella pneumoniae has developed multiple mechanisms to resist antibiotics. The efflux pump mechanism is a system based on a specific protein that is important in expelling antibiotics and some toxic substances out of the cell, where its work is with membrane permeability to reduce the concentrations of these substances (Effah et al., 2020; Ashwath and Sannejal, 2022). The RND family is one of the most important pump families according to its classification, as it includes Gram-negative bacteria, including K. pneumoniae, which contains the acrAB-tolc gene and the diffusion protein in the compartment, as well as a protein to the inside of the channel in the membrane, which releases materials into the environment outside the cell (Reza et al., 2019). Because efflux pumps have the ability to eliminate multiple medicines, including erythromycin, B-lactams, fluoroquinolones, and chloramphenicol, they are the most significant mechanism of antibiotic resistance (Li et al., 2021). However, it is well known that certain bacterial efflux pumps export host-derived antimicrobial compounds in addition to antibiotics and other materials like dyes and detergents (Islamieh et al., 2018; Swedan and Aldakhily, 2024). Many substances, notably antibiotics, are pumped out of bacterial cells via an ATP-dependent transport system (Reza et al., 2019). Antibiotic resistance is related to resistance to efflux pumps, and this has been studied (Kahlmeter et al., 2019). Many gram-negative bacteria also contribute to environmental pollution and thus the occurrence of diseases in society through their ease of transmission among people (Al-Shanawa, 2013). Klebsiella has mechanisms that cause multiple diseases in humans and animals, as it expresses on its surface a soft lipopolysaccharide (LPS with O antigen) and a capsular polysaccharide (K antigen) (Kadhum and Khudor, 2021). In this study, we isolated Klebsiella bacteria from samples of human and sheep with respiratory infection. The isolates were then identified using advanced bacteriological methods, given the critical importance of investigating pathogens that pose public health risks (Mazaal et al., 2021). This study aimed to show the efflux pump mechanism of clinical K. pneumoniae isolates and antibiotic resistance in samples collected from sheep and human respiratory tract infections in southern Iraq. Materials and MethodsSpecimens collection and bacterial identificationThree hundred specimens were collected from the AL-Nasiriya Teaching Hospital and Veterinary Hospital, and the specimens included 150 nasal swabs from sheep. Nasal swabs taken from sheep are a rapid, nonsurgical method that provides accuracy and effectiveness in diagnosing respiratory diseases, and 150 human sputum samples. From the patient records, information about each patient was collected. The samples were then taken and transferred to the laboratory in special containers for the purpose of culturing them on MacConkey agar and incubating them at 37°C for 24 hours. Bacterial cells were stained by Gram staining, and isolates were identified by p Vitek-2 techniques. Biochemical tests were employed to confirm K. pneumoniae as per the methods described by (Jamalludeen, 2020). Antibiotic susceptibility testingThe diffusion method was carried out according to the guidelines of the European Committee for Susceptibility Testing. The antibiotic discs were selected according to the scientific standards. Ten antibiotics were selected against the bacterial isolates after planting them on Mueller-Hinton agar medium. Then, the antibiotics were placed. After that, the plates containing the bacteria and antibiotics were incubated at 37°C for 24 hours. The results were compared with the latest standard tables in the Clinical Laboratory Standards Institute (CLSI) for the year 2023 (Ibraheim et al., 2023). Mechanism efflux pumpThe agar ethidium bromide cartwheel method (Gharbi et al., 2024). Ethidium bromide at concentrations of 0.5, 1, 1.5, and 2 mg/l was incorporated into Müller Hinton agar to evaluate efflux pump phenotypes in K. pneumoniae. Bacterial strains were streaked in a linear cartwheel formation adjacent to a negative control, followed by overnight incubation at 37°C. Efflux-mediated fluorescence was detected using a Uvitec Cambridge UV transilluminator (Sepehr et al., 2022). Molecular assayBy (Bromega/United States), we extracted genomic DNA from K. pneumoniae isolates according to the instructions and methodology of Gram-negative bacteria. Conventional PCR amplification of genes using universal primers (Table 1). The PCR program was performed as described in Table 2. Statistical analysisThe results of the present experiment were statistically analyzed using the Chi-square test, and the variances at p < 0.05 are significant statistically. Ethical approvalThe Laboratory Animal Ethics Committee granted the license and approval to conduct the research at the College of Veterinary Medicine, University of Basra. ResultsSample collection and identification of bacteriaOf 150 sputum human specimens and 150 nasal swabs from sheep were tested, 25 and 17 K. pneumoniae species isolates from patients and sheep, respectively, with respiratory infections were identified using cultural methods. Biochemical tests of the bacterial isolates gave negative results for the indole, methyl red, and oxidase tests, while the bacterial isolates produced catalase and urease enzymes and gave positive results for the Voges-Proskauer and citrate consumption tests. Suspected isolates were also confirmed using the Vitek2 system, which was supported by K. pneumoniae iron through phenotypic methods on culture media. The results of identifying all isolates were positive (100%). Molecular methods through PCR amplification of 16S rRNA using universal primers for 16S rRNA of K. pneumonia (Fig. 1). Table 1. Sequences and sizes of primers used to amplify the acrAB and tolC genes.
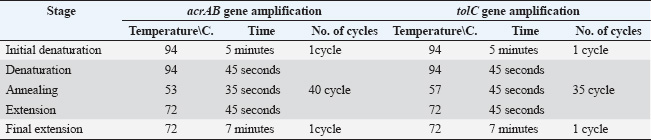
Table 2. Special program for acrAB, tolC, and 16sRNA gene amplification.
Antibiotic susceptibility profile for K. PneumoniaeThe resistance and sensitivity of bacteria to the selected group of antibiotics under study were determined by the Kirby-Bauer method. Then, the inhibition zone of K. pneumoniae bacteria was shown, where the area was measured in millimeters, compared to the standard zone of inhibition as stated in CLSI (2023). The findings revealed that. The majority of sensitive isolates from humans and sheep were for IMipenem. While there were 22% resistant isolates for Piperacillin antibiotics in humans, there were 14 resistant isolates (sheep) for rifampin, as well as some isolates were intermediate or sensitive (Tables 3 and 4). The percentage of resistance to antibiotics was revealed as follows in humans: the highest percentage of resistance to Piperacillin was (88%) and to ceftriaxone was (76 %) whereas the lowest percentage was (936%) to the antibiotic IMipenem and (1040%) to ciprofloxacin whereas the highest percentage of bacterial susceptibility to the antibiotic IMipenem was (1144%) and (936%) to Levofloxacin (Table 3). Regarding the bacterial isolates from sheep, the highest rate of resistance to Rifampin was (14/82.3%), as well as the highest rate of susceptibility to the antibodies IMipenem and Levofloxacin. The results revealed that the antibiotic susceptibility of K. pneumoniae isolated from human and sheep. The results showed a significant (p < 0.05) increase in resistance for antibiotics, while the results were obtained significant (p < 0.05) decreased in sensitive for antibiotics but the resultsnon-significant (p > 0.05) difference in intermediate for antibiotics (Tables 3 and 4). Mechanism efflux pumpThe bacterial isolates were tested to determine the phenotypic appearance of the efflux pump using the Cartwheel test, and the results showed that most of the bacteria, 39 isolates, 92.8% possess the efflux pump mechanism to resist antibiotics. The isolates containing the efflux pump exhibited fluorescent emission under a UV transilluminator (Fig. 2). The result of genotyping to determine the tolC and acrAB efflux pump genes showed that all isolates carried the genes. (Figs. 3 and 4). DiscussionKlebsiella pneumoniae has the ability to cause serious diseases in humans and animals because of its many virulence characteristics; thus, an immune response occurs as an immune reaction against the infection. As observed during our study, the incidence of K. pneumoniae in sheep was lower than in human (Karampatakis et al., 2023; Mahrous et al., 2023). The bacteria were grown on MaConkey agar, after which biochemical tests were performed. Then, the identity of the suspected K. pneumoniae was determined by the VITEK2 system, where the results were all K. pneumoniae, and this result was 100% consistent with previous studies, such as Su et al. (2023) and Al-Mousawi and Al-Daraghi (2022). Because some isolates are more resistant to piperacillin and ceftazidime, which indicates an increase in the expression of the beta-lactamase enzyme, most of the bacterial isolates were sensitive to imipenem, and this is what was agreed upon with Mohammed et al. (2023) and Hou et al. (2024). It was shown that K. pneumoniae isolates are highly susceptible to imipenem, followed by ceftazidime and ciprofloxacin. This result was in agreement with the study by Jasim et al. (2021) reported that bacteria are sensitive to ciprofloxacin. However, K. pneumoniae was resistant to tetracycline and ceftriaxone. Klebsiella pneumoniae bacteria isolated from sheep showed higher resistance to most antibiotics compared to human isolates, as they showed more resistance to rifampin (82.3%), ceftriaxone, and tetracycline (64.7%) Among these isolates, varying degrees of sensitivity were observed, as they were more sensitive to imipenem and levofloxacin, respectively (47%) and 35.2% (Oraibi et al., 2020; Yadav, 2020). The presence of high resistance in bacterial isolates of sheep may be due to the irresponsible use of antibiotics in livestock feed, which may lead to resistance that affects the health of animals and humans, as well as the direct exposure of animals to environmental factors such as environmental pollution resulting from the discharge of antibiotics into water and fertilizers that are not subject to health control, which gives bacteria greater resistance and thus increases bacterial resistance (Larsson and Flach, 2022).
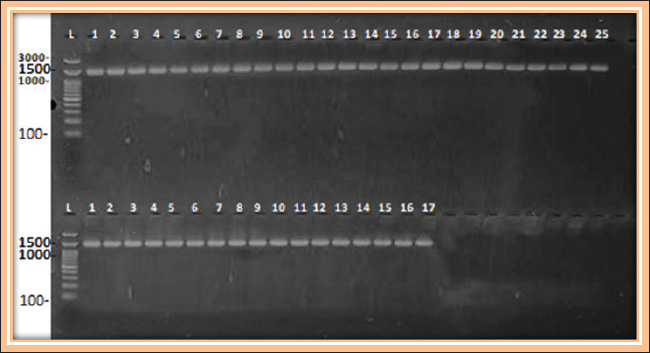
Fig. 1. Gel electrophoresis, for amplification of 16S rRNA 1500bp. of K. pneumoniae using conventional PCR, stained with RedSafe™ and visualized on a UV transilluminator. Marker (L): DNA ladder (100–3,000 bp), Lanes: (1–25) human sputum samples, (1–17) sheep nasal swabs samples. Table 3. Antibiotic susceptibility of K. pneumoniae isolates from human patients.
Table 4. Antibiotic susceptibility of K. pneumoniae isolated from sheep.
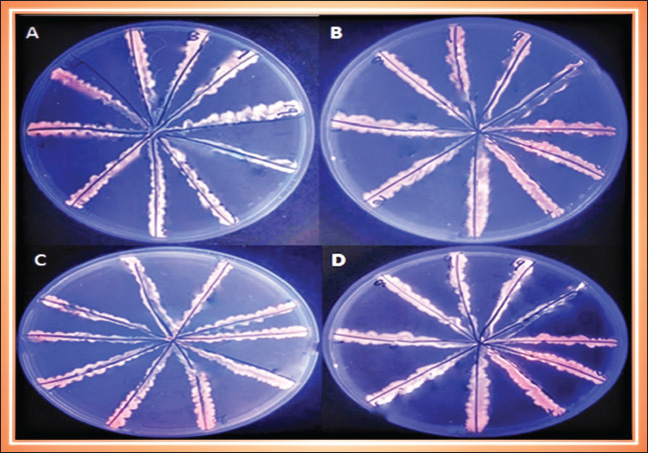
Fig. 2. Efflux pump phenotype using the Cartwheel test for K. pneumoniae in different EtBr concentrations. A: 0.5 mgl B: 1 mgl C: 1.5 mgl D: 2 mgl. The isolates containing the efflux pump exhibited fluorescent emission under a UV transilluminator. All of the K. pneumoniae isolates investigated in this study possess an active efflux pump. This could explain that the active flow pump plays a role in the level of antibiotic resistance (Szabo et al., 2018; Maurya et al., 2019), where the role of the efflux pump in quinolones was explained, resistance to β-lactam antibiotics in K. pneumoniae. During the current study, we found that all isolates possessed an efflux pump system, which is consistent with the study of Akinpelu et al. (2020). In this study, the efflux pumps acrAB and tolC genes were identified. In both human and sheep isolates, it was 100%. These results are consistence to other results recorded by Suresh and Pillai (2024) and Ferreira (2019).
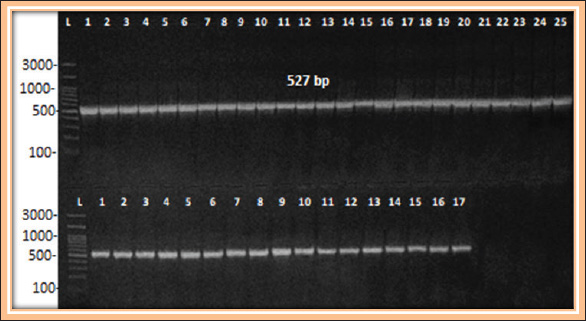
Fig. 3. Conventional PCR results. Gel electrophoresis for tolC gene size 527 bp with RedSafe™ and visualized on a UV transilluminator. Lane (L): 300 bp. ladder, lane (1–25) Positive result for human isolates, lane (1–17) positive result for sheep isolates.
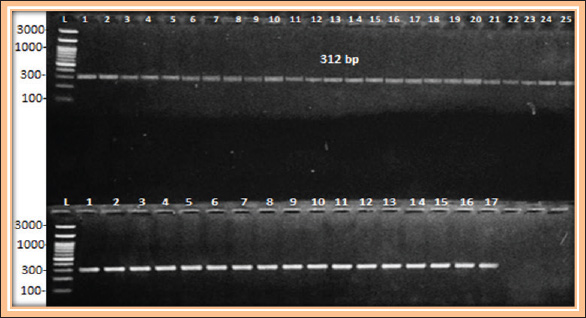
Fig. 4. Conventional PCR results. Gel electrophoresis for acrAB gene size 312 bp. with RedSafe™ and visualized on a UV transilluminator. Lane(L): 300 bp. ladder, lane (1–25) Positive result for human isolates, lane (1–17) positive result for sheep isolates. ConclusionMost K. pneumoniae isolates were isolated from human patients with respiratory infections, as well as from sheep with respiratory infections. In addition, all the isolates were resistant to antibiotics, as it was known that the isolates under study, most bacteria (39 isolates), 92.8% possess the efflux pump mechanism. All bacteria also have efflux pump genes, and this gives the bacteria more resistance against many antibiotics. AcknowledgmentsThis study was conducted to fulfill the requirements for the degree of Doctor of Philosophy in Science Zoonotic Disease between human and animals and Public Health, Faculty of Veterinary Medicine, University of Basrah, Iraq. Conflict of interestThe authors declare no conflict of interest. FundingThe authors received no financial support for the research, authorship, and/or publication of this article. Author’s contributionsAll authors have conceiving, designing, collecting data, analyzing and interpreting, composing, and preparing the first draft, writing, and revising the manuscript. All authors have reviewed and approved the published version of the work. Data availabilityAll the information is contained in the paper. ReferencesAbdulla, N.R., Abdullah, F.A., Kadhum, A.A., Ghanyem, H.S. and Abdulla, N.R. 2024. PCR detection of nontuberculous Mycobacteria 16s Rrna in cows and sheep subclinical mastitis. Adv. Anim. Vet. Sci. 12(10), 1969–1975. Ahmed, Z.S. and Mawlood, A.H. 2023. Molecular characterization of efflux pump and porin related genes in multidrug resistance Klebsiella pneumoniae isolates recovered from Erbil hospitals. J. Univ. Babylon P. Appl. Sci. 31(2), 115–127. Akinpelu, S., Ajayi, A., Smith, S.I. and Adeleye, A.I. 2020. Efflux pump activity, biofilm formation and antibiotic resistance profile of Klebsiella spp. isolated from clinical samples at Lagos University Teaching Hospital. BMC Res. Notes 13(1), 258. Al-Kanany, F., Al-Maliki, M.M., Othman, R.M. and Jaafar, R.S. 2023. The importance of Pseudomonas aeruginosa plasmids for the n-alkanes biodegradation ability. Mesopot. J. Marine Sci. 38(1), 47–54. AlKhafaji, M.H., Mohsin, R.H. and Faqri, A.M.A. 2024. Food additive mediated biosynthesis of AgNPs with antimicrobial activity against hypermucoviscous enterotoxigenic foodborne Klebsiella pneumoniae. Basrah J. Agric. Sci. 37(1), 278–295. Al-Mousawi, M.A.M. and Al-Daraghi, W.A.H. 2022. Molecular detection of Klebsiella pneumoniae and its relationship with multidrug resistance isolated from clinical samples. Biochem. Cell Arch. 22(1), 753. Al-Shanawa, M.A.A. 2023. Bacterial bio-detector for CdCl2 and NiCl2 heavy metal pollutants based on their optical properties. Basrah J. Sci. 41(1), 123–137. Al-Tememe T.M.K. and Abbas B.A. 2022. Molecular detection and phylogenetic analysis of Pseudomonas aeruginosa isolated from some infected and healthy ruminants in Basrah, Iraq. Arch. Razi. Inst. 77(2), 537–544. Ashwath, P. and Sannejal, A.D. 2022. The action of efflux pump genes in conferring drug resistance to Klebsiella species and their inhibition. J. Health Allied Sci. NU. 12(01), 24–31. Baqer, G.K., Adhi, K.S., Baqer, F.K., Baqer, L.K., and Abbas, B.A. 2023. Antibacterial activity and phytochemical screening of Linum usitatissimum L. on bacteria isolated from wound infections. Iranian J. War Public Health 15(4), 347–352. Carvalho, I., Chenouf, N.S., Carvalho, J.A., Castro, A.P., Silva, V., Capita, R., and Poeta, P. 2021. Multidrug-resistant Klebsiella pneumoniae harboring extended spectrum β-lactamase encoding genes isolated from human septicemias. PLoS One 16(5), e0250525. Clinical and Laboratory Standards Institute (CLSI) 2023. Performance standards for antimicrobial susceptibility testing; twenty-five informational supplement. M100-S21 31 (1), 1–163. Effah, C.Y., Sun, T., Liu, S. and Wu, Y. 2020. Klebsiella pneumoniae an increasing threat to public health. Ann. Clin. Microbiol. Antimicro. 19, 1–9. Ferreira, R.L., Da Silva, B.C., Rezende, G.S., Nakamura-Silva, R., Pitondo-Silva, A., Campanini, E.B. and Pranchevicius, M.C. 2019. High prevalence of multidrug-resistant Klebsiella pneumoniae harboring several virulence and β-lactamase encoding genes in a Brazilian intensive care unit. Front. Microbiol. 9, 3198. Gharbia, J.A.R., Kareem, A.A. and Treaf, M.F. 2024. Determination of acrab, tolc efflux pumps genes in strong biofilm former Klebsiella pneumoniae. Romanian J. Diab. Nutr. Metabol. Dis. 31(1), 130–140. Hou, G., Ahmad, S., Li, Y., Yan, D., Yang, S., Chen, S. and Qu, Y. 2024. Epidemiological, virulence, and antibiotic resistance analysis of Klebsiella pneumoniae, a major source of threat to livestock and poultry in some regions of Xinjiang, China. Animals 14(10), 1433. Ibraheim, H.K., Fayez, R.A., Jasim, A.S. and Gharban, H.A. 2023. Role of nuc gene in Staphylococcus aureus to phagocytic activity in different cattle infections. Open Vet. J. 13(8), 1021–1026. Islamieh, I.D., Afshar, D., Yousefi, M. and Esmaeili, D. 2018. Efflux pump inhibitors derived from natural sources as novel antibacterial agents against Pseudomonas aeruginosa: a review. Inter. J. Med. Res. 5(3), 94–105. Jamalludeen, N.M. 2020. Bacterial contamination associated with mobile phones used by students at Basrah Medical College, Basrah, Iraq. J. Basrah Univ. 38(1), 58–66. Jasim, H.A., Abbas, B.A., Farid, H.A. and Basim, M. 2021. Inhibitory effect of honey against some pathogenic bacterial species isolated from clinical specimens. J. Res. Med. Dental Sci. 9(8), 192–197. Kadhum, A.A. and Khudor, M.H. 2021. Phenotypic and molecular characteristics of biofilm and other virulence genes in E. coli and K. pneumoniae isolates from healthy dairy cow, human and environmental sources. Indian J. Forensic Med. Toxicol. 15(1), 2452–2458. Kahlmeter, G., Giske, C.G., Kirn, T.J., and Sharp, S.E. 2019. Point-counterpoint: differences between the European Committee on antimicrobial susceptibility testing and Clinical and Laboratory Standards Institute recommendations for reporting antimicrobial susceptibility results. J. Clin. Microbiol. 57(9), 1110–1128. Karampatakis, T., Tsergouli, K. and Behzadi, P. 2023. Carbapenem-resistant Klebsiella pneumoniae: virulence factors, molecular epidemiology and latest updates in treatment options. Antibiotics 12(2), 234. Larsson, D.J. and Flach, C.F. 2022. Antibiotic resistance in the environment. Nat. Rev. Microbiol. 20(5), 257–269. Li, Y., Wen, H. and Ge, X. 2021. Hormesis effect of berberine against Klebsiella pneumoniae is mediated by up regulation of the efflux pump KmrA. J. Nat. Prod. 84(11), 2885–2892. Mahrous, S.H., El-Balkemy, F.A., Abo-Zeid, N.Z., El-Mekkawy, M.F., El Damaty, H.M. and Elsohaby, I. 2023. Antibacterial and anti-biofilm activities of cinnamon oil against multidrug-resistant Klebsiella pneumoniae isolated from pneumonic sheep and goats. Pathogens 12(9), 1138. Maurya, N., Jangra, M., Tambat, R. and Nandanwar, H. 2019. Alliance of efflux pumps with β-lactamases in multidrug-resistant Klebsiella pneumoniae isolates. Microb. Drug Resist. 25(8), 1155–1163. Mazaal, M.A., Ibrahim, H.K. and Mater, A.D. 2021. Molecular detection of nuc and sea genes of Staphylococcus aureus isolated from cow and sheep meat in Basrah city. Basrah J. Vet. Res. 20(1), 138–149. Miftode, I.L., Nastase, E.V., Miftode, R.Ș., Miftode, E.G., Iancu, L.S., Luncă, C. and Dorneanu, O.S. 2021. Insights into multidrug-resistant K. pneumoniae urinary tract infections: from susceptibility to mortality. Experimen. Ther. Med. 22(4), 1086. Mohammed, A.N., Al-Rawi, D.F. and Buniya, H.K. 2023. Evaluation of the antibiotic resistance of Klebsiella pneumoniae isolated from patients in hospitals in Iraq. Acta Microbiol. Bulg. 39(04), 103–113. Monawer, A.T. 2025. Molecular analysis of Klebsiella pneumoniae isolates collected from sputum samples in Duhok, Iraq. Al-Nahrain J. Sci. 28(1), 91. Oraibi, H.M., Hasan, A.S. and Alazawy, A. 2020. Multiple drug resistant Klebsiella pneumoniae recovered from human and animal sources in Diyala province, Iraq. Plant Arch. 20(2), 7847–7853. Reza, A., Sutton, J.M. and Rahman, K.M. 2019. Effectiveness of efflux pump inhibitors as biofilm disruptors and resistance breakers in gram-negative (ESKAPEE) bacteria. Antibiotics 8(4), 229. Sepehr, A., Fereshteh, S. and Shahrokhi, N. 2022. Detection of efflux pump using ethidium bromide-agar cartwheel method in Acinetobacter baumannii clinical isolates. J. Med. Microbiol. Infect. Dis. 10(1), 36–41. Su, H.Y., Hussain, B., Hsu, B.M., Lee, K.H., Mao, Y.C., Chiang, L.C. and Chen, J.S. 2023. Bacterial community analysis identifies Klebsiella pneumoniae as a native symbiotic bacterium in the newborn Protobothrops mucrosquamatus. BMC Microbiol. 23(1), 213. Suresh, K. and Pillai, D. 2024. Prevalence of antimicrobial resistance, biofilm formation, efflux pump activity, and virulence capabilities in multi-drug-resistant Klebsiella pneumoniae isolated from freshwater fish farms. J. Water Health 22(4), 721–734. Swedan, S.F. and Aldakhily, D.B. 2024. Antimicrobial resistance, biofilm formation, and molecular detection of efflux pump and biofilm genes among Klebsiella pneumoniae clinical isolates from Northern Jordan. Heliyon 10(14), e34370. Szabo, O., Kocsis, B., Szabo, N., Kristof, K. and Szabo, D. 2018. Contribution of OqxAB efflux pump in selection of fluoroquinolone-resistant Klebsiella pneumoniae. Can. J. Infect. Dis. Med. Microbiol. 2018, 4271638. Thamer, M.A. and Shareef, A.A. 2022. Molecular detection of pathogenic vancomycin bacteria in Basrah Governorate. J. Basrah Res. 48(2), 27–34. Yadav, M.M. 2020. Multidrug resistance among Klebsiella pneumoniae passed from the gut of diarrheic goats of University farm, Maharashtra, India. J. Entomol. Zool. Stud. 8, 990–994. | ||
| How to Cite this Article |
| Pubmed Style Kadhum AA, Khudor MH, Ibraheim HK. Clinical Klebsiella pneumoniae isolates and their efflux pump mechanism for antibiotic resistance challenge. Open Vet. J.. 2025; 15(7): 3185-3192. doi:10.5455/OVJ.2025.v15.i7.29 Web Style Kadhum AA, Khudor MH, Ibraheim HK. Clinical Klebsiella pneumoniae isolates and their efflux pump mechanism for antibiotic resistance challenge. https://www.openveterinaryjournal.com/?mno=248397 [Access: January 12, 2026]. doi:10.5455/OVJ.2025.v15.i7.29 AMA (American Medical Association) Style Kadhum AA, Khudor MH, Ibraheim HK. Clinical Klebsiella pneumoniae isolates and their efflux pump mechanism for antibiotic resistance challenge. Open Vet. J.. 2025; 15(7): 3185-3192. doi:10.5455/OVJ.2025.v15.i7.29 Vancouver/ICMJE Style Kadhum AA, Khudor MH, Ibraheim HK. Clinical Klebsiella pneumoniae isolates and their efflux pump mechanism for antibiotic resistance challenge. Open Vet. J.. (2025), [cited January 12, 2026]; 15(7): 3185-3192. doi:10.5455/OVJ.2025.v15.i7.29 Harvard Style Kadhum, A. A., Khudor, . M. H. & Ibraheim, . H. K. (2025) Clinical Klebsiella pneumoniae isolates and their efflux pump mechanism for antibiotic resistance challenge. Open Vet. J., 15 (7), 3185-3192. doi:10.5455/OVJ.2025.v15.i7.29 Turabian Style Kadhum, Ali A., Mohammed H. Khudor, and Hanaa K. Ibraheim. 2025. Clinical Klebsiella pneumoniae isolates and their efflux pump mechanism for antibiotic resistance challenge. Open Veterinary Journal, 15 (7), 3185-3192. doi:10.5455/OVJ.2025.v15.i7.29 Chicago Style Kadhum, Ali A., Mohammed H. Khudor, and Hanaa K. Ibraheim. "Clinical Klebsiella pneumoniae isolates and their efflux pump mechanism for antibiotic resistance challenge." Open Veterinary Journal 15 (2025), 3185-3192. doi:10.5455/OVJ.2025.v15.i7.29 MLA (The Modern Language Association) Style Kadhum, Ali A., Mohammed H. Khudor, and Hanaa K. Ibraheim. "Clinical Klebsiella pneumoniae isolates and their efflux pump mechanism for antibiotic resistance challenge." Open Veterinary Journal 15.7 (2025), 3185-3192. Print. doi:10.5455/OVJ.2025.v15.i7.29 APA (American Psychological Association) Style Kadhum, A. A., Khudor, . M. H. & Ibraheim, . H. K. (2025) Clinical Klebsiella pneumoniae isolates and their efflux pump mechanism for antibiotic resistance challenge. Open Veterinary Journal, 15 (7), 3185-3192. doi:10.5455/OVJ.2025.v15.i7.29 |