| Research Article | ||
Open Vet. J.. 2025; 15(6): 2861-2874 Open Veterinary Journal, (2025), Vol. 15(6): 2861-2874 Research Article Genetic variation in the bovine heat shock protein gene in Indonesian Bali cattle (Bos javanicus): A marker candidate for heat stress tolerance traitRatih Dewi Hapsari1,2, Tristianto Nugroho3, Galih Pambuko1, Luthfi Adya Pradista4, Joko Riyanto1, Nuzul Widyas1 and Sigit Prastowo1*1Faculty of Animal Science, Universitas Sebelas Maret, Surakarta, Indonesia 2Bali Cattle Breeding Center, Denpasar, Indonesia 3Faculty of Animal Science, Universitas Gadjah Mada, Yogyakarta, Indonesia 4Animal Science Study Program, Vocational College, Universitas Sebelas Maret, Surakarta, Indonesia *Correspondence to: Sigit Prastowo. Faculty of Animal Science, Universitas Sebelas Maret, Surakarta, Indonesia. Email: prastowo [at] staff.uns.ac.id Submitted: 23/03/2025 Revised: 27/05/2025 Accepted: 28/05/2025 Published: 30/06/2025 © 2025 Open Veterinary Journal
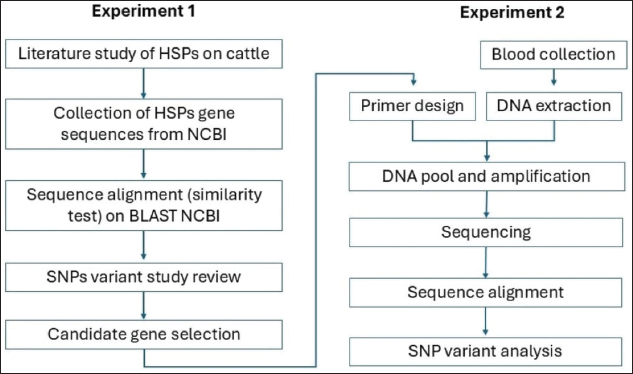
AbstractBackground: Heat stress has a significant impact on cattle performance, particularly in hot-humid climates like tropical regions. Bali cattle (Bos javanicus), an indigenous Indonesian breed, are widely known for their ability to adapt to and withstand heat stress. The heat shock protein (HSP) gene family is responsible for heat stress adaptation, and genetic diversity is linked to the ability of animals to cope with heat stress. Aims: The purpose of this study was to investigate the genetic variation of potential HSP gene candidates in Bali cattle by scanning single-nucleotide polymorphisms (SNPs) and comparing them to the available cattle genetic reference database. Methods: The research was divided into two experimental phases. The first step is to identify potential HSP gene candidates in silico. The selected genes were then mapped to the B. javanicus sequence (year 2023 version) to generate primers that were used to amplify all exons for SNP scanning. In the second experiment, a pool of DNA samples was amplified using a custom-designed primer, followed by sequencing and polymorphism analysis. The SNP variation was identified by comparing the nucleotide sequences with the existing Bos taurus and Bos indicus databases at the National Center for Biotechnology Information, which serve as genetic references. Genetic variations were then grouped, classified, and analyzed based on the type of nucleotide mutation and amino acid type. Results: The results revealed that among the HSP genes in B. javanicus, HSP90AB1 had the highest similarity to those in other breeds: 99.05% similarity with B. taurus and 99.05% with B. indicus. In total, 127 SNPs of the HSP90AB1 gene were identified across exons 2 to 6 of the Bali cattle and have the potential to affect protein function and expression related to cattle traits. Conclusion: HSP90AB1 can be a candidate gene for thermotolerance trait selection in Bali cattle but needs further study to be used as a selection marker. Keywords: Bali cattle, Bos javanicus, HSP gene family, Thermotolerance, Single nucleotide polymorphism (SNP).. IntroductionHeat stress is a condition in which environmental temperatures are higher than the animal’s thermoneutral zone. When cattle are exposed to heat stress, their feed intake is reduced, thereby reducing their metabolic rate. Heat stress exposure also activated several genes and affected the endocrine and immune systems (Shephard and Maloney, 2023). This condition has a negative effect on animal growth (Brown-Brandl, 2018) and meat quality by altering the rate and extension of postmortem muscle glycolysis and the resultant pH (Zhang et al., 2020). Heat stress is also affected in reproduction by reducing the developmental competence of oocyte, disturbs the hormonal balance, reduces the semen quality and sperm motility also will reduce the conception rate by 20%–30% (Khan et al., 2023). To mitigate these effects, cattle breeds differ in their adaptation to heat stress based on their evolutionary backgrounds. Bos indicus is more adapted to tropical climates than Bos taurus because B. taurus is less able to regulate its body temperature in hot conditions (Shephard and Maloney, 2023). On the other hand, there is a cattle breed, namely Bali cattle (Bos javanicus), that has been developed under hot conditions in the tropical country of Indonesia. This cattle are Indonesian indigenous breed that was domesticated from Banteng and has spread throughout the archipelago, mainly on the islands of Bali, Sumatra, Kalimantan, Java, East Nusa Tenggara, and West Nusa Tenggara (Widyas et al., 2017). Young Bali cattle are crimson red or reddish gold, with females retaining this color as they mature, while males change color into black at 12–18 months of age (Sutarno and Setyawan, 2016). Bali cattle are highly valued for their numerous advantages, including a high carcass yield of approximately 52.72%–57.6% (Jakaria et al., 2017) and excellent reproductive performance (Sutarno and Setyawan, 2016) compared to B. taurus and B. indicus. Moreover, it’s explained that they have exceptionally well-adapted to harsh environmental conditions, demonstrating the ability to survive and thrive in poor, dry climates. According to an earlier study, B. javanicus has a higher ranking in resistance to environmental stressors, especially heat, compared to B. taurus and has higher fertility compared to B. taurus and B. indicus (Burrow, 2019). The ability of Bali cattle to resist heat stress resistance and at the same time to maintain productivity will be increasingly valuable in the context of climate change and rising global temperatures. This makes them an ideal candidate for meat production in tropical regions. Heat stress resistance in Bali cattle is attributed to efficient thermoregulation mechanisms, including a lighter coat color in females, which reflects sunlight, and a compact body size that reduces heat retention (Martojo, 2012). Additionally, their ability to graze on low-quality forage and maintain body condition during periods of feed scarcity further enhances their suitability for tropical environments. In humid tropical regions where high temperatures and humidity can negatively impact livestock performance, Bali cattle’s natural resilience offers a sustainable solution for meat production. Their ability to thrive under such conditions reduces the need for costly interventions, such as artificial cooling systems or high-quality feed supplements, making these systems an economically viable option for smallholder farmers. However, Bali cattle also have disadvantages. Their growth is relatively slow and varies between individuals (Sutarno and Setyawan, 2016), presumably due to adaptations to the heat-stress environment in the tropics. In the view of genetic factors, heat stress conditions are crucial for cattle adaptation, and heat shock proteins (HSPa) are recognized for their role in heat stress alleviation. The HSP family consists of dozens of HSPs categorized by molecular weight, such as HSP40, HSP60, HSP70, and HSP90, which generally have a molecular weight near 70 kDa (Sejian et al., 2012), as well as small HSPs ranging from 12 to 43 kDa (Gu et al., 2023). Under heat stress, HSP synthesis is increased, whereas other proteins are downregulated (Sejian et al., 2012). As molecular chaperones, HSPs stabilize and refold damaged cellular proteins during stress (Hussain et al., 2023). Several studies have identified the role of the HSP gene in heat resistance in cattle. For example, HSP70 member 1A (HSPA1A) has a significant association with the physiological response of Bali cattle to heat stress (Suhendro et al., 2022). HSP90AB1 plays a role in regulating the response to heat stress and affects the production traits of Indian dairy cattle (Sajjanar et al., 2015). The role of the HSP family in Bali cattle has not been clearly defined. Although a few studies have been conducted to identify polymorphisms in Bali cattle using available reference sequences from B. taurus or B. indicus genomes. Nevertheless, the reference genome for B. javanicus was released at the end of 2023 National Center for Biotechnology Information (NCBI RefSeq assembly: GCF_032452875.1). Because B. javanicus is a distinct species (Sun et al., 2022) with a long-standing relationship with B. taurus and B. indicus (Mohamad et al., 2009), the HSP family nucleotide and amino acid sequences may differ. Therefore, this study aimed to explore variants of HSP family genes in B. javanicus to understand the thermotolerance mechanism in Bali cattle. Furthermore, the result will be used as a potential gene candidate for the selection process aiming to select fast-growing Bali cattle that are also adapted to the hot climate in tropical conditions. Materials and MethodsAnimal ethical clearanceThis study complied with the ethical guidelines for treating animals as stated in Indonesian Law Number 18, 2009, and all the protocols of sample collection that applied to the cattle were reviewed and accepted by the institutional board of ethical clearance of the Faculty of Animal Science, Universitas Sebelas Maret. During sample collection, a veterinarian handled the animals and ensured that no discomfort occurred throughout the process. Study pipelineThis study was designed to investigate the genetic variation of selected HSP genes, as candidates, through a structured pipeline, as shown in Figure 1. Two distinct experiments, Experiment 1 and 2, were conducted to address the research objectives.
Fig. 1. Study pipeline for HSP gene candidacy in B. javanicus. Experiment 1Experiment 1 performed in silico aimed to discover a potential candidate HSP gene in the HSP family that had characteristics among known cattle breeds. In this study, we used the HSP family data of B. taurus, B. indicus, and B. javanicus. The work pipeline (Fig. 1) was set by exploring the candidate genes by using a literature study to develop a list of HSP family genes that had previously been studied in cattle. We then obtained gene IDs, accession numbers, and locations for all three species from the NCBI at the URL In the second step, single nucleotide polymorphism (SNP) association studies were performed using a literature review of selected genes associated with heat resistance features in various cattle. The most highly linked SNP genes were selected for subsequent primer design experiments. Primers were designed using NCBI Primer-BLAST ( Experiment 2Experiment 2 sought to detect variations in genetic polymorphisms in the candidate HSP gene chosen in Experiment 1 using primers based on the new B. javanicus reference genome. Twenty blood samples (male and female) were randomly collected from Bali cattle reared at the Bali Cattle Breeding Center in Bali, Indonesia (–8.426461353848094, 114.86394424939012). Blood samples were drawn from the jugular vein using venoject needles and deposited in vacutainer containers containing EDTA to prevent coagulation. DNA extraction was performed using the Wizard® Genomic DNA Purification Kit (Promega, USA) without modification to the manufacturer’s protocol. Following extraction, DNA quality assessment was performed using gel electrophoresis to evaluate DNA integrity and a NanoDrop spectrophotometer to assess DNA purity (A260/A280 at 1.8–2.0) and concentration. The 20 DNA samples were then pooled into one DNA template by taking 200 ng from each sample and homogenizing it. The DNA fragment amplification was carried out using PCR, and each reaction consisted of 1 μl pooled DNA template (±100 ng), 10 μl Promega Green PCR Master Mix (Promega, USA), 7 μl nuclease-free water (Promega, USA), and 1 μl (1 μM) primer (Integrated DNA Technologies, Singapore) for each forward and reverse reaction. The total PCR reaction volume is 20 μl. The reactions were mixed in Axygen® 0.2 ml thin-wall 8 strips (Corning, USA), gently vortexed, then spun for 1 minute. The PCR reaction was performed using a Select Cycler™ II Thermal Cycler (Select Bioproduct, Taiwan) with settings of initial denaturation at 95° for 5 minutes, followed by 33 cycles of denaturation at 95° for 30 seconds, annealing at the primer’s annealing temperature for 30 seconds, extension at 72° for 30 seconds, and a final extension at 72° for 10 minutes. The products were subjected to electrophoresis for 30 minutes at 100 volts and observed on 2% agarose gel stained with DNA stain Diamond™ Nucleic Acid Dye (Promega, USA). Each primer pair was used in triplicate PCR reactions with the same DNA pool to ensure consistency across reactions. In each PCR reaction, a negative control (DNA template replaced with water) was set to ensure that no contamination occurred. Following PCR, the triplicate PCR products for each primer were pooled into a single sample, and the PCR products from all primer pairs were sequenced using the Sanger Method. The sequencing was performed by a company namely 1st BASE (Axil Scientific Pte Ltd-Singapore). The sequencing findings, including nucleotide sequences and chromatograms, were mapped to the new B. javanicus reference genome using Unipro UGENE ( ResultBali cattle (B. javanicus), native to Indonesia, exhibit distinct morphological and physiological traits that differentiate mature males from females, which are not only visually striking but also play a role in their adaptation to heat-stress environments. As depicted in Figure 2, the most noticeable difference lies in their coloration: mature males display a predominantly black coat, whereas mature females retain a reddish-brown hue.
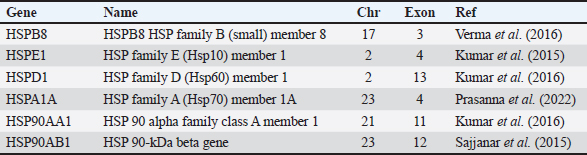
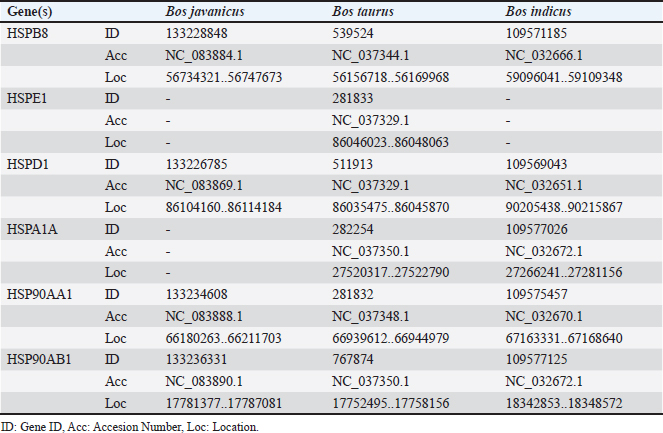
Fig. 2. Mature male (left) and female (right) Bali cattle (B. javanicus). Experiment 1Based on the in-silico analysis, in this study, we found six HSPs, namely HSPB8, HSPE1, HSPD1, HSPA1A, HSP90AA1, and HSP90AB1, that were previously explored in cattle (Table 1). In addition, information on genes including gene ID, location, sequence, etc was shown in Table 2. According to the analysis, all HSPs were available for B. taurus. However, HSPA1A was not available for B. javanicus, and HSPE1 was not available for B. javanicus and B. indicus (Table 2). Table 1. List of explored bovine HSP family.
Table 2. List of bovine HSP genes in the NCBI database.
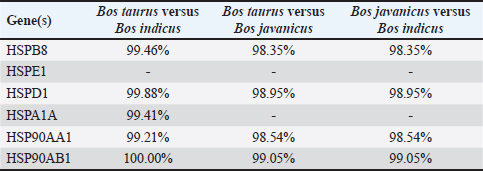
The nucleotides of each gene listed in Table 2 were then compared to assess similarity. A similarity test was conducted between the three cattle breeds: B. taurus versus B. indicus, B. taurus versus B. javanicus, and B. javanicus versus B. indicus. The similarity results (Table 3) showed that most HSPs exhibited high similarity between B. taurus and B. indicus (99.21%–100%). However, the similarity between B. javanicus and other breeds was more diverse (98.35%–99.42%). Among the HSPs in B. javanicus, HSP90AB1 had the highest similarity compared with the other breeds: 99.05% with B. taurus and 99.05% with B. indicus. Based on these results, HSP90AB1 was selected as a potential genetic marker in Bali cattle. Further experiments were conducted using that gene sequence to explore genetic variation. Table 3. Similarity analysis of HSP gene in three different cattle breeds.
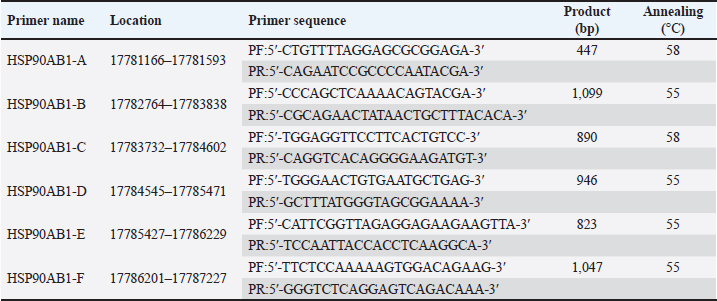
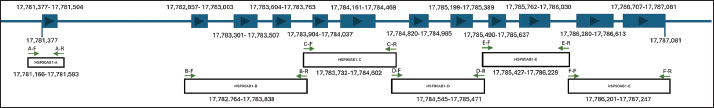
Following our analysis, we found that HSP90AB1 in the new B. javanicus sequence comprises 12 exons. To amplify all exon fragments, these 12 exons were divided into 6 pairs of primers, namely HSP90AB1-A, HSP90AB1-B, HSP90AB1-C, HSP90AB1-D, HSP90AB1-E, and HSP90AB1-F. The primer sequence, location, product size, and annealing are presented in Table 4, and the HSP90AB1 primer location scheme is presented in Figure 3. Table 4. List of primers for the HSP90AB1 gene in B. javanicus.
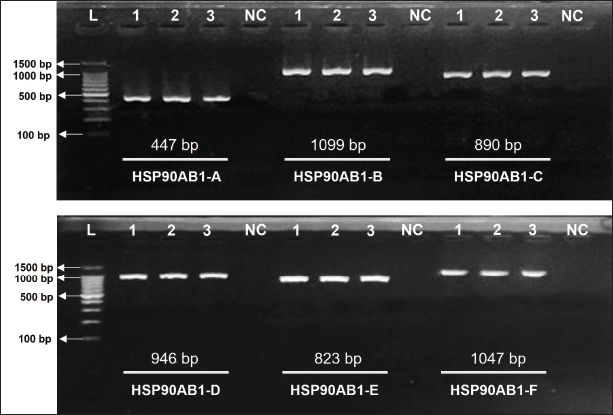
Fig. 3. Designed primer and its location in the exons of the HSP90AB1 gene. Experiment 2The six primer pairs listed in Table 4 were used to amplify the target sequences. Following amplification, the PCR products are presented in Figure 4. The PCR product was very clear, with product sizes ranging from 447 to 1,099 bp. The mutation analysis revealed a total of 127 SNPs across exons 2, 3, 4, 5, and 6.
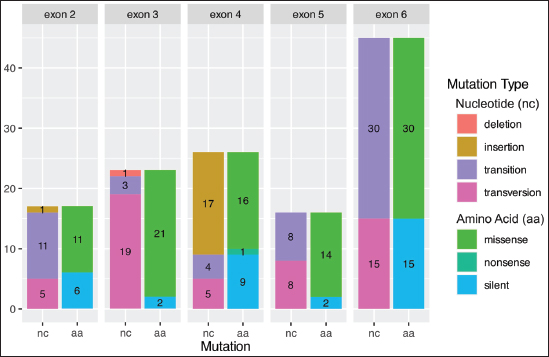
Fig. 4. The amplification results of the HSP90AB1 gene. HSP90AB1-A to HSP90AB1-F represent pairs of specific primers, L is the DNA ladder (100 bp), 1-3 are the DNA samples as templates, and NC is the negative control. Interestingly, no SNPs were found in exons 1, 7, 8, 9, 10, 11, and 12. The types of SNPs and mutations within these exons are shown in Figure 5, providing detailed insights into their specific positions. The mutations varied in type and distribution, with substitutions being the predominant type of mutation across all exons.
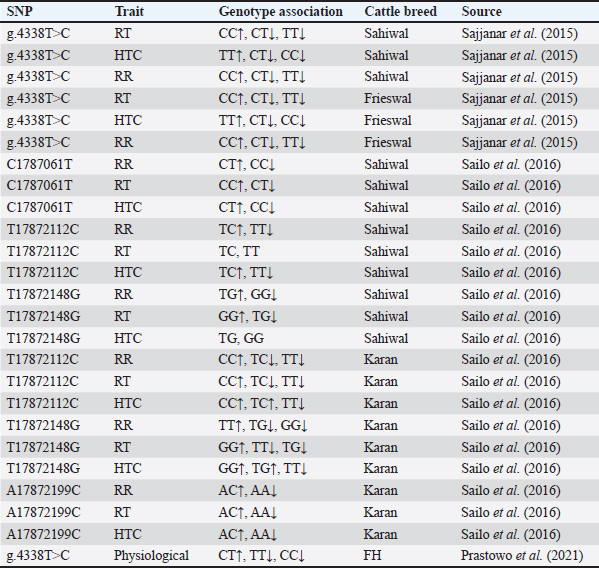
Fig. 5. Distribution of HSP90AB1 genetic variants in Bali cattle. Insertions were most frequent in exon 4, whereas exon 3 contained the only deletion observed in this study. The identification of 127 SNPs across five exons represents a significant finding and suggests that these regions of the gene may be particularly prone to genetic variation. The predominance of substitutions (106 out of 127 mutations) indicates that these exons may be susceptible to point mutations. Substitutions often have varied effects, ranging from silent mutations that do not affect protein function to nonsynonymous mutations that can alter the structure and function of proteins. DiscussionThe sexual dimorphism of Bali cattle in coloration (Fig. 2) between male and female are believed to be linked to hormonal differences and may also serve as a visual cue for social hierarchy and mating behavior within herds. Despite these differences, both sexes share a unique characteristic—the presence of white markings. These include white patches on all four legs and a distinctive white circle around the rump area, which are thought to aid in species recognition and possibly thermoregulation. In addition to their morphological differences, male and female Bali cattle exhibit variations in their performance and adaptation to heat stress, which is a critical factor given the tropical climate of their native habitat. Mature males, as shown in Figure 2, with darker coats, may absorb more solar radiation, potentially making them more susceptible to heat stress compared to females. However, their larger body size and muscular build may provide them with greater endurance and resilience in harsh environmental conditions. On the other hand, females, with their lighter reddish coats, may more readily reflect sunlight, thereby maintaining lower body temperatures. This adaptation is particularly advantageous for reproductive success because heat stress can significantly affect fertility. Both sexes have evolved physiological and behavioral adaptations to cope with high temperatures. For instance, Bali cattle seek shade during the hottest parts of the day and graze during cooler periods, such as the early morning and late afternoon. Their efficient sweating mechanisms and ability to conserve water further enhance their heat tolerance. One of the key genetic mechanisms underlying heat stress adaptation is the expression of HSPs, which play crucial roles in protecting cells from thermal damage by stabilizing proteins and facilitating their repair. Studies have suggested that variations in the HSP family, such as HSP70, are correlated with growth performance in Bali cattle, with certain genetic variants associated with better heat tolerance and productivity (Suhendro et al., 2024). These adaptive traits, combined with their distinct morphological features, make Bali cattle well-suited to thrive in tropical environments, ensuring their survival and productivity in regions prone to heat stress. Understanding these differences and adaptations is crucial for improving breeding programs and management practices to enhance the resilience of Bali cattle in the face of climate change. Many studies have indicated that HSP90AB1 is associated with thermotolerance. Research has shown that the HSP90AB1 gene plays a crucial role in thermotolerance and heat stress resistance in cattle (Deb et al., 2014). HSP like HSP90AB1 helps with protein folding, prevents aggregation, and repairs damaged proteins under stress. An earlier study indicated that HSP90AB1 expression increases significantly in cattle exposed to high temperatures, supporting its role in defending against heat stress. Polymorphisms in this gene (Banerjee et al., 2014) are linked to variations in heat tolerance across breeds, with certain variants offering better thermal stability. Additional research confirmed that HSP90AB1 enhances thermotolerance and overall cattle health in hot climates (Katiyatiya et al., 2017). These findings suggest that selecting favorable genetic variants of HSP90AB1 in breeding programs can improve heat stress resilience and cattle productivity. Several studies on the association of HSP90AB1 in cattle are presented in Table 5. The association of SNP g.4338T>C indicated that the TT genotype had significantly higher heat tolerance coefficient (HTC) and lower respiration rate (RR) values than the CC and CT genotypes in Sahiwal and Frieswal cattle (Sajjanar et al., 2015). In another study, the AA genotype at locus A17872199C was associated with lower RR compared with genotype AC in Karan Fries Cows (Sailo et al., 2016). In Sahiwal cows, the SNP at locus C1787061T has a significant association with rectal temperature (RT), and the CC genotype was higher than the CC genotype (Sailo et al., 2016). Therefore, HSP90AB1 polymorphism needs to be observed, especially in Bali cattle, which has been proven to have far genetic differences between existing cattle breeds (Mohamad et al., 2012). Table 5. Genetic variant association of HSP90AB1 with cattle heat stress tolerance traits.
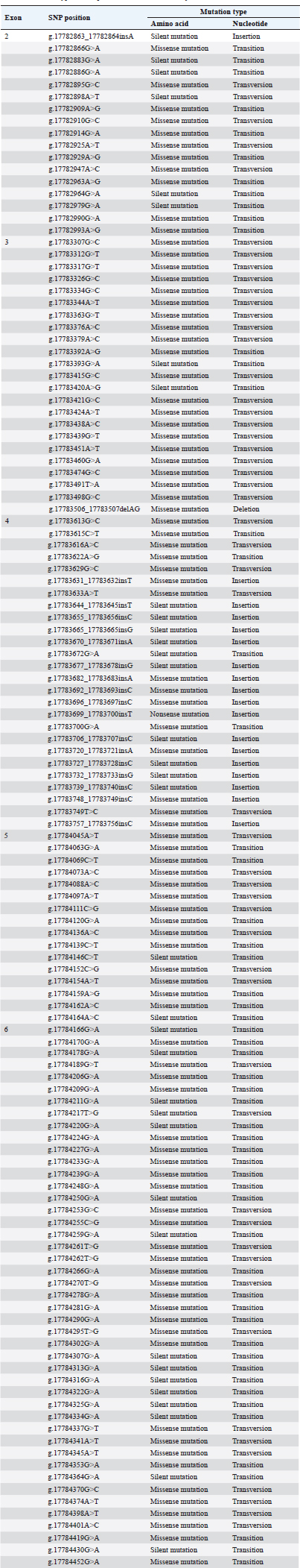
A study on the HSP90AB1 gene identified a total of 3 SNPs in exons 10 and 11 of Thai indigenous cattle (Charoensook et al., 2012), and SNPs were found in exons 8, 10, and 11 of Sahiwal and Karan Fries cows (Sailo et al., 2016). Additionally, SNPs in intron 9 were found to improve heat stress tolerance in Indian Dairy cattle (Bos taurus) (Sajjanar et al., 2015). In this study, no SNPs were detected at that location (Fig. 5). In exon 2 of the HSP90AB1 gene in Bali cattle, 17 SNPs were identified (Table 6) consisting of 16 substitutions and 1 insertion (Fig. 5). These mutations, based on amino acid changes, include 6 silent mutations and 11 missense mutations. From a nucleotide perspective, there are 1 insertion, 11 transitions, and 5 transversions. Interestingly, in other studies focusing on B. taurus (Sajjanar et al., 2015) and B. indicus cattle (Charoensook et al., 2012), no SNPs were found in this exon. This suggests the variability in the presence of SNPs across different cattle populations and the possible species-or breed-specific occurrence of these genetic mutations. Table 6. Table 6. Mutation type at SNP position of HSP90AB1 in B. javanicus.
In exon 3, 23 SNPs (Table 6) consist of 22 substitutions and 1 deletion (Fig. 5). At the amino acid level, these resulted in 2 silent mutations, and 21 missense mutations were observed. There were 3 insertions, 19 transversions, and 1 deletion in the nucleotide changes. This exon is unique in that it contains a deletion. The missense mutations identified in this region could interfere with essential domains of the HSP90AB1 protein, potentially affecting its function in ways that could be associated with disease (Petrosino et al., 2023). However, other studies on HSP90AB1 in cattle did not detect any SNPs in this exon, indicating variability across different research findings. In exon 4, 26 SNPs were identified (Table 6), consisting of 9 substitutions and 17 insertions (Fig. 5). At the amino acid level, these mutations lead to 9 silent mutations and 16 missense mutations. There were 17 insertions, 4 transitions, and 4 transversions in the nucleotide changes. Exon 4 exhibited a unique mutation pattern, with 17 out of 26 mutations being insertion. Insertions, depending on their location and size, can cause frameshift mutations, potentially leading to the production of truncated or nonfunctional proteins (Bartonek et al., 2020). This makes exon 4 a crucial region in which insertions might significantly impact gene function, warranting further study. A key mutation in this region is a nonsense mutation that introduces a premature stop codon. This could truncate the protein, leading to loss of function and possibly resulting in disease or other cellular abnormalities (Martins-Dias and Romão, 2021). In exon 5, the mutation rate is the lowest, with only 16 SNPs (Table 6), all of which are substitutions (Fig. 5). These include 2 silent mutations and 14 missense mutations at the amino acid level. Regarding nucleotide changes, there are 8 transitions and 8 transversions. This observation is consistent with earlier research conducted on Nigerian cattle breeds (B. indicus), where 9 SNPs were found in exons 5 and 6, involving both transitions and transversions (De Campos et al., 2024). This consistency reinforces the importance of exon 5 and its mutation patterns in maintaining functional stability in cattle, although the present missense mutations could still have implications for protein function depending on their specific effects. Exon 6 exhibited the highest number of mutations, with 45 substitutions (Table 6), showing an unusually high frequency of mutations. This may indicate that exon 6 is a hotspot for genetic variation, possibly due to its involvement in regions that are more tolerant of mutations or due to its exposure to mutagenic factors. Further functional studies are needed to assess how these substitutions affect the protein product and whether they have any association with economic traits in Bali cattle. Mutation analysis revealed 15 silent and 30 missense mutations at the amino acid level. In terms of nucleotide changes, there are 30 transitions and 15 transversions (Fig. 5). This observation is consistent with previous research on Nigerian cattle breeds (B. indicus), where 9 SNPs were identified across exons 5 and 6, which also consist of transitions and transversions (De Campos et al., 2024). Silent mutations were present across all exons, suggesting that some genetic alterations did not lead to changes in protein function. However, missense mutations are significantly more prevalent in exons 3, 4, and 6, where amino acid substitutions are likely to affect protein structure and function. These missense mutations could disrupt critical domains within the protein, potentially altering its function in ways that could be associated with disease (Petrosino et al., 2023). The concentration of mutations in these five exons (2, 3, 4, 5, and 6) of HSP90AB1 may indicate that the regions are either more exposed to mutagenic pressures or regions where variations are more tolerated. Future research should focus on characterizing the functional impact of these mutations, particularly those in exon 6 given their high mutation burden. Moreover, the difference sequence around the SNP position can be developed as a marker associated with economic traits (Hartatik et al., 2023), after the association study validates the result. ConclusionSix HSP gene families have been identified in cattle, although only four are represented in the new B. javanicus genome database. The B. javanicus HSP90AB1 gene was highly comparable to those of B. taurus and B. indicus. A total of 127 SNPs in the HSP90AB1 gene were discovered across exons 2 to 6 of the Bali cattle, with most of the SNPs resulting in missense changes, potentially affecting protein function and expression related to bovine attributes. Additional association studies are needed to determine the effects of these mutations on thermotolerance in Bali cattle. AcknowledgmentsThe authors are grateful to the Bali Cattle Breeding Center, Pulukan, Bali, Indonesia, for their support. Conflict of interestThe authors declare that there are no conflicts of interest related to the current study. FundingThis study was funded by the Fundamental Research Scheme provided by The Ministry of Education, Culture, Research and Technology Republic of Indonesia at fiscal year 2024 (contract number 1076.1/ UN27.22/PT.01.03/2024). Author contributionConceptualization: R.D.H., T.N., S.P.; sample collection: R.D., T.N., G.P.; data curation and analysis: R.D.H., T.N., G.P., L.A.P., N.Z., S.P.; data validation: S.P., T.N.; software analysis and visualization: G.P.; drafting the original draft: R.P., S.P., T.N.; supervision, review and editing the draft: J.R., N.Z., S.P.; project administration: L.A.; funding acquisition: S.P., N.Z., T.N. All authors have read and agreed to the publication of this paper. Data availabilityThe data supporting the findings of this study are available upon request to the corresponding author. ReferencesBanerjee, D., Upadhyay, R.C., Chaudhary, U.B., Kumar, R., Singh, S., Ashutosh, Mohanarao, J.G., Polley, S., Mukherjee, A., Das, T.K. and De, S. 2014. Seasonal variation in expression pattern of genes under HSP70: seasonal variation in expression pattern of genes under HSP70 family in heat-and cold-adapted goats (Capra hircus). Cell Stress Chaperones 19(3), 401–408; doi:10.1007/s12192-013-0469-0 Bartonek, L., Braun, D. and Zagrovic, B. 2020. Frameshifting preserves key physicochemical properties of proteins. Proc. Natl. Acad. Sci. USA 117, 5907–5912; doi:10.1073/pnas.1911203117 Brown-Brandl, T.M. 2018. Understanding heat stress in beef cattle. R. Bras. Zootec. 47, e20160414; doi:10.1590/rbz4720160414 Burrow, H. 2019. Strategies for increasing beef cattle production under dryland farming systems. Indonesian Bull. Anim. Vet. Sci. 29(4), 161; doi:10.14334/wartazoa.v29i4.2452 Charoensook, R., Gatphayak, K., Sharifi, A.R., Chaisongkram, C., Brenig, B. and Knorr, C. 2012. Polymorphisms in the bovine HSP90AB1 gene are associated with heat tolerance in Thai indigenous cattle. Trop. Anim. Health Prod. 44, 921–928; doi:10.1007/s11250-011-9989-8 Deb, R., Sajjanar, B., Singh, U., Kumar, S., Singh, R., Sengar, G. and Sharma, A. 2014. Effect of heat stress on the expression profile of HSP90 among Sahiwal (Bos indicus) and Frieswal (Bos indicus × Bos taurus) breeds of cattle: a comparative study. Gene 536, 435–440; doi:10.1016/j.gene.2013.11.086 De Campos, J.S., Onasanya, G.O., Ubong, A., Yusuff, A.T., Adenaike, A.S., Mohammed, A.A. and Ikeobi, C.O. 2024. Potentials of single nucleotide polymorphisms and genetic diversity studies at HSP90AB1 gene in Nigerian White Fulani, Muturu, and N’Dama cattle breeds. Trop. Anim. Health Prod. 56(2), 58; doi:10.1007/s11250-024-03909-z Gu, C., Fan, X. and Yu, W. 2023. Functional diversity of mammalian small heat shock proteins: a review. Cells 12(15), 1947; doi:10.3390/cells12151947 Hartatik, T., Chairunissa, F.A.Z., Bintara, S., Fadillah, F.J., Ningrum, N.P., Puspitasari, D. and Adiwimarta, K. 2023. Mutation analysis and restriction site mapping of GDF9 in Indonesian Bligon goat. Trop. Anim. Sci. J. 46, 163–171; doi:10.5398/ tasj.2023.46.2.163 Hussain, T., Qadri, R., Wajid, A. and Babar, M.E. 2023. Cattle be in two mind states: an overview of heat stress tolerance in cattle. Int. J. Agric. Biol. 29, 133–140; doi:10.17957/IJAB/15.2012 Jakaria, J., Khasanah, H., Priyanto, R., Baihaqi, M. and Ulum, M.F. 2017. Prediction of meat quality in Bali cattle using ultrasound imaging. J. Indonesian Trop. Anim. Agric. 42, 59–65; doi:10.14710/ jitaa.42.2.59-65 Katiyatiya, C.L.F., Bradley, G. and Muchenje, V. 2017. Thermotolerance, health profile and cellular expression of HSP90AB1 in Nguni and Boran cows raised on natural pastures under tropical conditions. J. Therm. Biol. 69, 85–94; doi:10.1016/j. jtherbio.2017.06.009 Khan, I., Mesalam, A., Heo, Y.S., Lee, S.H., Nabi, G. and Kong, I.K. 2023. Heat stress as a barrier to successful reproduction and potential alleviation strategies in cattle. Animals 13(14), 2359; doi:10.3390/ani13142359 Kumar, A., Ashraf, S., Goud, T.S., Grewal, A., Singh, S.V., Yadav, B.R. and Upadhyay, R.C. 2015. Expression profiling of major heat shock protein genes during different seasons in cattle (Bos indicus) and buffalo (Bubalus bubalis) under tropical climatic condition. J. Therm. Biol. 51, 55–64; doi:10.1016/j.jtherbio.2015.03.006 Kumar, R., Gupta, I.D., Verma, A., Singh, S.V., Verma, N., Vineeth, M.R., Magotra, A. and Das, R. 2016. Novel SNP identification in exon 3 of HSP90AA1 gene and their association with heat tolerance traits in Karan Fries (Bos taurus × Bos indicus) cows under tropical climatic condition. Trop. Anim. Health Prod. 48, 735–740; doi:10.1007/s11250-016-1016-7 Martins-Dias, P. and Romão, L. 2021. Nonsense suppression therapies in human genetic diseases. Cell. Mol. Life Sci. 78, 4677–4701; doi:10.1007/ s00018-021-03809-7 Martojo, H. 2012. Indigenous Bali cattle are most suitable for sustainable small farming in Indonesia. Reprod. Domest. Anim. 47, 10–14; doi:10.1111/ j.1439-0531.2011.01958.x Mohamad, K., Olsson, M., Andersson, G., Purwantara, B., van Tol, H.T.A., Rodriguez-Martinez, H., Colenbrander, B. and Lenstra, J.A. 2012. The origin of Indonesian cattle and conservation genetics of the Bali cattle breed. Reprod. Domest. Anim. 47, 18–20; doi:10.1111/j.1439-0531.2011.01960.x Mohamad, K., Olsson, M., van Tol, H.T.A., Mikko, S., Vlamings, B.H., Andersson, G., Rodríguez-Martínez, H., Purwantara, B., Paling, R.W., Colenbrander, B. and Lenstra, J.A. 2009. On the origin of Indonesian cattle. PLoS One 4, 1–6; doi:10.1371/journal.pone.0005490 Okonechnikov, K., Golosova, O. and Fursov, M. 2012. Unipro UGENE : a unified bioinformatics toolkit. Bioinformatics 28, 1166–1167; doi:10.1093/ bioinformatics/bts091 Petrosino, M., Novak, L., Pasquo, A., Turina, P., Capriotti, E., Minicozzi, V., Consalvi, V. and Chiaraluce, R. 2023. The complex impact of cancer-related missense mutations on the stability and on the biophysical and biochemical properties of MAPK1 and MAPK3 somatic variants. Hum. Genomics. 17, 95; doi:10.1186/s40246-023-00544-x Prasanna, J.S., Rao, S.T.V., Prakash, M.G., Rathod, S., Kalyani, P. and Reddy, B.R. 2022. Association of SSCP polymorphisms of HSP70 gene with physiological, production and reproduction performance in Sahiwal and crossbred cows. Asian J. Dairy Res. 41, 150–155; doi:10.18805/ajdfr.DR-1796 Prastowo, S., Adzdzakiy, M.M., Vanessa, R., Pambuko, G., Purwadi, Susilowati, A. and Sutarno. 2021. Polymorphism scanning of HSP90AB1 gene in local Friesian Holstein as molecular marker for heat stress resistance. E3S Web. Conf. 306, 05016. Sailo, L., Gupta, I.D., Verma, A., Das, R., Chaudhari, M.V. and Singh, S. 2016. Polymorphisms in Hsp90ab1 gene and their association with heat tolerance in Sahiwal and Karan Fries cows. Indian J. Anim. Res. 50, 856–861; doi:10.18805/ijar.v0iOF.6662 Sajjanar, B., Deb, R., Singh, U., Kumar, S., Brahmane, M., Nirmale, A., Bal, S.K. and Minhas, P.S. 2015. Identification of SNP in HSP90AB1 and its association with the relative thermotolerance and milk production traits in Indian dairy cattle. Anim. Biotechnol. 26, 45–50; doi:10.1080/10495398.2014.882846 Sejian, V., Naqvi, S.M.K., Ezeji, T., Lakritz, J. and Lal, R. 2012. Environmental stress and amelioration in livestock production. Berlin, Germany: Springer-Verlag Berlin Heidelberg. Shephard, R.W. and Maloney, S.K. 2023. A review of thermal stress in cattle. Aust. Vet. J. 101, 417–429; doi:10.1111/avj.13275 Suhendro, I., Jakaria, J., Priyanto, R., Manalu, W. and Noor, R.R. 2022. The association of single nucleotide polymorphism-69t> G HSPA1A gene with bali cattle heat tolerance. Trop. Anim. Sci. J. 45, 429–435; doi:10.5398/tasj.2022.45.4.429 Suhendro, I., Noor, R.R., Jakaria, J. Priyanto, R., Manalu, W. and Andersson, G. 2024. Association of heat-shock protein 70.1 gene with physiological and physical performance of Bali cattle. Vet. World 17, 17–25; doi:10.14202/vetworld.2024.17-25 Sun, X., Ciucani, M.M., Rasmussen, J.A., Gilbert, M.T.P. and Sinding, M.H.S. 2022. Genomic evidence refutes the hypothesis that the Bornean banteng is a distinct species. BMC Ecol. Evol. 22(1), 110; doi:10.1186/s12862-022-02062-1 Sutarno. and Setyawan, A.D. 2016. The diversity of local cattle in Indonesia and the efforts to develop superior indigenous cattle breeds. Biodiv. J. Biol. Divers. 17, 275–295; doi:10.13057/biodiv/d170139 Verma, N., Gupta, I.D., Verma, A., Kumar, R., Das, R. and Vineeth, M.R. 2016. Novel SNPs in HSPB8 gene and their association with heat tolerance traits in Sahiwal indigenous cattle. Trop. Anim. Health Prod. 48, 175–180; doi:10.1007/s11250-015-0938-9 Widyas, N., Nugroho, T. and Prastowo, S. 2017. Rooms for genetic improvement in Indonesian Bali cattle population. IOP Conf. Ser. Mater. Sci. Eng. 193, 12037. Zhang, M., Dunshea, F.R., Warner, R.D., Digiacomo, K., Osei-Amponsah, R. and Chauhan, S.S. 2020. Impacts of heat stress on meat quality and strategies for amelioration: a review. Int. J. Biometeorol. 64(9), 1616–1628; doi: 10.1007/s00484-020-01929-6 | ||
| How to Cite this Article |
| Pubmed Style Hapsari RD, Nugroho T, Pambuko G, Pradista LA, Riyanto J, Widyas N, Prastowo S. Genetic variation in the bovine heat shock protein gene in Indonesian Bali cattle (Bos javanicus): A marker candidate for heat stress tolerance trait. Open Vet. J.. 2025; 15(6): 2861-2874. doi:10.5455/OVJ.2025.v15.i6.55 Web Style Hapsari RD, Nugroho T, Pambuko G, Pradista LA, Riyanto J, Widyas N, Prastowo S. Genetic variation in the bovine heat shock protein gene in Indonesian Bali cattle (Bos javanicus): A marker candidate for heat stress tolerance trait. https://www.openveterinaryjournal.com/?mno=248870 [Access: January 25, 2026]. doi:10.5455/OVJ.2025.v15.i6.55 AMA (American Medical Association) Style Hapsari RD, Nugroho T, Pambuko G, Pradista LA, Riyanto J, Widyas N, Prastowo S. Genetic variation in the bovine heat shock protein gene in Indonesian Bali cattle (Bos javanicus): A marker candidate for heat stress tolerance trait. Open Vet. J.. 2025; 15(6): 2861-2874. doi:10.5455/OVJ.2025.v15.i6.55 Vancouver/ICMJE Style Hapsari RD, Nugroho T, Pambuko G, Pradista LA, Riyanto J, Widyas N, Prastowo S. Genetic variation in the bovine heat shock protein gene in Indonesian Bali cattle (Bos javanicus): A marker candidate for heat stress tolerance trait. Open Vet. J.. (2025), [cited January 25, 2026]; 15(6): 2861-2874. doi:10.5455/OVJ.2025.v15.i6.55 Harvard Style Hapsari, R. D., Nugroho, . T., Pambuko, . G., Pradista, . L. A., Riyanto, . J., Widyas, . N. & Prastowo, . S. (2025) Genetic variation in the bovine heat shock protein gene in Indonesian Bali cattle (Bos javanicus): A marker candidate for heat stress tolerance trait. Open Vet. J., 15 (6), 2861-2874. doi:10.5455/OVJ.2025.v15.i6.55 Turabian Style Hapsari, Ratih Dewi, Tristianto Nugroho, Galih Pambuko, Luthfi Adya Pradista, Joko Riyanto, Nuzul Widyas, and Sigit Prastowo. 2025. Genetic variation in the bovine heat shock protein gene in Indonesian Bali cattle (Bos javanicus): A marker candidate for heat stress tolerance trait. Open Veterinary Journal, 15 (6), 2861-2874. doi:10.5455/OVJ.2025.v15.i6.55 Chicago Style Hapsari, Ratih Dewi, Tristianto Nugroho, Galih Pambuko, Luthfi Adya Pradista, Joko Riyanto, Nuzul Widyas, and Sigit Prastowo. "Genetic variation in the bovine heat shock protein gene in Indonesian Bali cattle (Bos javanicus): A marker candidate for heat stress tolerance trait." Open Veterinary Journal 15 (2025), 2861-2874. doi:10.5455/OVJ.2025.v15.i6.55 MLA (The Modern Language Association) Style Hapsari, Ratih Dewi, Tristianto Nugroho, Galih Pambuko, Luthfi Adya Pradista, Joko Riyanto, Nuzul Widyas, and Sigit Prastowo. "Genetic variation in the bovine heat shock protein gene in Indonesian Bali cattle (Bos javanicus): A marker candidate for heat stress tolerance trait." Open Veterinary Journal 15.6 (2025), 2861-2874. Print. doi:10.5455/OVJ.2025.v15.i6.55 APA (American Psychological Association) Style Hapsari, R. D., Nugroho, . T., Pambuko, . G., Pradista, . L. A., Riyanto, . J., Widyas, . N. & Prastowo, . S. (2025) Genetic variation in the bovine heat shock protein gene in Indonesian Bali cattle (Bos javanicus): A marker candidate for heat stress tolerance trait. Open Veterinary Journal, 15 (6), 2861-2874. doi:10.5455/OVJ.2025.v15.i6.55 |