| Research Article | ||
Open Vet. J.. 2025; 15(6): 2782-2788 Open Veterinary Journal, (2025), Vol. 15(6): 2782-2788 Research Article Pasteurella multocida genome assembly of Indonesian local isolates using long-read Oxford Nanopore Technology sequencingSri Suryatmiati Prihandani1,2, I. Wayan Teguh Wibawan1*, Susan Maphilindawati Noor2, Safika Safika1, Aswin Rafif Khairullah2, Susanti Susanti2, Sutiastuti Wahyuwardani2, Andriani Andriani2, Sumarningsih Sumarningsih2, Heri Kurnianto2, Tati Ariyanti2, Yudi Adinata3, Harimurti Nuradji2 and Alif Rahman Rohim Puarada41Division of Medical Microbiology, School of Veterinary Medicine and Biomedical, Bogor Agricultural University (IPB University), Bogor, Indonesia 2Research Center for Veterinary Science, National Research and Innovation Agency (BRIN), Bogor, Indonesia 3Research Center for Animal Husbandry, National Research and Innovation Agency (BRIN), Bogor, Indonesia 4Division of Clinic and Laboratory, Pethut Group, Tangerang, Indonesia *Corresponding Author: I. Wayan Teguh Wibawan. Division of Medical Microbiology, School of Veterinary Medicine and Biomedical, Bogor Agricultural University (IPB University), Bogor, Indonesia. Email: wayanwi [at] apps.ipb.ac.id Submitted: 29/03/2025 Revised: 10/05/2025 Accepted: 14/05/2025 Published: 30/06/2025 © 2025 Open Veterinary Journal
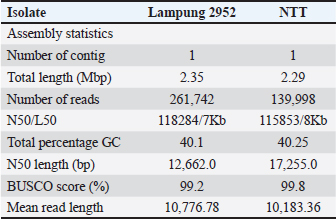
AbstractBackground: Pasteurella multocida is a Gram-negative bacterium responsible for various diseases in animals, including hemorrhagic septicemia (HS). Oxford Nanopore Technologies’ (ONT) platform streams the sequencing data immediately, allowing for on-the-fly analysis. This capability enables researchers to monitor experiments in real time and make prompt decisions, which is particularly beneficial for clinical diagnostics and outbreak surveillance. Aim: This study presents the genome assembly of two P. multocida isolates using long-read sequencing technology provided by ONT. Methods: This method involves passing DNA strands through nanopores and detecting changes in electrical current to determine nucleotide sequences. The long-read capability of ONT technology allows for the sequencing of extensive DNA fragments, which is particularly beneficial for resolving complex genomic regions and achieving high-contiguity genome assemblies. Results: The study generated high-quality single-contig genomes with sizes of 2.35 and 2.29 Mbp and completeness scores exceeding 99% (BUSCO). These assemblies provide valuable resources for functional analyses, enhancing the understanding of virulence factors and the pathogenesis of HS in Indonesian livestock. Conclusion: This study successfully assembled the genomes of two P. multocida isolates from Lampung (2,952) and Kupang, Nusa Tenggara Timur, Indonesia, using Oxford Nanopore Technologies’ long-read sequencing. The resulting single-contig genomes, measuring 2.35 and 2.29 Mbp, respectively, exhibited high completeness scores (BUSCO: >99%). These high-quality assemblies enhance genomic contiguity, providing valuable resources for functional analyses aimed at understanding the virulence factors and pathogenesis of HS in Indonesian livestock. Keywords: Genome assembly, Hemorrhagic septicemia, Oxford Nanopore Technologies, P. multocida, Whole-genome sequencing.. IntroductionPasteurella multocida is a Gram-negative pathogen bacterial of veterinary significance that affects livestock and causes substantial economic losses worldwide (Lestari et al., 2025). P. multocida is a major causative agent of hemorrhagic septicemia (HS), a severe, often fatal disease affecting cattle and buffalo (Hasnan et al., 2022). HS outbreaks have significant economic impacts on smallholder farmers and the livestock industry throughout the Indonesian archipelago, particularly in regions such as Sumatra and Nusa Tenggara (Yohanes and Theresia, 2012). This is because both locations are livestock warehouses in Indonesia and HS-endemic areas. This disease has long been recognized in Indonesia as a strategic infectious animal disease. Young livestock are usually more susceptible to disease than adult animals. Buffalo are more susceptible to HS than cattle (Almoheer et al., 2022). This disease attacks various organs, causing bleeding in the digestive system, under the skin, and even the respiratory tract (Wei et al., 2025). Clinical signs usually begin with fever, lethargy, lacrimation, excessive salivation, subcutaneous edema, and nasal discharge, followed by respiratory distress, snoring, and septic shock with extensive bleeding (Lestari et al., 2025). In addition, swelling and edema can be seen on the head, lower chest, legs, or base of the tail (Doyle- Baker et al., 2020). Lesions in the esophagus cause shortness of breath and difficulty swallowing. Affected animals appear depressed. Death may occur 1-3 days after clinical signs begin (Shivachandra et al., 2011). Antibiotic treatment can be effective in the early stages, but because acute clinical signs are usually very rapid, mortality is almost 100% (Almoheer et al., 2022). Effective and efficient prevention of HS is achieved through vaccination (Gowrakkal et al., 2014). Proper genome assembly is essential for identifying antimicrobial resistance genes and virulence factors (Zankari et al., 2012). Traditional short-read sequencing methods often produce fragmented assemblies, complicating downstream genomic analysis. Long-read sequencing technologies, such as Oxford Nanopore Technology (ONT), offer improved genome contiguity and completeness by resolving repetitive regions that typically fragment short-read assemblies (Wick et al., 2019). In this study, we generated genome assemblies for local Indonesian isolates of P. multocida from Lampung, Sumatera (labeled 2952) Nusa Tenggara Timur (NTT), and Kupang NTT (labeled NTT) using ONT sequencing. This approach aims to provide high-quality reference genomes to better understand the genetic basis of the virulence and pathogenicity of local P. multocida strains, which is crucial for developing improved diagnostic tools and control strategies specific to the Indonesian context. Materials and MethodsBacterial strains and DNA extractionPasteurella multocida 2952 was collected from clinical cases of bovine HS in Lampung, Sumatera Island, and P. multocida NTT was collected from Nusa Tenggara Timur, Indonesia. Both P. multocida isolates were grown on a brain heart infusion (BHI) medium overnight in an incubator at 37°C, and DNA was extracted using Qiagen QIAamp DNA Mini Kits, following the manufacturer’s protocol. To continue DNA extraction, a series of tests were conducted to ensure the feasibility of whole-genome sequencing (WGS). The test included polymerase chain reaction targeting on the km gene, a specific marker for P. multocida in confirming the identity of the isolates. Initial DNA quantification and purity assessment were performed using an Implen NanoPhotometer N50/N60, followed by visualization via agarose gel electrophoresis. Precise DNA quantification was performed using a QubitFlex Fluorometer (Thermo Scientific), and DNA integrity was assessed using an Agilent TapeStation 4150. Library preparation and sequencingA library preparation kit from ONT was used to prepare whole-genome DNA libraries from P. multocida isolates NTT and 2952. The prepared libraries were quantified using a Qubit Fluorometer before being loaded onto the flow cell. DNA repair was performed using an end-prep enzyme mix, resulting in 5'-phosphorylated and 3'-dA- tailed ends. The repaired DNA was then ligated with ONT-compatible adapters. Sequencing was conducted using the PromethION 2 Solo platform (ONT) until the required sequencing yield was achieved. The sequencing process was managed using MinKNOW software, and base calling was performed using Dorado in high-accuracy mode (HAC). The sequencing run was configured with the following parameters: Flow cell type FLO-PRO114M, Kit type SQK-NBD114-24, MinKNOW version 24.02.16, Bream version 7.9.8, Configuration 5.9.18, Dorado version 7.3.11, and MinKNOW Core version 5.9.12. Data analysisFiltering was performed using Filtlong (https://github. com/rrwick/Filtlong), and data transformation was performed using Nanoplot, Canu, and Flye. The read quality was evaluated using Nanoplot (De Coster et al., 2018). Reads correction and assembly were performed using Canu and Flye (Koren et al., 2017). The assembled sequence was polished four times with Racon (Luan et al., 2024) and three times with Medaka (https://github.com/nanoporetech/medaka) for further refinement. Racon was chosen for initial polishing due to its efficiency in correcting errors from long-read assemblies, while Medaka was used for final polishing because it is specifically optimized for Oxford Nanopore data. The polishing was mapped using minimap2 v2.2-r1122 (Li et al., 2021). The quality of the assembled sequence was determined using Quast v5.2.0 and Qualimap v2.2.2. Gene annotation was conducted using NCBI’s Prokaryotic Genome Annotation Pipeline (PGAP) (Li et al., 2021), and genome visualization was performed using Circos v0.69-9. Taxonomy check verification was performed using dfast_qc (Tanizawa et al., 2018), as shown in Table 3 and 4. Genome completeness was evaluated using BUSCO v5.4.3 (Levy and Boone, 2019) and CheckM (integrated in dfast_qc) (Simão et al., 2015). Ethical approvalThis research was approved by the ethics committee for animal care and use of the National Research and Innovation Agency (BRIN) (approval number: 139/ KE.02/SK/07/2023. ResultSequencing output and assembly statisticsThe two isolates yielded approximately 2.82 and 1.43 Gb of sequencing data for isolates 2952 and NTT, respectively. The mean read length exceeded 10 kb (10,776.78 bp for 2952 and 10,183.36 bp for NTT), facilitating high-contiguity assemblies. Both genomes were assembled into single, complete circular chromosomes with no gaps or ambiguities. Assembly metricsBoth isolates were successfully assembled into single-contig genomes with high completeness scores (BUSCO: 99.2% and 99.8%), indicating high-quality assemblies. The genome sizes (2.34 Gb for 2952 and 2.29 Gb for NTT; 2.35 Mbp for 2952 and 2.29 Mbp for NTT) and GC content (40.1% and 40.25%) were consistent with other P. multocida strains reported in the literature. The mean read length exceeded 10 kb, facilitating high-contiguity assemblies (Table 1). Table 1. Summary of recent genome assembly statistics of bovine P. multocida.
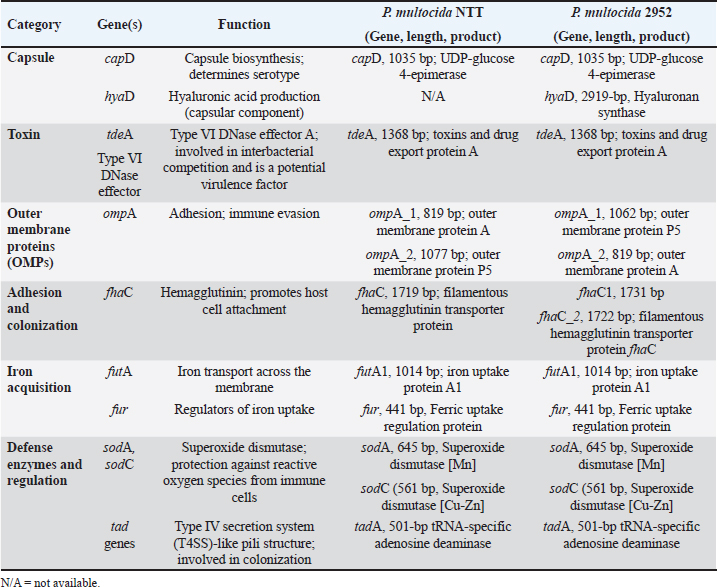
Genome annotation featuresAnnotation of the genomes using NCBI’s PGAP identified 2,104 coding sequences (CDS), 19 rRNAs (5S, 16S, 23S), and 57 tRNAs in isolate 2952, and 2,065 CDS, 19 rRNAs, and 56 tRNAs in isolate NTT. The genomes of NTT and 2952 contained genes associated with virulence, including capsular biosynthesis genes (capD), outer membrane proteins (ompA_1 and ompA_2), iron acquisition systems (futA1 and fur), and adhesions (fhaC), as shown in Table 2. Table 2. Virulence-associated genes identified in P. multocida NTT and 2952 isolates using the Prokka annotation tool.
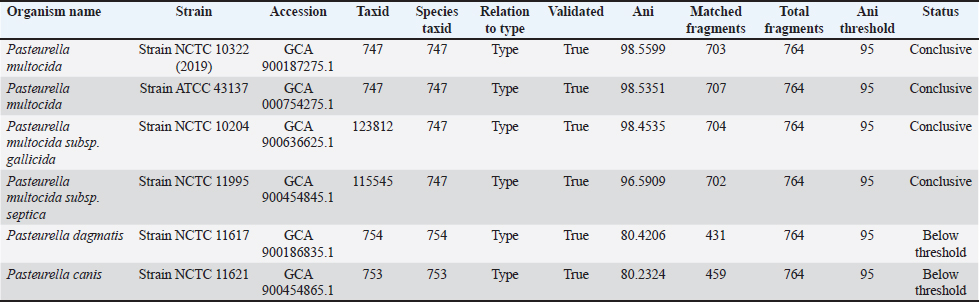
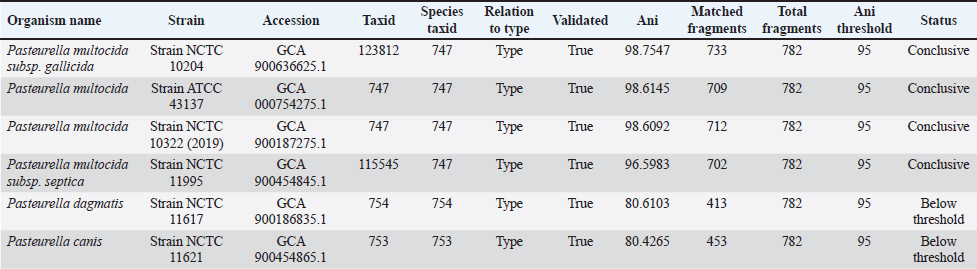
Taxonomy check based on the percentage of sequence similarity between the sample and reference genomes. The strain most closely related to the P. multocida NTT genome was NCTC 10322 (Table 3). According to the ANI analysis, the genome of P. multocida Lampung 2952 shared the highest similarity (98.754%) with P. multocida subsp. gallicida NCTC 10204 (Table 4). Long-read sequencing effectively resolved repetitive regions, enabling improved genome assemblies compared with short-read methods. The sequencing produced complete circular chromosomes (2.35 Mbp with 261,742 reads) with 100% mapped reads for both isolates, demonstrating the effectiveness of the ONT platform for bacterial genome assembly. Table 3. Comparison of the P. multocida NTT isolate with several other P. multocida strains available in GenBank.
Table 4. compArison of the P. multocida Lampung 2952 isolate with several other P. multocida strains available in GenBank.
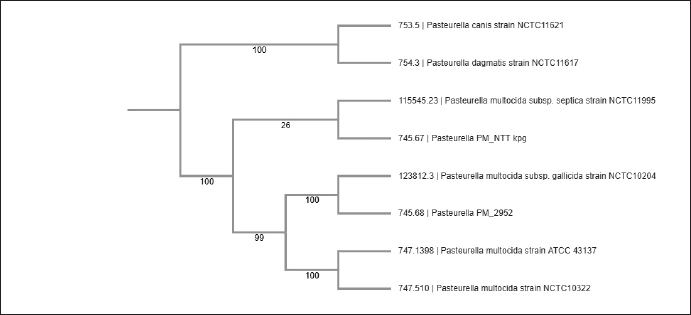
The whole genome dendrogram representation of P. multocida genomes illustrates a compArative analysis between our two isolates, one from East Nusa Tenggara (NTT) and the other from Lampung (2952) against reference isolates obtained from GenBank conducted in PATRIC (Wattam et al., 2017), as shown in Figure 1.
Fig. 1. Phylogenetic tree of 8 species reconstructed from a compArison of whole-genome sequences using PATRIC. DiscussionHigh-quality genome assemblies of P. multocida isolates from Indonesia provide valuable genomic resources for understanding the molecular basis of virulence and host adaptation in these local strains. The use of long- read ONT sequencing allowed us to achieve complete single-contig assemblies, overcoming the limitations of short-read sequencing in resolving repetitive regions that are commonly present in bacterial genomes (Li, 2021; Hall et al., 2024). This approach is particularly valuable for P. multocida genomics because accurate assembly enables precise identification of virulence determinants (Wick et al., 2019). The genome sizes and GC contents of both isolates were consistent with previously reported P. multocida genomes, supporting the accuracy of our assemblies (Boyce et al., 2012). The slight differences in genome size between the two isolates (2.35 Mbp vs. 2.29 Mbp) suggest potential genomic variations associated with adaptation to different geographical regions within Indonesia. Such variations could influence virulence, host specificity, and environmental persistence and warrant further investigation (Tarigan et al., 2010). A preliminary analysis of the annotated genomes revealed the presence of genes associated with virulence, including those involved in capsule biosynthesis, lipopolysaccharide production, and iron acquisition (Harper et al., 2006; Hatfaludi et al., 2010). These virulence factors are likely important for the pathogenesis of HS in Indonesian cattle (Mariana and Hirst, 2000). Capsule biosynthesis genes are of particular interest because capsule type correlates with host specificity and disease manifestation in P. multocida (Hatfaludi et al., 2010; Harper et al., 2012). The lipopolysaccharide structure, which varies among strains, also plays a crucial role in host–pathogen interactions and immune evasion (Harper and Boyce, 2017). The high BUSCO scores (>99%) indicate that our assemblies capture nearly complete gene sets, providing a robust foundation for compArative genomics and functional studies (Levy and Boone, 2019). Future work will focus on an in-depth analysis of virulence factors, antimicrobial resistance determinants, and compArative genomics with isolates from other geographical regions (Zankari et al., 2012; Boyce et al., 2012). To date, >1,800 genomes of Pasteurella spp. have been deposited in public genomic repositories. P. multocida NTT isolate exhibits an ANI of 98.56% with P. multocida NCTC10322 (GCA_900187275.1), as shown in Table 3. The P. multocida Lampung 2952 strain had an ANI of 98.76%, whereas the P. multocida subsp. gallicida strain NCTC10204 (GCA_900636625.1) as presented in Table 4. Indonesia’s diverse geography and livestock management practices may influence the evolution and spread of P. multocida strains (Tarigan et al., 2010; Peng et al., 2018). The availability of complete genomes will facilitate molecular epidemiological studies to track transmission patterns and evolutionary relationships among Indonesian isolates (OIE Terrestrial Manual, 2021). This information is crucial for developing targeted control strategies specific to the Indonesian context (Benkirane and de Alwis, 2002), where HS remains a significant economic concern for the livestock industry (Kawasaki et al., 2015). ConclusionThis study presents high-quality complete genome assemblies of P. multocida isolates from Lampung and Kupang, Indonesia. The assemblies, achieved through long-read Oxford Nanopore sequencing, provide valuable resources for studying the molecular epidemiology and pathogenesis of HS in Indonesian cattle. The single-contig assemblies with high completeness scores demonstrate the effectiveness of ONT sequencing for bacterial genome assembly. AcknowledgmentsThe authors thanks to the National Research and Innovation Agency. Author’s contributionsSSP, IWTW, SMN, and SS: Conceived, designed, and coordinated the study. SSP, SS, IWTW, and SW: Designed data collection tools, and supervised the field sample and data collection, laboratory work, and data entry. ARK, SS, and YA: Validation, supervision, and formal analysis. AA, SS, HN, TA, and SS: Contributed reagents, materials, and analysis tools. SSP, SS, HK, ARK, and ARRP: Statistical analysis and interpretation and preparation of the manuscript. All authors have read, reviewed, and approved the final manuscript. Conflict of interestThe authors declare no conflict of interest. FundingWe thank the collaborators who assisted in the sample collection. This work was supported by the Research Organization for Health and the National Research and Innovation Agency. Data availabilityThe research is ongoing and several datasets from the WGS results are still in use; therefore, not all data are available in the manuscript. Once the study has been completed, we will promptly submit the data to GenBank. ReferencesAlmoheer, R., Abd Wahid, M.E., Zakaria, H.A., Jonet, M.A.B., Al-Shaibani, M.M., Al-Gheethi, A. and Addis, S.N.K. 2022. Spatial, temporal, and demographic patterns in the prevalence of hemorrhagic septicemia in 41 countries in 2005–2019: a systematic analysis with special focus on the potential development of a new-generation vaccine. Vaccines (Basel) 10(2), 315. Benkirane, A. and de Alwis, M.C.L. 2002. Haemorrhagic septicaemia, its significance, prevention and control in Asia. Vet. Med-Czech. 47(8), 234–240. Boyce, J.D., Seemann, T., Adler, B. and Harper, M. 2012. Pathogenomics of Pasteurella multocida. Curr. Top. Microbiol. Immunol. 361(1), 23–38. De Coster, W., D’Hert, S., Schultz, D.T., Cruts, M. and Van Broeckhoven, C. 2018. NanoPack: visualizing and processing long-read sequencing data. Bioinformatics 34(15), 2666–2669. Doyle-Baker, D., Ngeleka, M., Janzen, E., Briggs, R.E. and Davies, J.L. 2020. Septicemic pasteurellosis causing peracute death and necrotizing myositis in a beef heifer calf (Bos taurus) in Alberta, Canada. Can. Vet. J. 61(12), 1303–1306. Gowrakkal, M., Chandrashekar, M., Bhajantri, S., Satav, J., Chandakala, G., Mayanna, A., Byregowda, S. and Renukaprasad, C. 2014. Evaluation of immuno efficiency of hemorrhagic septicemia vaccine strain (vaccine seed). Asian Pac. J. Trop. Biomed. 4(Suppl 1), S263–S267. Hall, M.B., Wick, R.R., Judd, L.M., Nguyen, A.N., Steinig, E.J., Xie, O., Davies, M., Seemann, T., Stinear, T.P. and Coin, L. 2024. Benchmarking reveals superiority of deep learning variant callers on bacterial nanopore sequence data. Elife 13(1), RP98300. Harper, M. and Boyce, J.D. 2017. The myriad properties of Pasteurella multocida Lipopolysaccharide. Toxins (Basel) 9(8), 254. Harper, M., Boyce, J.D. and Adler, B. 2006. Pasteurella multocida pathogenesis: 125 years after Pasteur. FEMS Microbiol. Lett. 265(1), 1–10. Harper, M., Boyce, J.D. and Adler, B. 2012. The key surface components of Pasteurella multocida: capsule and lipopolysaccharide. Curr. Top. Microbiol. Immunol. 361(1), 39–51. Hasnan, Q., Puspitasari, Y., Othman, S., Zamri- Saad, M. and Salleh, A. 2022. Phagocytosis and intracellular killing of Pasteurella multocida B:2 by macrophages: a comparative study between buffalo and cattle. Vet. World. 15(2), 275–280. Hatfaludi, T., Al-Hasani, K., Boyce, J.D. and Adler, B. 2010. Outer membrane proteins of Pasteurella multocida. Vet. Microbiol. 144(1–2), 1–17. Kawasaki, M., Young, J.R., Suon, S., Bush, R.D. and Windsor, P.A. 2015. The socioeconomic impacts of clinically diagnosed haemorrhagic septicaemia on smallholder large ruminant farmers in cambodia. Transbound. Emerg. Dis. 62(5), 535–548. Koren, S., Walenz, B.P., Berlin, K., Miller, J.R., Bergman, N.H. and Phillippy, A.M. 2017. Canu: scalable and accurate long-read assembly via adaptive k-mer weighting and repeat separation. Genome Res. 27(5), 722–736. Lestari, T.D., Khairullah, A.R., Damayanti, R., Mulyati, S., Rimayanti, R., Hernawati, T., Utama, S., Wardhani, B.W.K., Wibowo, S., Kurniasih, D.A.A., Ma’ruf, I.F., Moses, I.B., Lisnanti, E.F., Ahmad, R.Z., Fauziah, I., Amalia, N., Fauzia, K.A. and Kusala, M.K.J. 2025. Hemorrhagic septicemia: a major threat to livestock health. Open Vet. J. 15(2), 519–532. Levy, S.E. and Boone, B.E. 2019. Next-generation sequencing strategies. Cold Spring Harb. Perspect. Med. 9(7), a025791. Li, H. 2021. New strategies to improve minimap2 alignment accuracy. Bioinformatics 37(23), 4572–4574. Luan, T., Commichaux, S., Hoffmann, M., Jayeola, V., Jang, J.H., Pop, M., Rand, H. and Luo, Y. 2024. Benchmarking short and long read polishing tools for nanopore assemblies: achieving near-perfect genomes for outbreak isolates. BMC Genomics 25(1), 679. Mariana, S. and Hirst, R. 2000. The immunogenicity and pathogenicity of Pasteurella multocida isolated from poultry in Indonesia. Vet. Microbiol. 72(1–2), 27–36. OIE Terrestrial Manual. 2021. Haemorrhagic Septicaemia. Chapter 3.4.10. Peng, Z., Liang, W., Wang, F., Xu, Z., Xie, Z., Lian, Z., Hua, L., Zhou, R., Chen, H. and Wu, B. 2018. Genetic and phylogenetic characteristics of Pasteurella multocida isolates from different host species. Front. Microbiol. 9(1), 1408. Shivachandra, S.B., Viswas, K.N. and Kumar, A.A. 2011. A review of hemorrhagic septicemia in cattle and buffalo. Anim. Health Res. Rev. 12(1), 67–82. Simão, F.A., Waterhouse, R.M., Ioannidis, P., Kriventseva, E.V. and Zdobnov, E.M. 2015. BUSCO: assessing genome assembly and annotation completeness with single-copy orthologs. Bioinformatics 31(19), 3210–3212. Tanizawa, Y., Fujisawa, T. and Nakamura, Y. 2018. DFAST: a flexible prokaryotic genome annotation pipeline for faster genome publication. Bioinformatics 34(6), 1037–1039. Tarigan, S., Suwarno, S. and Priadi, A. 2010. Characterization of Pasteurella multocida local isolates from cattle in Indonesia. Asian-Australas. J. Anim. Sci. 23(10), 1442–1449. Wattam, A.R., Davis, J.J., Assaf, R., Boisvert, S., Brettin, T., Bun, C., Conrad, N., Dietrich, E.M., Disz, T., Gabbard, J.L., Gerdes, S., Henry, C.S., Kenyon, R.W., Machi, D., Mao, C., Nordberg, E.K., Olsen, G.J., Murphy-Olson, D.E., Olson, R., Overbeek, R., Parrello, B., Pusch, G.D., Shukla, M., Vonstein, V., Warren, A., Xia, F., Yoo, H. and Stevens, R.L. 2017. Improvements to PATRIC, the all-bacterial bioinformatics database and analysis resource center. Nucleic Acids Res. 45(D1), D535– D542. Wei, B., Liu, C., Zhu, J., Zou, X. and Zhang, Z. 2025. Pasteurella multocida infection: a differential retrospective study of 482 cases of P. multocida infection in patient of different ages. BMC Infect. Dis. 25(1), 313. Wick, R.R., Judd, L.M. and Holt, K.E. 2019. Performance of neural network basecalling tools for Oxford Nanopore sequencing. Genome Biol. 20(1), 129. Yohanes, T.R.M.R.S. and Theresia, F.I.M.D.M. 2012. The Occurrence(s) of Septicaema epizootica in Bali Cattle at Kupang Regency in 2005 – 2011. J. Sain Vet. 30(2), 53–60. Zankari, E., Hasman, H., Cosentino, S., Vestergaard, M., Rasmussen, S., Lund, O., Aarestrup, F.M. and Larsen, M.V. 2012. Identification of acquired antimicrobial resistance genes. J. Antimicrob. Chemother. 67(11), 2640–2644. | ||
| How to Cite this Article |
| Pubmed Style Prihandani SS, Wibawan IWT, Noor SM, Safika S, Khairullah AR, Susanti S, Wahyuwardani S, Andriani A, Sumarningsih S, Kurnianto H, Ariyanti T, Adinata Y, Nuradji H, Puarada ARR. Pasteurella multocida genome assembly of Indonesian local isolates using long-read Oxford Nanopore Technology sequencing. Open Vet. J.. 2025; 15(6): 2782-2788. doi:10.5455/OVJ.2025.v15.i6.46 Web Style Prihandani SS, Wibawan IWT, Noor SM, Safika S, Khairullah AR, Susanti S, Wahyuwardani S, Andriani A, Sumarningsih S, Kurnianto H, Ariyanti T, Adinata Y, Nuradji H, Puarada ARR. Pasteurella multocida genome assembly of Indonesian local isolates using long-read Oxford Nanopore Technology sequencing. https://www.openveterinaryjournal.com/?mno=250015 [Access: January 25, 2026]. doi:10.5455/OVJ.2025.v15.i6.46 AMA (American Medical Association) Style Prihandani SS, Wibawan IWT, Noor SM, Safika S, Khairullah AR, Susanti S, Wahyuwardani S, Andriani A, Sumarningsih S, Kurnianto H, Ariyanti T, Adinata Y, Nuradji H, Puarada ARR. Pasteurella multocida genome assembly of Indonesian local isolates using long-read Oxford Nanopore Technology sequencing. Open Vet. J.. 2025; 15(6): 2782-2788. doi:10.5455/OVJ.2025.v15.i6.46 Vancouver/ICMJE Style Prihandani SS, Wibawan IWT, Noor SM, Safika S, Khairullah AR, Susanti S, Wahyuwardani S, Andriani A, Sumarningsih S, Kurnianto H, Ariyanti T, Adinata Y, Nuradji H, Puarada ARR. Pasteurella multocida genome assembly of Indonesian local isolates using long-read Oxford Nanopore Technology sequencing. Open Vet. J.. (2025), [cited January 25, 2026]; 15(6): 2782-2788. doi:10.5455/OVJ.2025.v15.i6.46 Harvard Style Prihandani, S. S., Wibawan, . I. W. T., Noor, . S. M., Safika, . S., Khairullah, . A. R., Susanti, . S., Wahyuwardani, . S., Andriani, . A., Sumarningsih, . S., Kurnianto, . H., Ariyanti, . T., Adinata, . Y., Nuradji, . H. & Puarada, . A. R. R. (2025) Pasteurella multocida genome assembly of Indonesian local isolates using long-read Oxford Nanopore Technology sequencing. Open Vet. J., 15 (6), 2782-2788. doi:10.5455/OVJ.2025.v15.i6.46 Turabian Style Prihandani, Sri Suryatmiati, I. Wayan Teguh Wibawan, Susan Maphilindawati Noor, Safika Safika, Aswin Rafif Khairullah, Susanti Susanti, Sutiastuti Wahyuwardani, Andriani Andriani, Sumarningsih Sumarningsih, Heri Kurnianto, Tati Ariyanti, Yudi Adinata, Harimurti Nuradji, and Alif Rahman Rohim Puarada. 2025. Pasteurella multocida genome assembly of Indonesian local isolates using long-read Oxford Nanopore Technology sequencing. Open Veterinary Journal, 15 (6), 2782-2788. doi:10.5455/OVJ.2025.v15.i6.46 Chicago Style Prihandani, Sri Suryatmiati, I. Wayan Teguh Wibawan, Susan Maphilindawati Noor, Safika Safika, Aswin Rafif Khairullah, Susanti Susanti, Sutiastuti Wahyuwardani, Andriani Andriani, Sumarningsih Sumarningsih, Heri Kurnianto, Tati Ariyanti, Yudi Adinata, Harimurti Nuradji, and Alif Rahman Rohim Puarada. "Pasteurella multocida genome assembly of Indonesian local isolates using long-read Oxford Nanopore Technology sequencing." Open Veterinary Journal 15 (2025), 2782-2788. doi:10.5455/OVJ.2025.v15.i6.46 MLA (The Modern Language Association) Style Prihandani, Sri Suryatmiati, I. Wayan Teguh Wibawan, Susan Maphilindawati Noor, Safika Safika, Aswin Rafif Khairullah, Susanti Susanti, Sutiastuti Wahyuwardani, Andriani Andriani, Sumarningsih Sumarningsih, Heri Kurnianto, Tati Ariyanti, Yudi Adinata, Harimurti Nuradji, and Alif Rahman Rohim Puarada. "Pasteurella multocida genome assembly of Indonesian local isolates using long-read Oxford Nanopore Technology sequencing." Open Veterinary Journal 15.6 (2025), 2782-2788. Print. doi:10.5455/OVJ.2025.v15.i6.46 APA (American Psychological Association) Style Prihandani, S. S., Wibawan, . I. W. T., Noor, . S. M., Safika, . S., Khairullah, . A. R., Susanti, . S., Wahyuwardani, . S., Andriani, . A., Sumarningsih, . S., Kurnianto, . H., Ariyanti, . T., Adinata, . Y., Nuradji, . H. & Puarada, . A. R. R. (2025) Pasteurella multocida genome assembly of Indonesian local isolates using long-read Oxford Nanopore Technology sequencing. Open Veterinary Journal, 15 (6), 2782-2788. doi:10.5455/OVJ.2025.v15.i6.46 |