| Review Article | ||
Open Vet. J.. 2025; 15(10): 4814-4833 Open Veterinary Journal, (2025), Vol. 15(10): 4814-4833 Review Article Cryptosporidiosis: A global threat to human and animal healthWimbuh Tri Widodo1*, Aswin Rafif Khairullah2, Bima Putra Pratama3, Rahmania Ambarika4, Abdul Hadi Furqoni5,6, Sonny Kristianto1, Bantari Wisynu Kusuma Wardhani7,8, Widoretno Widoretno5, Khariri Khariri5, Luluk Hermawati9, Auliyani Andam Suri10, Ikechukwu Benjamin Moses11, Alifiani Kartika Putri12, Dea Anita Ariani Kurniasih13, Masri Sembiring Maha5, Andi Thafida Khalisa8, Riza Zainuddin Ahmad2 and Syahputra Wibowo141Master of Forensic Science, Postgraduate School, Universitas Airlangga, Surabaya, Indonesia 2Research Center for Veterinary Science, National Research and Innovation Agency (BRIN), Bogor, Indonesia 3Research Center for Agroindustry, National Research and Innovation Agency (BRIN), South Tangerang, Indonesia 4Universitas Strada Indonesia, Kediri, Indonesia 5Center for Biomedical Research, National Research and Innovation Agency (BRIN), Bogor, West Java, Indonesia 6Research Center on Global Emerging and Re-Emerging Infectious Diseases, Institute of Tropical Disease, Universitas Airlangga, Surabaya, Indonesia 7Research Center for Pharmaceutical Ingredients and Traditional Medicine, National Research and Innovation Agency (BRIN), Bogor, Indonesia 8Faculty of Military Pharmacy, Universitas Pertahanan, Bogor, Indonesia 9Department of Molecular Biology, Faculty of Medicine and Health Sciences, Universitas Sultan Ageng Tirtayasa, Banten, Indonesia 10Departement of Physiology, Faculty of Medicine, UIN Syarif Hidayatullah Jakarta, Banten, Indonesia 11Department of Applied Microbiology, Faculty of Science, Ebonyi State University, Abakaliki, Nigeria 12Muhammadiyah Hospital Tuban, Tuban, Indonesia 13Research Center for Public Health and Nutrition, National Research and Innovation Agency (BRIN), Bogor, Indonesia 14Eijkman Research Center for Molecular Biology, National Research and Innovation Agency (BRIN), Bogor, Indonesia *Corresponding Author: Wimbuh Tri Widodo. Postgraduate School of Forensic Science, Universitas Airlangga, Surabaya, Indonesia. Email: wimbuh.tri [at] pasca.unair.ac.id Submitted: 25/05/2025 Revised: 30/08/2025 Accepted: 10/09/2025 Published: 31/10/2025 © 2025 Open Veterinary Journal
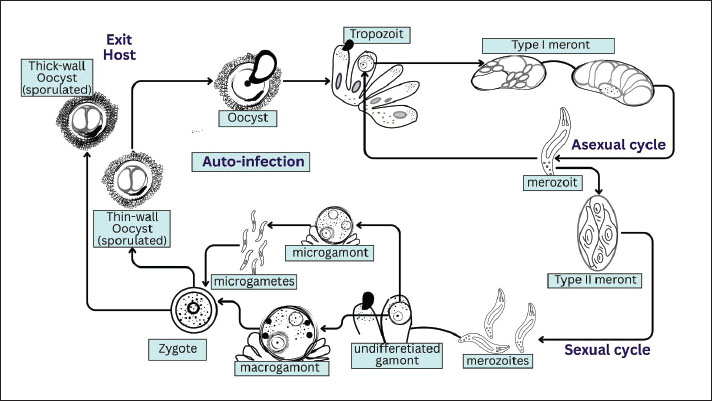
AbstractCryptosporidiosis is a significant zoonotic illness that infects both humans and animals. The protozoan parasite that causes this illness is a member of the genus Cryptosporidium, a eukaryote in the phylum Apicomplexa. The parasite Cryptosporidium is monoxenic, meaning it has only one host. Oocyte-to-oocyte development occurs in the host organism without an intermediate host. Developing nations have a far greater prevalence of Cryptosporidium infections because many people there still lack access to basic sanitation and clean water. Both innate and adaptive immune system components are involved in the host immunological response to Cryptosporidium infection; both pathways contribute to the defense against Cryptosporidiosis. Globally, cryptosporidiosis is estimated to cause tens of millions of cases each year, with a prevalence of 10%–20% among children under five in sub-Saharan Africa and South Asia, whereas approximately 750,000 cases occur annually in the United States. The disease is more common in developing countries due to limited access to safe water and sanitation. Cryptosporidium can be detected in the digestive tract, lungs, and conjunctiva; however, the intestines are most susceptible to cryptosporidiosis. Nowadays, most people agree that Cryptosporidium is a frequent parasite that causes diarrheal illness. Furthermore, infections caused by Cryptosporidium have spread to humans, primarily affecting individuals with immunological problems, such as those with AIDS. Cryptosporidiosis is typically diagnosed by looking for parasite eggs, oocyte antigen, or oocyte DNA in stool samples. Consuming food or beverages containing the oocysts of these protozoa can infect both humans and animals. Most diseased individuals and animals with robust immune systems can heal themselves without medical intervention. The main strategy for preventing cryptosporidiosis is to reduce or eradicate environmental contamination with infectious oocysts, as there is currently no effective treatment for the disease. Keywords: Cryptosporidiosis, Diarrhea, Immunity, Oocyst, Parasite, Prevention. IntroductionCryptosporidiosis is a significant zoonotic disease that affects the intestinal tract of both humans and animals (Gerace et al., 2019; Alarcón-Zapata et al., 2023). It is caused by Cryptosporidium, a protozoan parasite belonging to the phylum Apicomplexa (Bouzid et al., 2013). All Cryptosporidium species are obligate intracellular parasites that depend on host cells to complete their life cycle (Arrowood, 2002). The parasite undergoes three major developmental phases—schizogony, gametogony (sexual reproduction), and merogony (asexual reproduction)—followed by sporogony (sporulation) (Dragomirova, 2022). An encysted oocyst stage is excreted in the feces of the host, allowing transmission to new hosts (Leitch and He, 2012). Cryptosporidium is a small protozoan, measuring 4–6 µm, and typically inhabits the microvilli of the mucosal epithelium in many vertebrates, including humans (Xiao et al., 2004a). Tyzzer first described and named Cryptosporidium in 1907 (Tzipori and Widmer, 2008). However, its pathogenic role was not recognized until 1955, when it was associated with gastrointestinal disease in turkeys (Leitch and He, 2012). In the early 1980s, Cryptosporidium was identified as a major cause of diarrheal illness in ruminants (O'Hara and Chen, 2011) and later became established as an important human pathogen, especially among individuals with immunodeficiency, such as those with AIDS (Hunter and Nichols, 2002). Today, Cryptosporidium is widely acknowledged as a common cause of acute diarrhea in healthy individuals and a severe, sometimes life-threatening infection in young children and immunocompromised patients (Prabakaran et al., 2023). Transmission occurs mainly through the ingestion of food or water contaminated with infectious oocysts (Pumipuntu and Piratae, 2018). Prevalence is particularly high in developing countries, where sanitation and clean water access remain limited (Mahmoudi et al., 2017). Oocysts are resistant to many disinfectants, including chlorine, enabling their persistence in drinking water, swimming pools, and recreational facilities (Ali et al., 2024a). Human-to-human transmission has been reported in hospitals, households, day-care centers, and through sexual contact (Ramirez et al., 2004). Zoonotic transmission is also well documented, involving livestock, pets, wild animals, and veterinary workers (El-Alfy and Nishikawa, 2020). In livestock, cryptosporidiosis causes economic losses by increasing veterinary costs, reducing growth, and contributing to mortality (Santin, 2020). Pets and wild animals may also serve as reservoir hosts, further increasing the human infection risk (Pal et al., 2021). Globally, the disease remains a serious health concern, ranking second only to rotavirus as the leading cause of diarrhea and mortality in children (Mafokwane et al., 2023). Currently, there is no effective cure for cryptosporidiosis. Cryptosporidiosis is a global issue with serious implications for food safety, veterinary medicine, and public health, given its impact on both human and animal health (LeJeune and Kersting, 2010). This article reviews the key aspects of cryptosporidiosis, including its epidemiology, pathogenesis, clinical symptoms, diagnostic methods, and current treatment and prevention approaches. It also highlights existing knowledge gaps and future research needs to support better control and management of this important disease. EtiologyCryptosporidium oocysts are among the smallest within coccidia, typically ovoid to spherical, and fully sporulated. Their size averages about 4.5 μm in C. parvum and 5.6 μm in C. muris (Helmy and Hafez, 2022; Shehata et al., 2024). Each sporulated oocyst contains four sporozoites and a residuum composed of small granules and a membrane-bound globule (O'Hara and Chen, 2011). Cryptosporidium lacks morphological traits, such as polar granules and micropyles, which are commonly present in coccidian oocysts (Bruno et al., 2006). The oocyst wall is smooth, colorless, and approximately 50 nm thick, consisting of two electron-dense layers separated by a thin electron-lucent gap (Nageeb et al., 2024). A faint suture line may sometimes be observed at one pole of the oocyst under light microscopy (Ruecker et al., 2007). Cryptosporidium belongs to the phylum Apicomplexa, class Sporozoa, subclass Coccidia, order Eucoccidiorida, and family Cryptosporidiidae (Mamedova and Karanis, 2025). Historically, species differentiation has been based on host specificity, endogenous site location, and morphological features (Gerace et al., 2019). However, oocyst morphology alone is unreliable for species identification because many species have overlapping size ranges, leading to misclassification (Tosini et al., 2010). Early taxonomic attempts often relied on the host origin. For example, C. agni was described from sheep (Barker and Carbonell, 1974), and C. garnhami or C. enteritis from humans (Wilmsmeyer et al., 1993). Nevertheless, these species names are not considered valid due to the absence of sufficient molecular, biological, and morphological data to distinguish them from recognized species (Bones et al., 2019). Recent findings also show that many Cryptosporidium species can infect multiple hosts, and some researchers suggest a broader species concept (Rideout et al., 2024). The morphological differentiation of oocysts is only reliable at extreme sizes. Cryptosporidium muris with large oocysts typically infects the gastric mucosa of mammals, whereas C. parvum with small oocysts infects the intestinal mucosa (Abeywardena et al., 2015). Cryptosporidium has been reported in several vertebrates, including birds, fish, reptiles, and mammals. Valid recognized species include C. andersoni, C. parvum, C. hominis, C. galli, C. baileyi, C. meleagridis, C. molnari, C. canis, C. muris, C. felis, C. saprophilum, and C. wrairi (Alves et al., 2003; Xiao et al., 2007; García-Livia et al., 2020; Scorza et al., 2022). Life cycleCryptosporidium is monoxenous (its direct life cycle is completed within a single host). After ingestion or inhalation of sporulated oocysts, excystation releases four sporozoites (~5 μm) that invade epithelial cells of the gastrointestinal tract—and occasionally the respiratory tract—at the brush border within an intracellular but extracytoplasmic niche. Endogenous development proceeds through three major phases: merogony (asexual replication, also termed schizogony), gametogony (differentiation into microgamonts and macrogamonts), and sporogony (zygote formation and in-host sporulation) (Hijjawi, 2004; Heo et al., 2018; Tandel et al., 2019 ; English et al., 2022). Two types of oocysts are produced: thin-walled oocysts that enable autoinfection and thick-walled oocysts that are immediately infectious upon excretion and mediate fecal–oral transmission (Helmy and Hafez, 2022; Hussain et al., 2025). Exogenous development involves the persistence of thick-walled, fully sporulated oocysts shed in the feces of infected hosts (Leitch and He, 2012). Oocysts are excised following ingestion or inhalation, releasing motile sporozoites that invade epithelial cells (Vanathy et al., 2017). Within the host cell, Cryptosporidium resides in a parasitophorous vacuole located just beneath the apical membrane; this compartment is intracellular yet outside the host cytoplasm, bounded by a host-derived membrane and a parasite-derived interface (including a feeder organelle) (Xu et al., 2024). Oocyst shedding typically lasts 7–30 days, depending on host immunity, and thin-walled oocysts drive endogenous reinvasion (autoinfection) (Leitch and He, 2012; Balendran et al., 2024). The host stage of the biological life cycle of Cryptosporidium spp. is illustrated in Figure 1.
Fig. 1. Host stage of the biological life cycle of Cryptosporidium spp. It involves two developmental phases: intracellular asexual and sexual reproduction through Type I and Type II meront, which produce gamont and zygotes. Environmental transmission occurs when zygotes develop into thick-walled oocysts, and auto-infection is possible through the production of thin-walled oocysts, which preserves the parasite and supports its spread to animals and humans. At the genomic and metabolic levels, Cryptosporidium has a compact genome (~9.2 Mb across eight chromosomes) and shows reductions in several organelles and core metabolic pathways typical of Apicomplexa and other eukaryotes (Widmer and Sullivan, 2012; Ryan and Hijjawi, 2015). Consequently, due to the loss of key pathways (e.g., oxidative phosphorylation and many biosyntheses), Cryptosporidium species depend extensively on the host for nutrients; energy generation relies mainly on glycolysis and substrate-level phosphorylation. This dependency is reflected in an expanded repertoire of transporters for nutrient acquisition (Xu et al., 2004; Mazurie et al., 2013; Li et al., 2021b). HistoryIn 1895, Clarke (1895) reported spores of what he termed “free-encapsulated coccidia” in the cardiac glands of rat stomachs. Experimental mice that ingested these spores became ill within a week. Later, the prominent American parasitologist Edward Ernest Tyzzer studied similar organisms and, in 1910–1912, described them as C. muris, which infected the gastric epithelium of several strains of laboratory mice (Tyzzer, 1910; Tyzzer, 1912). Tyzzer also described a second species, C. parvum, inhabiting the small intestine of rats. Unlike typical coccidia, Cryptosporidium oocysts lack sporocysts surrounding the sporozoites, which is reflected in the genus name (Tzipori and Widmer, 2008; Pumipuntu and Piratae, 2018). Although the life cycle stages of Cryptosporidium were only faintly visible under light microscopy, Tyzzer (1907) was able to identify and order them, noting that the oocyst sporulates while still attached to the host cell, enabling autoinfection (Tyzzer, 1907). He concluded that the parasite obtains nutrients from the host via a specialized attachment organelle, later termed the feeder organelle. In 1986, Current et al. (1986) expanded Tyzzer’s work using electron microscopy, which confirmed schizogony with multiple generations and established the now widely accepted model of the life cycle. Electron microscopy and freeze-fracture techniques further demonstrated the intracellular but extracytoplasmic localization of Cryptosporidium (Tzipori and Widmer, 2008). Between 1961 and 1986, morphological studies led to the identification of nearly 20 additional Cryptosporidium species from fish, birds, mammals, and reptiles (Robinson et al., 2010). However, only a limited number, such as C. muris and C. parvum, are now considered valid, as many earlier names lacked sufficient molecular and biological support. Slavin (1955) was the first to link C. meleagridis infection to clinical disease when he reported acute diarrhea and mortality in young turkeys, later attributed to C. meleagridis. In 1971, C. parvum was recognized as a cause of diarrhea in calves, raising its veterinary importance (Roblin et al., 2023). C. parvum is a major cause of neonatal diarrhea in ruminants (Resnhaleksmana et al., 2021; Sawant et al., 2021; Sheoran et al., 2022; Veshkini et al., 2024). Additional species of clinical significance were later identified, including C. baileyi, a cause of respiratory disease in poultry (Zaheer et al., 2021; Lin et al., 2022). The first human case of Cryptosporidium infection was reported in 1976. C. parvum and C. hominis have been established as important human pathogens, causing acute self-limiting diarrhea in immunocompetent individuals and severe, chronic disease in immunocompromised patients, especially those with AIDS (Rossle and Latif, 2013). EpidemiologyAlthough the global incidence of cryptosporidiosis is estimated to be approximately three cases per 100,000 population, the true prevalence is likely much higher—up to 100 times greater—because many cases are underreported or misdiagnosed due to non-specific clinical symptoms and limited diagnostic capacity (Gerace et al., 2019). The burden is disproportionately higher in developing countries, where the lack of access to safe water and adequate sanitation promotes widespread transmission (Ahmed and Karanis, 2020). Cryptosporidiosis is a major cause of childhood morbidity in these settings. Accounting for 10%–15% of severe diarrheal episodes in undernourished children aged 5 years (Helmy and Hafez, 2022). Outbreaks linked to contaminated drinking water, recreational water, and swimming pools have been documented in many countries, highlighting the resilience and global distribution of the parasite (Mahmoudi et al., 2017; Ali et al., 2024b). Although more than 30 Cryptosporidium species have been described, only a few are commonly associated with human infection, notably C. hominis, C. parvum, C. meleagridis, C. felis, and C. canis (Šlapeta, 2013; Ryan et al., 2014; Ayinmode et al., 2018; Peng et al., 2024). C. hominis is considered anthroponotic, infecting only humans, whereas C. parvum is primarily zoonotic, circulating between humans and ruminants. Consequently, C. parvum infections are frequently linked to livestock contact and animal husbandry practices (Hunter and Thompson, 2005; Dixon et al., 2011; Ehsan et al., 2015). Zoonotic transmission from less common hosts has also been reported, although it is relatively rare. Sporadic human cases have been associated with exposure to sheep (Dessì et al., 2020), horses (Li et al., 2019), goats (Yang et al., 2021), and rats (Suprihati et al., 2024). Other species adapted to companion animals, such as C. canis in dogs and C. felis in cats, occasionally infect humans, usually immunocompromised individuals, although transmission from household pets to humans remains infrequent (Scorza et al., 2022; Wang et al., 2022; Dărăbuș et al., 2025). This epidemiological pattern highlights the dual importance of anthroponotic species (such as C. hominis) and zoonotic species (such as C. parvum) in sustaining human-to-human transmission and bridging infections between animals and humans, respectively. Together, these factors contribute to the widespread and persistent burden of cryptosporidiosis across diverse settings. At the global level, the World Health Organization estimates that tens of millions of cryptosporidiosis cases occur annually, with the highest burdens reported in sub-Saharan Africa and South Asia, where infection rates in children under five can reach 10%–20% (Kotloff et al., 2013; Khalil et al., 2018) . In the United States, approximately 7,500 laboratory-confirmed cases are reported each year, although the true number is believed to exceed 750,000 due to underdiagnosis (CDC, 2023) . In Europe, surveillance data indicate more than 14,000 cases annually, with the United Kingdom, Germany, and Ireland being the most affected countries (EFSA, 2022) . Such regional disparities underscore the global significance of cryptosporidiosis as a public health and veterinary concern. PathogenesisThe pathogenic mechanisms that cause Cryptosporidium to cause diarrhea, malabsorption, and wasting are still not well understood. The first host-parasite interaction of invasion and attachment is a crucial step in pathogenesis, regardless of the method used. Numerous elements impacting attachment and the ultrastructural features of invasion and attachment have been described. However, the precise host and parasite chemicals involved in this process are poorly understood (Smith et al., 2005). It is crucial to understand these compounds to understand the harmful mechanisms that this parasite employs. Many parasite ligands and host receptors are involved in the first host-parasitic interaction, which takes the form of attachment, invasion, and the creation of the parasitophorous vacuole (O’Hara and Chen, 2011). This interaction has been thoroughly investigated in Apicomplexans, including Plasmodium, Eimeria, and Toxoplasma (Janouškovec et al., 2019). The apical complex, which consists of specialized secretory organelles such as micronemes, dense granules, and rhoptry, is present in the invasive “zoite” stage of the Apicomplexa (Gubbels and Duraisingh, 2012). These organelles release proteins that aid in adhesion, invasion, and the development of the parasitophorous vacuole during the first host-parasite interaction (Bouzid et al., 2013). Some micronemal proteins express distinct domains, whereas others have sticky “modules” that are conserved throughout Apicomplexan parasites (Sanderson et al., 2008). The discovery of surface and/or apical complex proteins [including circumsporozoite-like (CSL), GP900, p23/27, TRAP C1, GP15, CP 15, CP60/15, cp47, gp40/45, and gp15/Cp17] that share characteristics with other Apicomplexans and mediate these interactions is a result of the growing recognition of Cryptosporidium as an emerging human pathogen (Tzipori and Widmer, 2008). A large number of these proteins have already been reviewed (Theodos, 1998; Tomley and Soldati, 2001). Monoclonal antibodies (mAbs) against repetitive carbohydrate epitopes are used to identify the CSL antigen, a highly glycosylated 1,300 kDa glycoprotein (Riggs, 2002). This mAb causes a reaction similar to that of a circumsporozoite, where the antigen is moved posteriorly along the sporozoite pellicle, causing the infection to stop spreading. In vitro and in vivo, this antibody neutralizes infection in a mouse model of cryptosporidiosis (Riggs et al., 1997). CSL is found on the surface of sporozoites and merozoites as well as in the micronema and dense granules of the apical complex (Langer and Riggs, 1999). The purified protein exhibits saturable and dose-dependent binding to host cells (Paluszynski et al., 2014). According to a recent study, CSL attaches to the 85-kDa receptor of intestinal epithelial cells (Langer et al., 2001). Although the molecular structure of this protein is unknown, these results collectively suggest that CSL is a ligand driving adhesion and invasion. Similar to CSL, the apical microneme complex synthesizes GP900, a highly glycosylated high molecular weight glycoprotein that is secreted onto the surface of the invasive zoite stage and released in trail form during gliding movement (Wanyiri and Ward, 2006). According to an analysis of the deduced amino acid sequence of the gene, the gene encoding GP900 is a multidomain protein with a transmembrane domain, a cytoplasmic tail, and cysteine-rich and mucin-like domains (Bhalchandra et al., 2013). Both O-linked and N-linked glycosylation are widely present in GP900 (Rider and Zhu, 2010). Purified natural GP900, the recombinant protein’s cysteine-rich domain, and antibodies to this domain bind to intestinal epithelial cells and competitively inhibit C. parvum infection in vitro (Cevallos et al., 2000a). Our findings imply that GP900 mediates invasion and attachment as well. Whether the GP900 and CSL are related in any way remains unknown. Micronemal proteins, such as TRAP, CTRP, CS, Etp100, and MIC-2 from the Apicomplexans linked to Plasmodium falciparum, Eimeria tenella, and Toxoplasma gondii, respectively, are homologous to the thrombospondin-related adhesive protein of Cryptosporidium-1 (TRAP C1) (Boucher and Bosch, 2015). These proteins mediate attachment to host cells and feature a conserved thrombospondin domain that is distinguished by the presence of multiple TRMs. The deduced amino acid sequence of TRAP C1 includes an N-terminal signal sequence, a polyserenin domain, a thrombospondin domain with six TRMs, a transmembrane domain, and a cytoplasmic tail (Bhalchandra et al., 2013). Although TRAP C1 is localized to the apical region of sporozoites by antibodies to recombinant TRAP C1, there is no experimental proof that TRAP C1 plays a role in invasion or attachment. Another mucin-like O-glycosylated glycoprotein, gp40, has recently been identified. It is secreted from the parasite surface and is found on the surface and apical region of C. parvum’s invasive stages (Dąbrowska et al., 2023). Native C. parvum gp40 binds selectively to host cells, suggesting that the protein is involved in adhesion and invasion, and gp40-specific antibodies block infection in vitro. The gp40-coding gene, Cpgp40/15, has been cloned and sequenced (Cevallos et al., 2000b). A polyserine domain with many anticipated mucin-type O-glycosylation sites, an N-terminal signal peptide, and a hydrophobic region at the C-terminal end that is compatible with that needed for glycosylphosphatidylinositol (GPI) anchor addition were all found in the inferred amino acid sequence of this gene (Tran and Hagen, 2013). Cpgp40/15 not only encodes gp40 but also a 15/17-kDa immunodominant glycoprotein that is present on the invasive stage’s surface and participates in host-parasite interactions (Preidis et al., 2007). Gp40 and gp15 are generated by post-translational processing of precursor glycoproteins that are expressed in the parasite’s internal stages and encoded by Cpgp40/15 (Wanyiri et al., 2007). The C-terminal part of the gp40/15 precursor, gp15, and the soluble N-terminal segment, gp40, appear to remain connected following PTP. Thus, gp15, which is attached to the parasite surface by GPI linkage, may serve as a “stalk” to attach gp40 (Winter et al., 2000). The Cpgp40/15 gene has an unparalleled degree of polymorphism, which is significantly higher than that of any other gene that has been examined in Cryptosporidium to date (Leav et al., 2002). The length of the N-terminal polyserine domain was the primary source of variance in genotype 2 isolates. Nonetheless, at least four allelic subgroups in genotype 1 isolates are defined by a large number of single-nucleotide and single-amino acid polymorphisms (Priest et al., 2001). The discovery of widespread polymorphism in the Cpgp40/15 locus indirectly supports the role of this glycoprotein in facilitating infection, which is consistent with its gene product being a surface-associated virulence determinant that may be under host immunological pressure (Widmer and Lee, 2010). Both isolate genotypes have a single copy of the Cpgp40/15 gene, which is expressed as numerous transcripts produced by alternate polyadenylation (O’Connor et al., 2002). The predicted signal sequence, GPI anchor attachment site, proteolytic processing site, predicted O-glycosylation site in the polyserine domain, and 3′ UTR are conserved among isolates despite the broad polymorphism in the Cpgp40/15 coding sequence, indicating that these regions play a significant role in structure and function (Cevallos et al., 2000b). Cp 47 is a membrane-associated protein that binds to the surface of intestinal epithelial cells and is localized to the apical region of sporozoites (Nesterenko et al., 1999). However, no clone of the gene encoding this protein has been made. According to experimental or indirect data, the proteins mentioned above appear to play a role in facilitating adhesion and invasion. However, the inability to grow C. parvum in vitro and the lack of appropriate transient or stable DNA transfection systems, such as those created for other Apicomplexan parasites, such as Toxoplasma, have significantly impeded efforts to definitively determine the functional role of this protein (Nesterenko et al., 1999). Immune responseInnate and adaptive immune system components are involved in the host immunological response to Cryptosporidium infection; both processes contribute to the defense against Cryptosporidiosis. The initial line of defense is intestinal epithelial cells, followed by the recruitment of innate immune cells, such as mast cells, dendritic cells, natural killer cells, and macrophages (Ludington and Ward, 2015). The primary basis for diagnosis is the detection of oocysts from fecal material, and cell-mediated immunity is a crucial component of the immunological response to infection (Chalmers and Katzer, 2013). Intestinal epithelial cells are a crucial part of gastrointestinal mucosal defense to protect the intestinal mucosa from commensal microbes and pathogenic organism invasion (He et al., 2021). Furthermore, because Cryptosporidium produces parasitophorous vacuoles in infected host cells and releases antimicrobial peptides, these cells are crucial for the start, control, and resolution of innate and adaptive immune responses to Cryptosporidium infection (Stoyanova and Pavlov, 2019). Additionally, chemokines and inflammatory cytokines stimulate immune effector cells to the infection site, and nitric oxide can kill and stop C. parvum from growing (He et al., 2021). Immunoevasion mechanism of cryptosporidiosis in infected epithelial cells through NF-κB signaling activation to initiate antiapoptotic cell death signals in infected cells (Hussain et al., 2025). IFN-γ-dependent gene transactivation in the intestinal epithelium may be suppressed as a result of the decrease of signal transducer and activator of transcription 1α (STAT1α), a crucial transcription factor in IFN-γ signaling (Nava et al., 2010). The expression of the antiparasitic cytokine C-C motif chemokine ligand 20 (CCL20) is suppressed when host epithelial cells are infected (Li et al., 2024). CD3+/CD4+ cells are essential for the recovery of the immune system from cryptosporidiosis (Korbel et al., 2011). CD4+ T cells are crucial after antigen stimulation because they secrete cytokines, including IL-2, IFN-α, and IFN-γ from Th1 cells and IL-4 and IL-6 from Th2 cells. The IFN-γ also prevents Cryptosporidium disease invasion (Borad and Ward, 2010). Sporozoites cause dendritic cells and macrophages to release IL-12 during the infection’s acute phase. IL-12, in turn, combines with TNF-α and IL-18 to activate NK cells (Mead, 2023). Furthermore, certain immunocompetent cells generate proinflammatory cytokines (IL-1, IL-6) that have protective effects, and TNF-α stops Cryptosporidium infection in enterocytes (Cohn et al., 2022). This implies that host immunological factors play a significant role in the regulation of cryptosporidiosis. PathologyCryptosporidiosis can be detected in the digestive tract, lungs, and conjunctiva; however, the intestines sustain most of the damage from cryptosporidiosis (Pardy et al., 2024). Swelling of the mesenteric lymph nodes occurs rarely (Perez-Cordon et al., 2014). The pathophysiology of this disease may involve the toxic effects and sensitization of the parasite through its metabolic products and toxins in endogenous development (Gerace et al., 2019). Intestinal damage includes degenerative alterations in the lamina propria, which is rich in neutrophils, eosinophils, lymphocytes, and macrophages, as well as villous atrophy and epithelial hyperplasia in the villous crypts (Lauwers et al., 2018). Consequently, the absorption surface area and small intestine enzyme activity are decreased. The signs of malabsorption syndrome include protein metabolism, water and electrolyte balance, lactose and enzyme shortages, and excessive diarrhea (Kelly et al., 1996). Ascending intestinal parasites can harm the urogenital tract (Hechenbleikner and McQuade, 2015). Additionally, hematogenous spread is conceivable. Approximately 107 oocytes can infect (Hadfield et al., 2011). Infected humans and animals shed most oocytes in the first week (Gerace et al., 2019). Data on C. parvum and C. hominis indicate that oocytes may continue to shed for weeks beyond the cessation of diarrhea (Shirley et al., 2012). Nonetheless, oocyte excretion has been shown in immunocompetent people infected with C. muris for 7 months (Ali et al., 2024a). The establishment of the genomes of C. parvum and C. hominis, along with the description of over 25 potential virulence factors identified by diverse immunological and molecular approaches, led to significant advancements in the identification of putative virulence factors (Bouzid et al., 2013; Ayan et al., 2024). The immune system lowers the number of thin-walled oocytes and the rate at which type 1 merozoites develop (O’Hara and Chen, 2011). This reduces the chance of autoinfection. A prior infection may lessen the intensity of the illness and the quantity of oocytes produced, but it also lowers resistance to subsequent infections in individuals with a sound immune system (Chappell et al., 1996). Clinical symptomsIn humansThe most frequent intestinal infection species are C. hominis and C. parvum, which cause symptoms such as watery diarrhea, stomach pain, vomiting, nausea, dehydration, and weight loss, despite the fact that 21 species have been linked to human infection (Pal et al., 2021). People with CD4 T cell counts below 150/ml who are exposed to C. parvum suffer from a chronic infection that causes severe and occasionally fatal diarrhea (Khan and Witola, 2023). Those with strong immune systems typically experience self-limiting symptoms, whereas those with compromised immune systems experience the worst symptoms of dehydration and diarrhea (Liu et al., 2023). Furthermore, this illness can be lethal in AIDS patients with compromised immune systems (Sinyangwe et al., 2020). C. hominis produces more severe clinical symptoms in humans than C. parvum, which exhibits oocyte shedding and a longer duration of symptoms (Shirley et al., 2012). In animalsThe animal’s immune system determines the primary symptoms. Cryptosporidiosis is most common in calves younger than 6 weeks (Shaw et al., 2020). The most common symptoms are paste-like to watery diarrhea, followed by fever, dehydration, tiredness, appetite loss, and poor health (Gerace et al., 2019). Most illnesses go away on their own after a few days, although there are wide variations in how animals react to and recover from infections (Ryan et al., 2016). Infection may be lethal in certain situations. Cryptosporidiosis has been linked to elevated rates of morbidity and mortality in sheep and lambs (Ulutaş and Voyvoda, 2004). Common symptoms include diarrhea that ranges from paste-like to watery, yellow, and foul-smelling, as well as anorexia, apathy/depression, and abdominal pain (Sparks et al., 2015). However, animals have also been reported to experience conjunctivitis, pneumonia, sinusitis, dyspnea, and nasal discharge (Robertson et al., 2013). Subclinical infections typically affect animals older than 1 month, while younger animals may also be affected (Santín, 2013). The disease can still affect production, which can result in slower growth, lower carcass weights, lower slaughter percentages, and lower body condition scores (Kifleyohannes et al., 2022). DiagnosisCryptosporidiosis is typically diagnosed by the detection of parasite oocysts, parasite antigen, or parasite DNA in stool samples (Mergen et al., 2020). Since watery diarrhea is the most typical sign of cryptosporidiosis, bacterial, viral, and parasitic enteric infections linked to acute diarrhea—such as rotavirus, coronavirus, Escherichia coli, and Salmonella spp.—are included in the differential diagnosis for Cryptosporidium (Pawlowski et al., 2009). However, non-infectious causes of gastrointestinal problems, such as inflammatory bowel disease, can also occur in humans (Alsaady, 2024). Cryptosporidiosis is often diagnosed by microscopically detecting oocysts with a diameter of 4–6 μm in an affected subject’s stool (Leitch and He, 2012). However, to rule out Cryptosporidium infection in individuals with severe diarrhea, three stool samples taken on successive days should be microscopically analyzed for oocysts because Cryptosporidium oocyst detection can be challenging (Ramirez et al., 2004). Additionally, samples must be concentrated using the formalin-ether sedimentation method prior to microscopic analysis to identify oocysts in feces (Pacheco et al., 2013). The oocysts of Cryptosporidium can also be observed by phenol-auramine staining of unconcentrated stool smears or acid-fast staining (a modified Ziehl–Neelsen method), where they appear red and brilliant yellow, respectively (Khurana et al., 2012). Oocysts may also appear as “ghost” forms; therefore, this staining needs to be performed carefully (de Oliveira Lemos et al., 2012). Furthermore, although the oocysts of Cryptosporidium are approximately half the size of those of Cyclospora cayetanensis—another coccidian protozoan parasite that infects the human intestine and causes acute diarrhea (approximately 4–5 μm in diameter vs. 9–10 μm)—stool samples must be evaluated with extreme caution because both parasites’ oocysts are autofluorescent and acid-fast (Quintero-Betancourt et al., 2002). Immediate person-to-person fecal–oral transmission is unlikely for C. cayetanensis because its oocysts are not sporulated or infectious when expelled in the feces, despite sharing general life-cycle features with Cryptosporidium (Almeria et al., 2019). Microscopic detection of oocysts in stool smears is the standard method for routine diagnosis of cryptosporidiosis; nevertheless, despite its ease of use and low cost, this method has a low sensitivity (≤30%) (Mittal et al., 2014). Furthermore, the proficiency of the microscopist is crucial for the precise diagnosis of cryptosporidiosis using this method (O’Leary et al., 2021). The sensitivity can be increased using a modified acid-fast stain, which has been linked to a ~55% sensitivity and is frequently used when a structure appears suspect for Cryptosporidium (Garcia et al., 2017). However, this technique is unable to distinguish between distinct Cryptosporidium species. Apart from the previously mentioned techniques, loose or mushy stools can be analyzed for laboratory diagnosis of cryptosporidiosis using a variety of methods, including immunochromatographic assays and enzyme-linked immunosorbent assays (ELISA), which have good sensitivity and specificity for detecting Cryptosporidium antigens (Ghoshal et al., 2018). Although commercial kits are generally more sensitive and specific than microscopic techniques (≈58%–95%), prior research has demonstrated that these antigen/antibody-based detection techniques become less reliable at low parasite burdens (Hawash, 2014). Furthermore, the polymerase chain reaction (PCR)—currently recognized by the majority of laboratories as the gold standard for identifying this parasite in stool—is more expensive than antigen-based approaches but offers superior sensitivity and species-level identification. Prior research has demonstrated that immunochromatography assays, microscopy, and ELISA are more time-consuming and less convenient in terms of overall performance (cost, sensitivity, and specificity) compared with PCR (Laude et al., 2016; Autier et al., 2018; Friesen et al., 2018). Accessibility to these molecular approaches is restricted in certain laboratories and nonexistent in others, despite significant advancements in diagnostic tools (such as multiplex PCR assays for the detection of intestinal protozoa). Furthermore, the expense and requirement for technical know-how have restricted the use of these methods, especially in developing nations with high prevalence (Lenière et al., 2024). Figure 2 summarizes the various diagnostic modalities used to detect Cryptosporidium spp. in fecal samples.
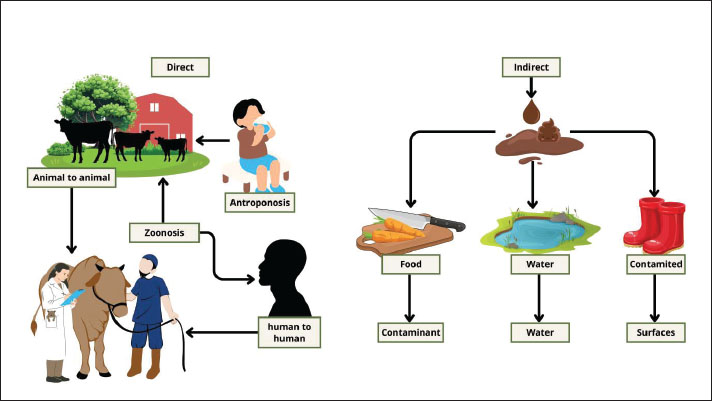
Fig. 2. Diagnostic modalities for fecal detection of Cryptosporidium spp. (A) Acid-fast: adjusted microscopy, highlighting oocyst morphology; (B) quick antigen detection immunochromatographic test; (C) Lugol iodine staining for the identification of oocyst morphology; (D) PCR with sensitive molecular detection; and (E) enzyme-linked immunosorbent assay (ELISA) detecting specific antibodies or antigens. Various diagnostic methods contain various levels of sensitivity, specificity, and practicality in the field and clinical use. TransmissionThis parasite can spread in two ways: direct transmission and indirect transmission. Cryptosporidium oocysts are excreted in the feces, and they are directly transmitted through inadvertent intake via the fecal–oral pathway (Pumipuntu and Piratae, 2018). Human-to-human transmission typically occurs in swimming pools, water parks, child care facilities, hospitals, and during sexual practices involving oral–anal contact (Hellard, 2003; Ahmed and Karanis, 2020). Transmission can also occur between animals and humans (zoonosis) or vice versa (anthroponosis or reverse zoonosis) (Messenger et al., 2014; Javed and Alkheraije, 2023). Furthermore, direct exposure to infected animals can occur when veterinarians or animal researchers—who are at high risk of contacting infected animals—come into contact with infected calves (Berhanu et al., 2022). Indirect transmission can occur when food supplies, food ingredients, drinking water, and various items, such as clothing and footwear, used on farms or in wildlife parks come into contact with infected human or animal excrement (Ali et al., 2024b). The parasite is transmitted via feces and can infect and reside on the intestinal epithelium surface in humans and other vertebrates (Di Genova and Tonelli, 2016). It can then contaminate soil and water sources, including ponds, rivers, wastewater, sewage, or slurry, and even many water containers, particularly public water supplies that are not adequately treated (Bilal et al., 2024). Distribution and transmission can increase following flooding and intense rainfall (Wamsley et al., 2025). Ingestion of an oocyst containing four sporozoites typically initiates infection in both humans and animals. These complex zoonotic and environmental transmission pathways are illustrated in Figure 3.
Fig. 3. Zoonotic and environmental transmission pathways of Cryptosporidium spp. Direct transmission occurs through animal-to-animal contact, from animals to humans (zoonosis), from humans to animals (reverse zoonosis), and between humans. Indirect transmission occurs by ingestion of oocysts via food, water, or surfaces. These pathways highlight the importance of hygiene, environmental management, and cross-species surveillance in the control of cryptosporidiosis. Public health importanceCryptosporidiosis is a serious public health concern because it may cause diarrhea in both humans and animals, and if drinking water is contaminated, it can rapidly cause outbreaks (Ahmed and Karanis, 2020; Ryan et al., 2021). It can lead to major difficulties in individuals with compromised immune systems and is spread by the fecal–oral route, frequently through tainted water (Rossle and Latif, 2013; Zhang et al., 2022). Keeping an eye on and managing cryptosporidiosis in the area is crucial to gain a better understanding of the epidemiology in terms of species, subtype distribution, and trends, even though the EU/EEA notification level is quite low (Lebbad et al., 2021). This necessitates more thorough reporting, species and subtype identification, and enhanced laboratory testing for parasites (Dąbrowska et al., 2023). This knowledge is frequently essential to epidemic investigation and source tracing because outbreak control relies on an understanding of infection dynamics. Cryptosporidiosis is underdiagnosed, and there is currently no proven cure or vaccine (Sparks et al., 2015). Additionally, the public should be made aware of ways to reduce the risk of cryptosporidiosis, such as washing, peeling, and boiling fruits and vegetables as needed, as well as maintaining good hand hygiene (Mraz et al., 2023). Public health officials should spread the word about these preventative measures, particularly to families with small children who could visit zoos or farms and to those who have direct contact with farm animals (such as farm workers and specialized veterinarians) (Rahman et al., 2020). Awareness regarding the risk of swallowing contaminated water in swimming pools or other recreational waters, or when participating in mass sporting events involving water or mud, should also be increased (Ryan et al., 2017). Economic impactThe high morbidity and occasional fatality of Cryptosporidium infection in livestock have a substantial financial impact on producers (Roblin et al., 2023). Additionally, Cryptosporidium infections have a larger economic impact due to detrimental effects on growth and reduced feed conversion (Innes et al., 2020). Cryptosporidium-infected animals develop diarrhea and may die, resulting in financial losses (Vermeulen et al., 2017). Young animals are more prone to infection than adults, whereas adults can infect young animals and act as healthy carriers (Murnik et al., 2022). In addition, Cryptosporidium infection can cause animal death, restrict the growth of affected animals, increase the cost of medicines and veterinary care, and increase staff labor (Aboelsoued and Megeed, 2022). According to a Brazilian study, affected animals died about a week after the onset of diarrhea. The animal did not recover from the infection despite receiving gentamicin and oral and intravenous fluid therapy. Numerous Cryptosporidium oocysts were found in the sample, according to a fecal smear of a dead animal. This study demonstrates how the death of sick animals and the wasted medication expenses caused by Cryptosporidium illness have a detrimental effect on the economy (Cappellaro et al., 2023). TreatmentBased on past evidence, most sick humans and animals with a robust immune response can recover on their own without any therapy (Borad and Ward, 2010). Some supportive treatments, such as fluid and electrolyte replacement, anti-nausea or antiemetic medications, or analgesic medications, can help control some clinical symptoms, such as watery fever, vomiting, nausea, abdominal cramping, and dehydration (Sparks et al., 2015). These medications can alleviate cryptosporidiosis symptoms; however, antiprotozoal therapy is required in certain situations. The only anti-cryptosporidial medication authorized by the US Food and Drug Administration to treat cryptosporidiosis in humans is nitazoxanide, which is also the most effective medication for treating people infected with Cryptosporidium spp. (Khan and Witola, 2023). Nevertheless, this medication is currently neither extensively utilized nor commercially available. Furthermore, the effective use of nitazoxanide in immunocompromised individuals is contingent on a robust host immunological response (Cohn et al., 2022). According to a small number of reports examining its effects on clinical Cryptosporidium infections in animals, nitazoxanide may lessen the excretion of Cryptosporidium oocysts in animals (Ollivett et al., 2009). However, the use of this medication in animals is still uncommon. VaccinationCurrently, there is no vaccine to prevent human or animal Cryptosporidium infections (Korpe, 2021). Vaccine development is urgently needed, especially for high-risk populations such as immunocompromised individuals, malnourished populations, and children. Vaccination of cows against various viruses that cause diarrhea, such as rotavirus, coronavirus, and E. coli, can shield calves from Cryptosporidium infection through colostrum, assisting the calves in fending off illness in the first few weeks of life (Innes et al., 2011). Understanding the host immune response to infection, host-parasite interactions, and both innate and adaptive host responses is necessary for developing a successful vaccine (Mead, 2014). The nature of this response remains unknown, though, and more research is needed. Numerous studies have attempted to create a vaccine that effectively prevents cryptosporidiosis (Gilbert et al., 2023). MicroRNAs (miRNAs) are crucial for controlling the levels of miRNA expression in epithelial cells and protecting host cells from Cryptosporidium (Kirkpatrick et al., 2006). Mannose-binding lectin (MBL), particularly in youngsters and immunocompromised adults with MBL deficiency, may offer protection against cryptosporidiosis (Carmolli et al., 2009). Moreover, a number of antigens, including gp15, cp15, and cp23, are being developed as potential vaccines. Significant cross-reactivity exists between C. parvum and C. hominis, indicating that the gp15 antigen is mainly conserved between the two species. In contrast, cp23 is conserved among C. parvum isolates and is present in both sporozoites and merozoites (Lucio-Forster et al., 2010). Pregnant goats that receive the cp15 vaccine are protected as progeny (Sagodira et al., 1999). Goats that received the vaccine had a brief decrease in Cryptosporidium in their stools, but they were not entirely immune to infection (Roche et al., 2013). Interestingly, vaccinations with several dominant antigens might improve infection resistance. For instance, in mice, the divalent cp23 + cp15 vaccine decreased oocyte release and extended the prepatent period compared with cp23 vaccination alone (Liu et al., 2010). Additionally, immunocompetent people infected with Cryptosporidium are protected against diarrhea by serum antibodies against cp23 and gp15 (Frost et al., 2005). All things considered, the ideal vaccine should prevent the spread of Cryptosporidium, offer permanent immunity to the vaccinated population, and offer protection against Cryptosporidium species and subtypes to guarantee cross-protection against the most frequently infected species. One Health approachThe “One Health” concept is a global strategy that aims to avoid infections at the human-animal-environment interface to improve health and decrease the incidence of zoonotic diseases (Rodriguez, 2024). Collaboration among various health sectors, including public health operators, occupational health physicians, and veterinarians, can aid in infection control by enhancing administrative structures, education systems, attitudes, and laws (Yassi et al., 2011). The One Health approach has been proposed in the past to address cryptosporidiosis and other zoonotic diseases because there is a critical need for One Health-oriented interactions among professionals working in a variety of fields, including doctors, veterinarians, diagnosticians, epidemiologists, public health experts, ecologists, economists, social scientists, governments, decision makers, and the pharmaceutical industry (Innes et al., 2020). Through an understanding of disease pathogenesis, life cycle, genomics, epidemiology, past outbreaks, transmission sources and dynamics, host spectrum, risk factors, high-risk groups, disease in humans and animals, diagnosis, treatment, and control, and the potential for an effective anti-Cryptosporidium vaccine, we propose the use of a One Health approach as a preventive measure for Cryptosporidium infections in humans, animals, and the environment. The One Health approach consists of educating the public about cryptosporidiosis and its transmission methods, disrupting the parasite cycle, conducting epidemiological studies to determine risk factors, implementing regular surveillance, treating infected animals to prevent human outbreaks, and hiring qualified professionals (Ali et al., 2024a). ControlThe main strategy for preventing cryptosporidiosis is to reduce or eradicate environmental contamination with infectious oocysts, as there is currently no effective treatment for the disease (Helmy and Hafez, 2022). Moving animals to a dry, clean location and disinfecting the polluted area are advised, but this is typically not feasible on farms with many animals (Silverlås and Blanco-Penedo, 2013). If contaminated locations are continuously disinfected, person-to-person transmission in homes and institutions will be decreased (Ahmed and Karanis, 2020). Oocyte infectious power and survival duration will typically be recovered at low temperatures (5°C) and augmented at temperatures above 15°C for 3 months (Hagen et al., 2014). Cryptosporidium oocytes are often susceptible to some physical stressors, such as drying, pressure, heat, cold, and radiation (Bergwerff and Debast, 2021). The carbohydrate energy reserves and waste products of sporozoites, such as amylopectin granules, which facilitate burrowing and host cell invasion, are what cause C. parvum oocytes to get infected at varying temperatures. These chemicals are quickly used up at higher temperatures (Jaskiewicz et al., 2020). The oocytes become noninfectious when the temperature is increased to 64.2°C or higher for 5 minutes and 72.4°C for 1 minute. C. parvum oocytes may last extended lengths of time at −20°C, even with cryoprotectants present, but not at −70°C or lower (Jaskiewicz et al., 2018). However, Cryptosporidium oocytes may become non-infectious when exposed to ultraviolet radiation (Takahashi et al., 2020). Disinfectants containing hydrogen peroxide, ammonia, or chlorine dioxide are the most effective against Cryptosporidium oocysts (Quilez et al., 2005). High concentrations and prolonged exposure to substances linked to chlorine, bromine, and iodine can decrease the infectivity of oocytes; however, the practical use of these compounds is limited (Coleman et al., 2023). One of the best chemical disinfectants for Cryptosporidium is ozone, which may also be used to kill Cryptosporidium oocysts in water (Nasser, 2016). Additionally, C. parvum oocytes can be consumed by predatory protozoa and rotifers, which live in ponds, rivers, lakes, and seas (Stott et al., 2003). Rotifers can be used to regulate Cryptosporidium oocysts in water because some of them have been shown to expel oocytes in boluses that contain a combination of other ingested ingredients (Fayer et al., 2000). ConclusionCryptosporidiosis is a critical zoonotic disease affecting humans and animals, with significant implications for public health, especially in vulnerable populations. Food and water contamination poses a risk, emphasizing the need for increased prevention and greater awareness among caregivers and policy makers to effectively control its spread. AcknowledgmentsIrjen. Pol. (Purn.) Dr. Dra. Juansih, S.H., M.Hum as the head of the Center of Women Empowerment In Law Enforcement, Postgraduate School, Airlangga University. Author’s contributionsWTW, KK, AKP, and ARK drafted the manuscript. BPP, IBM, DAAK, WW, and AHF revise and edit the manuscript. RA, RZA, LH, and SK prepared and critically checked the manuscript. ATK, SW, AAS, and BWKW edit the references. All authors have read and approved the final version of the manuscript. Conflict of interestThe authors declare no conflict of interest. FundingThis research was funded by the Center of Women Empowerment In Law Enforcement Research Fund Postgraduate School University Airlangga. Contract Number: 3904/UN3.SPS/PT.01.03/2023. Data availabilityAll references are open access, so data can be obtained from the internet. ReferencesAbeywardena, H., Jex, A.R. and Gasser, R.B. 2015. A perspective on Cryptosporidium and Giardia, with an emphasis on bovines and recent epidemiological findings. Adv. Parasitol. 88(1), 243–301. Aboelsoued, D. and Abdel Megeed, K.N. 2022. Diagnosis and control of cryptosporidiosis in farm animals. J. Parasit. Dis. 46(4), 1133–1146. Ahmed, S.A. and Karanis, P. 2020. Cryptosporidium and Cryptosporidiosis: the Perspective from the Gulf Countries. Int. J. Environ. Res. Public Health 17(18), 6824. Alarcón-Zapata, M.A., Romero-Salas, D., Chaparro-Gutiérrez, J.J., GonzálezHernández, M.O.-C. and Serrano-Solís Arturo, M.M. 2023. Frequency of Giardia spp. and Cryptosporidium spp. in domestic and captive wild animals in the north of Veracruz, Mexico. Pak. Vet. J. 43(4), 814–818. Ali, M., Ji, Y., Xu, C., Hina, Q., Javed, U. and Li, K. 2024a. Food and Waterborne Cryptosporidiosis from a One Health Perspective: a Comprehensive Review. Animals 14(22), 3287. Ali, M., Xu, C., Wang, J., Kulyar, M.F.E.A. and Li, K. 2024b. Emerging therapeutic avenues against Cryptosporidium: a comprehensive review. Vet. Parasitol. 331(1), 110279. Almeria, S., Cinar, H.N. and Dubey, J.P. 2019. Cyclospora cayetanensis and Cyclosporiasis: an Update. Microorganisms 7(9), 317. Alsaady, I.M. 2024. Cryptosporidium and irritable bowel syndrome. Trop. Parasitol. 14(1), 8–15. Alves, M., Xiao, L., Sulaiman, I., Lal, A.A., Matos, O. and Antunes, F. 2003. Subgenotype analysis of Cryptosporidium isolates from humans, cattle, and zoo ruminants in Portugal. J. Clin. Microbiol. 41(6), 2744–2747. Arrowood, M.J. 2002. In vitro cultivation of Cryptosporidium species. Clin. Microbiol. Rev. 15(3), 390–400. Autier, B., Belaz, S., Razakandrainibe, R., Gangneux, J.P. and Robert-Gangneux, F. 2018. Comparison of three commercial multiplex PCR assays for the diagnosis of intestinal protozoa. Parasite 25(1), 48. Ayan, A., Celik, B.A., Celik, O.Y., Akyildiz, G., Kilinc, O.O., Ayan, O.O., Oguz, F.E., Goz, Y., Yuksek, N., Yilmaz, A.B., Myrzhiyeva, A., Akhmetzhanova, A. and Uslu, U. 2024. First report of zoonotic Cryptosporidium parvum subtype IIaA15G2R1 in dogs in Türkiye. Pak. Vet. J. 44(4), 1263–1268. Ayinmode, A.B., Oliveira, B.C.M., Obebe, O.O., Dada-Adgebola, H.O., Ayede, A.I. and Widmer, G. 2018. Genotypic Characterization of Cryptosporidium Species in Humans and Peri-Domestic Animals in Ekiti and Oyo States, Nigeria. J. Parasitol. 104(6), 639–644. Balendran, T., Iddawela, D. and Lenadora, S. 2024. Cryptosporidiosis in a Zoonotic Gastrointestinal Disorder Perspective: present Status, Risk Factors, Pathophysiology, and Treatment, Particularly in Immunocompromised Patients. J. Trop. Med. 1(1), 6439375. Barker, I.K. and Carbonell, P.L. 1974. Cryptosporidium agni sp.n. from lambs, and Cryptosporidium bovis sp.n. from a calf, with observations on the oocyst. Z. Parasitenkd. 44(4), 289–298. Bergwerff, A.A. and Debast, S.B. 2021. Modernization of Control of Pathogenic Micro-Organisms in the Food-Chain Requires a Durable Role for Immunoaffinity-Based Detection Methodology—A Review. Foods 10(4), 832. Berhanu, K., Ayana, D., Megersa, B., Ashenafi, H. and Waktole, H. 2022. Cryptosporidium in human-animal-environment interphase at Adama and Asella areas of Oromia regional state, Ethiopia. BMC. Vet. Res. 18(1), 402. Bhalchandra, S., Ludington, J., Coppens, I. and Ward, H.D. 2013. Identification and characterization of Cryptosporidium parvum Clec, a novel C-type lectin domain-containing mucin-like glycoprotein. Infect. Immun. 81(9), 3356–3365. Bilal, H., Li, X., Iqbal, M.S., Tulcan, R.X.S. and Chhetri, M.T. 2024. Unveiling the Dynamics of Cryptosporidium in Urban Surface Water: a Quantitative Microbial Risk Assessment and Insights into Climatic and Seasonal Influences. Water 16(10), 1352. Bones, A.J., Jossé, L., More, C., Miller, C.N., Michaelis, M. and Tsaousis, A.D. 2019. Past and future trends of Cryptosporidium in vitro research. Exp. Parasitol. 196(1), 28–37. Borad, A. and Ward, H. 2010. Human immune responses in cryptosporidiosis. Future Microbiol. 5(3), 507–519. Boucher, L.E. and Bosch, J. 2015. The Apicomplexan glideosome and adhesins - Structures and function. J. Struct. Biol. 190(2), 93–114. Bouzid, M., Hunter, P.R., Chalmers, R.M. and Tyler, K.M. 2013. Cryptosporidium pathogenicity and virulence. Clin. Microbiol. Rev. 26(1), 115–134. Bruno, D., Nowak, B. and Elliott, D. 2006. Guide to the identification of fish protozoan and metazoan parasites in stained tissue sections. Dis. Aquat. Organ. 70(1), 1–36. Cappellaro, V., Matzembacker, B., Chitolina, M.B., Knorst, C.R., Girardini, L.K., Prestes, A.M., Mortari, A.P.G., Fernandes, F.D., Vogel, F.S.F. and Camillo, G. 2023. Cryptosporidium infection in diarrheal bovine dairy calves: occurrence and risk factors in Santa Catarina, Brazil. Semina. Ciênc. Agrár. Londrina 44(1), 317–328. Carmolli, M., Duggal, P., Haque, R., Lindow, J., Mondal, D., Petri WA Jr., Mourningstar, P., Larsson, C.J., Sreenivasan, M., Khan, S. and Kirkpatrick, B.D. 2009. Deficient serum mannose-binding lectin levels and MBL2 polymorphisms increase the risk of single and recurrent Cryptosporidium infections in young children. J. Infect. Dis. 200(10), 1540–1547. CDC. 2023. Cryptosporidiosis National Notifiable Disease Surveillance System (NNDSS) summary report for 2022. Centers for Disease Control and Prevention. Accessed 11 September 2023. Cevallos, A.M., Bhat, N., Verdon, R., Hamer, D.H., Stein, B., Tzipori, S., Pereira, M.E., Keusch, G.T. and Ward, H.D. 2000a. Mediation of Cryptosporidium parvum infection in vitro by mucin-like glycoproteins defined by a neutralizing monoclonal antibody. Infect. Immun. 68(9), 5167–5175. Cevallos, A.M., Zhang, X., Waldor, M.K., Jaison, S., Zhou, X., Tzipori, S., Neutra, M.R. and Ward, H.D. 2000b. Molecular cloning and expression of a gene encoding Cryptosporidium parvum glycoproteins gp40 and gp15. Infect. Immun. 68(7), 4108–4116. Chalmers, R.M. and Katzer, F. 2013. Looking for Cryptosporidium: the application of advances in detection and diagnosis. Trends Parasitol. 29(5), 237–251. Chappell, C.L., Okhuysen, P.C., Sterling, C.R. and DuPont, H.L. 1996. Cryptosporidium parvum: intensity of infection and oocyst excretion patterns in healthy volunteers. J. Infect. Dis. 173(1), 232–236. Clarke, J.J. 1895. A study of coccidia met with in mice. J. Microsc. Soc. 37(147), 277–302. Cohn, I.S., Henrickson, S.E., Striepen, B. and Hunter, C.A. 2022. Immunity to Cryptosporidium: lessons from Acquired and Primary Immunodeficiencies. J. Immunol. 209(12), 2261–2268. Coleman, C.K., Kim, J., Bailey, E.S., Abebe, L.S., Brown, J., Simmons, O.D. and Sobsey, M.D. 2023. Bromine and Chlorine Disinfection of Cryptosporidium parvum Oocysts, Bacillus atrophaeus Spores, and MS2 Coliphage in Water. Environ. Sci. Technol. 57(47), 18744–18753. Current, W.L., Upton, S.J. and Haynes, T.B. 1986. The Life Cycle of Cryptosporidium baileyi n. sp. (Apicomplexa, Cryptosporidiidae) Infecting Chickens. J. Protozool. 33(2), 289–296. Dąbrowska, J., Sroka, J. and Cencek, T. 2023. Investigating Cryptosporidium spp. Using Genomic, Proteomic and Transcriptomic Techniques: current Progress and Future Directions. Int. J. Mol. Sci. 24(16), 12867. Dărăbuș, R.G., Imre, M., Dărăbuș, G., Ilie, M.S., Olariu, A.T., Dărăbuș, D.M., Lăzureanu, V., Roșca, O. and Olariu, T.R. 2025. First Detection of Cryptosporidium canis and Occurrence of Cryptosporidium spp. in Hospitalized Patients in Romania. Microorganisms 13(4), 931. De Oliveira Lemos, F., Almosny, N.P., Soares, A.M. and Alencar, N.X. 2012. Cryptosporidium species screening using Kinyoun technique in domestic cats with diarrhea. J. Feline Med. Surg. 14(2), 113–117. Dessì, G., Tamponi, C., Varcasia, A., Sanna, G., Pipia, A.P., Carta, S., Salis, F., Díaz, P. and Scala, A. 2020. Cryptosporidium infections in sheep farms from Italy. Parasitol. Res. 119(12), 4211–4218. Di Genova, B.M. and Tonelli, R.R. 2016. Infection Strategies of Intestinal Parasite Pathogens and Host Cell Responses. Front. Microbiol. 7(1), 256. Dixon, B., Parrington, L., Cook, A., Pintar, K., Pollari, F., Kelton, D. and Farber, J. 2011. The potential for zoonotic transmission of Giardia duodenalis and Cryptosporidium spp. from beef and dairy cattle in Ontario, Canada. Vet. Parasitology 175(1–2), 20–26. Dragomirova, P.V. 2022. Cryptosporidiosis: history, Etiology, Biology, Pathogenesis and Pathoanatomy - A Review. J. Biomed. Clin. Res. 15(1), 22–29. EFSA. 2022. IMPACT: standardising molecular detection methods to improve risk assessment capacity for foodborne protozoan parasites, using Cryptosporidium in ready-to-eat salad as a model. European Food Safety Authority. Accessed 14 March 2022. Ehsan, A.M., Geurden, T., Casaert, S., Parvin, S.M., Islam, T.M., Ahmed, U.M., Levecke, B., Vercruysse, J. and Claerebout, E. 2015. Assessment of zoonotic transmission of Giardia and Cryptosporidium between cattle and humans in rural villages in Bangladesh. PLos One 10(2), 118239. El-Alfy, E.S. and Nishikawa, Y. 2020. Cryptosporidium species and cryptosporidiosis in Japan: a literature review and insights into the role played by animals in its transmission. J. Vet. Med. Sci. 82(8), 1051–1067. English, E.D., Guérin, A., Tandel, J. and Striepen, B. 2022. Live imaging of the Cryptosporidium parvum life cycle reveals direct development of male and female gametes from type I meronts. PLos Biol. 20(4), e3001604. Fayer, R., Trout, J.M., Walsh, E. and Cole, R. 2000. Rotifers Ingest Oocysts of Cryptosporidium parvum. J. Eukar. Microbiol. 47(2), 161–163. Friesen, J., Fuhrmann, J., Kietzmann, H., Tannich, E., Müller, M. and Ignatius, R. 2018. Evaluation of the Roche LightMix Gastro parasites multiplex PCR assay detecting Giardia duodenalis, Entamoeba histolytica, cryptosporidia, Dientamoeba fragilis, and Blastocystis hominis. Clin. Microbiol. Infect. 24(12), 1333–1337. Frost, F.J., Tollestrup, K., Craun, G.F., Fairley, C.K., Sinclair, M.I. and Kunde, T.R. 2005. Protective immunity associated with a strong serological response to a Cryptosporidium-specific antigen group, in HIV-infected individuals. J. Infect. Dis. 192(4), 618–621. Garcia, L.S., Arrowood, M., Kokoskin, E., Paltridge, G.P., Pillai, D.R., Procop, G.W., Ryan, N., Shimizu, R.Y. and Visvesvara, G. 2017. Practical Guidance for Clinical Microbiology Laboratories: laboratory Diagnosis of Parasites from the Gastrointestinal Tract. Clin. Microbiol. Rev. 31(1), e00025. García-Livia, K., Martín-Alonso, A. and Foronda, P. 2020. Diversity of Cryptosporidium spp. in wild rodents from the Canary Islands, Spain. Parasit. Vectors 13(1), 445. Gerace, E., Lo Presti, V.D.M. and Biondo, C. 2019. Cryptosporidium Infection: epidemiology, Pathogenesis, and Differential Diagnosis. Eur. J. Microbiol. Immunol. (Bp). 9(4), 119–123. Ghoshal, U., Jain, V., Dey, A. and Ranjan, P. 2018. Evaluation of enzyme linked immunosorbent assay for stool antigen detection for the diagnosis of cryptosporidiosis among HIV negative immunocompromised patients in a tertiary care hospital of northern India. J. Infect. Public Health 11(1), 115–119. Gilbert, I.H., Vinayak, S., Striepen, B., Manjunatha, U.H., Khalil, I.A. and Van Voorhis, W.C. 2023. Safe and effective treatments are needed for cryptosporidiosis, a truly neglected tropical disease. BMJ Glob. Health 8(8), 12540. Gubbels, M.J. and Duraisingh, M.T. 2012. Evolution of Apicomplexan secretory organelles. Int. J. Parasitol. 42(12), 1071–1081. Hadfield, S.J., Robinson, G., Elwin, K. and Chalmers, R.M. 2011. Detection and differentiation of Cryptosporidium spp. in human clinical samples by use of real-time PCR. J. Clin. Microbiol. 49(3), 918–924. Hagen, R.M., Loderstaedt, U. and Frickmann, H. 2014. An evaluation of the potential use of Cryptosporidium species as agents for deliberate release. J. R. Army Med. Corps 160(4), 289–294. Hawash, Y. 2014. Evaluation of an immunoassay-based algorithm for screening and identification of giardia and Cryptosporidium antigens in human faecal specimens from Saudi Arabia. J. Parasitol. Res. 1(1), 213745. He, W., Li, J., Gong, A.Y., Deng, S., Li, M., Wang, Y., Mathy, N.W., Feng, Y., Xiao, L. and Chen, X.M. 2021. Cryptosporidial Infection Suppresses Intestinal Epithelial Cell MAPK Signaling Impairing Host Anti-Parasitic Defense. Microorganisms 9(1), 151. Hechenbleikner, E.M. and McQuade, J.A. 2015. Parasitic colitis. Clin. Colon. Rectal. Surg. 28(2), 79–86. Hellard, M. 2003. Risk factors leading to Cryptosporidium infection in men who have sex with men. Sex. Transm. Infect. 79(5), 412–414. Helmy, Y.A. and Hafez, H.M. 2022. Cryptosporidiosis: from Prevention to Treatment, a Narrative Review. Microorganisms 10(12), 2456. Heo, I., Dutta, D., Schaefer, D.A., Iakobachvili, N., Artegiani, B., Sachs, N., Boonekamp, K.E., Bowden, G., Hendrickx, A.P.A., Willems, R.J.L., Peters, P.J., Riggs, M.W., O’Connor, R. and Clevers, H. 2018. Modelling Cryptosporidium infection in human small intestinal and lung organoids. Nat. Microbiol. 3(7), 814–823. Hijjawi, N. 2004. Complete development of Cryptosporidium parvum in host cell-free culture. Int. J. Parasitol. 34(7), 769–777. Hunter, P.R. and Nichols, G. 2002. Epidemiology and clinical features of Cryptosporidium infection in immunocompromised patients. Clin. Microbiol. Rev. 15(1), 145–154. Hunter, P.R. and Thompson, R.C.A. 2005. The zoonotic transmission of Giardia and Cryptosporidium. Int. J. Parasitol. 35(11–12), 1181–1190. Hussain, S., Ain, Q.U., Aamir, M., Alsyaad, K.M., Ahmed, A.E., Zakai, J.G., Zakai, H.A. and Hou, Y. 2025. Deciphering Host–Pathogen Interactions: role of Cryptosporidium in Tumorigenesis. Pathogens 14(3), 208. Innes, E.A., Bartley, P.M., Rocchi, M., Benavidas-Silvan, J., Burrells, A., Hotchkiss, E., Chianini, F., Canton, G. and Katzer, F. 2011. Developing vaccines to control protozoan parasites in ruminants: dead or alive?. Vet. Parasitol. 180(1–2), 155–163. Innes, E.A., Chalmers, R.M., Wells, B. and Pawlowic, M.C. 2020. A One Health Approach to Tackle Cryptosporidiosis. Trends Parasitol. 36(3), 290–303. Lejeune, J. and Kersting, A. 2010. Zoonoses: an occupational hazard for livestock workers and a public health concern for rural communities. J. Agric. Saf. Health 16(3), 161–179. Janouškovec, J., Paskerova, G.G., Miroliubova, T.S., Mikhailov, K.V., Birley, T., Aleoshin, V.V. and Simdyanov, T.G. 2019. Apicomplexan-like parasites are polyphyletic and widely but selectively dependent on cryptic plastid organelles. Elife 8(1), e49662. Jaskiewicz, J.J., Sandlin, R.D., Swei, A.A., Widmer, G., Toner, M. and Tzipori, S. 2018. Cryopreservation of infectious Cryptosporidium parvum oocysts. Nat. Commun. 9(1), 2883. Jaskiewicz, J.J., Sevenler, D., Swei, A.A., Widmer, G., Toner, M., Tzipori, S. and Sandlin, R.D. 2020. Cryopreservation of infectious Cryptosporidium parvum oocysts achieved through vitrification using high aspect ratio specimen containers. Sci. Rep. 10(1), 11711. Javed, K. and Alkheraije, K.A. 2023. Cryptosporidiosis: a foodborne zoonotic disease of farm animals and humans. Pak. Vet. J. 43(2), 213–223. Kelly, P., Thillainayagam, A.V., Smithson, J., Hunt, J.B., Forbes, A., Gazzard, B.G. and Farthing, M.J.G. 1996. Jejunal water and electrolyte transport in human cryptosporidiosis. Digestive Dis. Sci. 41(10), 2095–2099. Khalil, I.A., Troeger, C., Rao, P.C., Blacker, B.F., Brown, A., Brewer, T.G., Colombara, D.V., De Hostos, E.L., Engmann, C., Guerrant, R.L., Haque, R., Houpt, E.R., Kang, G., Korpe, P.S., Kotloff, K.L., Lima, A.A.M., Petri, W.A. Jr, Platts-Mills, J.A., Shoultz, D.A., Forouzanfar, M.H., Hay, S.I., Reiner, R.C. Jr. and Mokdad, A.H. 2018. Morbidity, mortality, and long-term consequences associated with diarrhoea from Cryptosporidium infection in children younger than 5 years: a meta-analysis study. Lancet Global Health 6(7), e758–e768; doi:10.1016/S2214-109X(18)30283-3 Khan, S.M. and Witola, W.H. 2023. Past, current, and potential treatments for cryptosporidiosis in humans and farm animals: a comprehensive review. Front. Cell. Infect. Microbiol. 13(1), 1115522. Khurana, S., Sharma, P., Sharma, A. and Malla, N. 2012. Evaluation of Ziehl-Neelsen staining, auramine phenol staining, antigen detection enzyme linked immunosorbent assay and polymerase chain reaction, for the diagnosis of intestinal cryptosporidiosis. Trop. Parasitol. 2(1), 20–23. Kifleyohannes, T., Nødtvedt, A., Debenham, J.J., Terefe, G. and Robertson, L.J. 2022. Cryptosporidium and Giardia in Livestock in Tigray, Northern Ethiopia and Associated Risk Factors for Infection: a Cross-Sectional Study. Front. Vet. Sci. 8(1), 825940. Kirkpatrick, B.D., Huston, C.D., Wagner, D., Noel, F., Rouzier, P., Pape, J.W., Bois, G., Larsson, C.J., Alston, W.K., Tenney, K., Powden, C., O’Neill, J.P. and Sears, C.L. 2006. Serum mannose-binding lectin deficiency is associated with cryptosporidiosis in young Haitian children. Clin. Infect. Dis. 43(3), 289–294. Korbel, D.S., Barakat, F.M., Di Santo, J.P. and McDonald, V. 2011. CD4+ T cells are not essential for control of early acute Cryptosporidium parvum infection in neonatal mice. Infect. Immun. 79(4), 1647–1653. Kotloff, K.L., Nataro, J.P., Blackwelder, W.C., Nasrin, D., Farag, T.H., Panchalingam, S., Wu, Y., Sow, S.O., Sur, D., Breiman, R.F., Faruque, A.S.G., Zaidi, A.K.M., Saha, D., Alonso, P.L., Tamboura, B., Sanogo, D., Onwuchekwa, U., Manna, B., Ramamurthy, T., Kanungo, S., Ochieng, J.B., Omore, R., Oundo, J.O., Hossain, A., Das, S.K., Ahmed, S., Qureshi, S., Quadri, F., Adegbola, R.A., Antonio, M., Hossain, M.J., Akinsola, A., Mandomando, I., Nhampossa, T., Acácio, S., Biswas, K., O’Reilly, C.E., Mintz, E.D., Berkeley, L.Y., Muhsen, K., Sommerfelt, H., Robins-Browne, R.M. and Levine, M.M. (2013). Burden and aetiology of diarrhoeal disease in infants and young children in developing countries (the Global Enteric Multicenter Study, GEMS): a prospective, case-control study. Lancet 382(9888), 209–222. Korpe, P.S. 2021. The Silent Reservoir of Cryptosporidiosis. Clin. Infect. Dis. 72(8), 1367–1368. Langer, R.C. and Riggs, M.W. 1999. Cryptosporidium parvum apical complex glycoprotein CSL contains a sporozoite ligand for intestinal epithelial cells. Infect. Immun. 67(10), 5282–5291. Langer, R.C., Schaefer, D.A. and Riggs, M.W. 2001. Characterization of an intestinal epithelial cell receptor recognized by the Cryptosporidium parvum sporozoite ligand CSL. Infect. Immun. 69(3), 1661–1670. Laude, A., Valot, S., Desoubeaux, G., Argy, N., Nourrisson, C., Pomares, C., Machouart, M., Le Govic, Y., Dalle, F., Botterel, F., Bourgeois, N., Cateau, E., Leterrier, M., Le Pape, P. and Morio, F. 2016. Is real-time PCR-based diagnosis similar in performance to routine parasitological examination for the identification of Giardia intestinalis, Cryptosporidium parvum/Cryptosporidium hominis and Entamoeba histolytica from stool samples? Evaluation of a new commercial multiplex PCR assay and literature review. Clin. Microbiol. Infect. 22(2), 190.e1–190.e8. Lauwers, G.Y., Mino-Kenudson, M. and Kradin, R.L. 2018. Infections of the Gastrointestinal Tract. Diagn. Pathol. Infect. Dis. 1(1), 232–271. Leav, B.A., Mackay, M.R., Anyanwu, A., O’ Connor, R.M., Cevallos, A.M., Kindra, G., Rollins, N.C., Bennish, M.L., Nelson, R.G. and Ward, H.D. 2002. Analysis of sequence diversity at the highly polymorphic Cpgp40/15 locus among Cryptosporidium isolates from human immunodeficiency virus-infected children in South Africa. Infect. Immun. 70(7), 3881–3890. Lebbad, M., Winiecka-Krusnell, J., Stensvold, C.R. and Beser, J. 2021. High Diversity of Cryptosporidium Species and Subtypes Identified in Cryptosporidiosis Acquired in Sweden and Abroad. Pathogens 10(5), 523. Leitch, G.J. and He, Q. 2012. Cryptosporidiosis-an overview. J. Biomed. Res. 25(1), 1–16. LeJeune, J. and Kersting, A. 2010. Zoonoses: an occupational hazard for livestock workers and a public health concern for rural communities. J. Agric. Saf. Health. 16(3), 161–179. Lenière, A.C., Vlandas, A. and Follet, J. 2024. Treating cryptosporidiosis: a review on drug discovery strategies. Int. J. Parasitol. Drugs Drug Resist. 25(1), 100542. Li, F., Su, J., Chahan, B., Guo, Q., Wang, T., Yu, Z., Guo, Y., Li, N., Feng, Y. and Xiao, L. 2019. Different distribution of Cryptosporidium species between horses and donkeys. Infect. Genet. Evol. 75(1), 103954. Li, J., Guo, Y., Roellig, D.M., Li, N., Feng, Y. and Xiao, L. 2021b. Cryptosporidium felis differs from other Cryptosporidium spp. in codon usage. Microb. Genom. 7(12), 711. Li, Y., Geng, W.L., Li, C.C., Wu, J.H., Gao, F. and Wang, Y. 2024. Progress of CCL20-CCR6 in the airways: a promising new therapeutic target. J. Inflamm. (Lond). 21(1), 54. Lin, X., Xin, L., Qi, M., Hou, M., Liao, S., Qi, N., Li, J., Lv, M., Cai, H., Hu, J., Zhang, J., Ji, X. and Sun, M. 2022. Dominance of the zoonotic pathogen Cryptosporidium meleagridis in broiler chickens in Guangdong, China, reveals evidence of cross-transmission. Parasites & Vectors 15(1), 188. Liu, K., Zai, D., Zhang, D., Wei, Q., Han, G., Gao, H. and Huang, B. 2010. Divalent Cp15-23 vaccine enhances immune responses and protection against Cryptosporidium parvum infection. Parasite Immunol. 32(5), 335–344. Liu, X., Wang, J., Liu, J., Li, X., Guan, Y., Qian, S. and Jia, X. 2023. Cryptosporidiosis diagnosed using metagenomic next-generation sequencing in a healthy child admitted to pediatric intensive care unit: a case report. Front. Cellular Infect. Microbiol. 13(1), 1269963. Lucio-Forster, A., Griffiths, J.K., Cama, V.A., Xiao, L. and Bowman, D.D. 2010. Minimal zoonotic risk of cryptosporidiosis from pet dogs and cats. Trends Parasitol. 26(4), 174–179. Ludington, J.G. and Ward, H.D. 2015. Systemic and Mucosal Immune Responses to Cryptosporidium-Vaccine Development. Curr. Trop. Med. Rep. 2(3), 171–180. Mafokwane, T., Djikeng, A., Nesengani, L.T., Dewar, J. and Mapholi, O. 2023. Gastrointestinal Infection in South African Children under the Age of 5 years: a Mini Review. Gastroenterol. Res. Pract. 1(1), 1906782. Mahmoudi, M.R., Ongerth, J.E. and Karanis, P. 2017. Cryptosporidium and cryptosporidiosis: the Asian perspective. Int. J. Hyg. Environ. Health 220(7), 1098–1109. Mamedova, S. and Karanis, P. 2025. Coccidia (Apicomplexa: eucoccidiorida) of Freshwater Fish. Microorganisms 13(2), 347. Mazurie, A.J., Alves, J.M., Ozaki, L.S., Zhou, S., Schwartz, D.C. and Buck, G.A. 2013. Comparative genomics of Cryptosporidium. Int. J. Genomics 1(1), 832756. Mead, J.R. 2014. Prospects for immunotherapy and vaccines against Cryptosporidium. Hum. Vac. Immun. 10(6), 1505–1513. Mead, J.R. 2023. Early immune and host cell responses to Cryptosporidium infection. Front. Parasitol. 2(1), 1113950. Mergen, K., Espina, N., Teal, A. and Madison-Antenucci, S. 2020. Detecting Cryptosporidium in Stool Samples Submitted to a Reference Laboratory. Am. J. Trop. Med. Hyg. 103(1), 421–427. Messenger, A.M., Barnes, A.N. and Gray, G.C. 2014. Reverse zoonotic disease transmission (zooanthroponosis): a systematic review of seldom-documented human biological threats to animals. PLos One 9(2), e89055. Mittal, S., Sharma, M., Chaudhary, U. and Yadav, A. 2014. Comparison of ELISA and Microscopy for detection of Cryptosporidium in stool. J. Clin. Diagn. Res. 8(11), DC07–DC08. Mraz, A.L., Mutyala, N., Cleary, S. and Seals, B.F. 2023. Is Personal Protective Equipment Worth the Hassle? Annual Risk of Cryptosporidiosis to Dairy Farmers and How Personal Protective Equipment and Handwashing Can Mitigate It. Microorganisms 11(10), 2413. Murnik, L.C., Daugschies, A. and Delling, C. 2022. Cryptosporidium infection in young dogs from Germany. Parasitol. Res. 121(10), 2985–2993. Nageeb, M.M., Mohamed El Fakahany, A.F., Abd El Maaboud, A.I., Youssif, S.H., Hegab, F.A. and Omar, G.H. 2024. Efficacy of Atorvastatin Loaded on Nano Particles on Cryptosporidium parvum in Experimentally Infected Mice. Benha Med. J. 41(6), 87–100. Nasser, A.M. 2016. Removal of Cryptosporidium by wastewater treatment processes: a review. J. Water Health 14(1), 1–13. Nava, P., Koch, S., Laukoetter, M.G., Lee, W.Y., Kolegraff, K., Capaldo, C.T., Beeman, N., Addis, C., Gerner-Smidt, K., Neumaier, I., Skerra, A., Li, L., Parkos, C.A. and Nusrat, A. 2010. Interferon-gamma regulates intestinal epithelial homeostasis through converging beta-catenin signaling pathways. Immunity 32(3), 392–402. Nesterenko, M.V., Woods, K. and Upton, S.J. 1999. Receptor/ligand interactions between Cryptosporidium parvum and the surface of the host cell. Biochim. Biophys. Acta. 1454(2), 165–173. O’Connor, R.M., Thorpe, C.M., Cevallos, A.M. and Ward, H.D. 2002. Expression of the highly polymorphic Cryptosporidium parvum Cpgp40/15 gene in genotype I and II isolates. Mol. Biochem. Parasitology 119(2), 203–215. O’Hara, S.P. and Chen, X.M. 2011. The cell biology of Cryptosporidium infection. Microbes Infect. 13(8–9), 721–730. O’Leary, J.K., Sleator, R.D. and Lucey, B. 2021. Cryptosporidium spp. diagnosis and research in the 21st century. Food Waterborne Parasitol. 24(1), 131. Ollivett, T.L., Nydam, D.V., Bowman, D.D., Zambriski, J.A., Bellosa, M.L., Linden, T.C. and Divers, T.J. 2009. Effect of nitazoxanide on cryptosporidiosis in experimentally infected neonatal dairy calves. J. Dairy Sci. 92(4), 1643–1648. Pacheco, F.T., Silva, R.K., Martins, A.S., Oliveira, R.R., Alcântara-Neves, N.M., Silva, M.P., Soares, N.M. and Teixeira, M.C. 2013. Differences in the detection of Cryptosporidium and Isospora (Cystoisospora) oocysts according to the fecal concentration or staining method used in a clinical laboratory. J. Parasitol. 99(6), 1002–1008. Pal, M., Shuramo, M.Y., Gutama, K.P. and Garedaghi, Y. 2021. Cryptosporidiosis: an Emerging Zoonotic Disease of Global Public Health Concern. Med. Parasitol. Epidemiol. Sci. 2(4), 81–85. Paluszynski, J., Monahan, Z., Williams, M., Lai, O., Morris, C., Burns, P. and O’Connor, R. 2014. Biochemical and functional characterization of CpMuc4, a Cryptosporidium surface antigen that binds to host epithelial cells. Mol. Biochem. Parasitology 193(2), 114–121. Pardy, R.D., Wallbank, B.A., Striepen, B. and Hunter, C.A. 2024. Immunity to Cryptosporidium: insights into principles of enteric responses to infection. Nat. Rev. Immunol. 24(2), 142–155. Pawlowski, S.W., Warren, C.A. and Guerrant, R. 2009. Diagnosis and treatment of acute or persistent diarrhea. Gastroenterology 136(6), 1874–1886. Peng, S., Xu, C., Saleem, M.U., Babar, W., Idrees, A. and Li, K. 2024. Epidemiological investigation of Cryptosporidium infection in yaks in Chamdo, China. Pak. Vet. J. 44(2), 535–538. Perez-Cordon, G., Yang, G., Zhou, B., Nie, W., Li, S., Shi, L., Tzipori, S. and Feng, H. 2014. Interaction of Cryptosporidium parvum with mouse dendritic cells leads to their activation and parasite transportation to mesenteric lymph nodes. Pathog. Dis. 70(1), 17–27. Prabakaran, M., Weible, L., Champlain, J., Jiang, R., Biondi, K., Weil, A., Van Voorhis, W. and Ojo, K. 2023. The Gut-Wrenching Effects of Cryptosporidiosis and Giardiasis in Children. Microorganisms 11(9), 2323. Preidis, G.A., Wang, H.C., Lewis, D.E., Castellanos-Gonzalez, A., Rogers, K.A., Graviss, E.A., Ward, H.D. and White, A.C. 2007. Seropositive human subjects produce interferon gamma after stimulation with recombinant Cryptosporidium hominis gp15. Am. J. Trop. Med. Hyg. 77(3), 583–585. Priest, J.W., Xie, L.T., Arrowood, M.J. and Lammie, P.J. 2001. The immunodominant 17-kDa antigen from Cryptosporidium parvum is glycosylphosphatidylinositol-anchored. Mol. Biochem. Parasitol. 113(1), 117–126. Pumipuntu, N. and Piratae, S. 2018. Cryptosporidiosis: a zoonotic disease concern. Vet. World 11(5), 681–686. Quilez, J., Sanchez-Acedo, C., Avendaño, C., Del Cacho, E. and Lopez-Bernad, F. 2005. Efficacy of two peroxygen-based disinfectants for inactivation of Cryptosporidium parvum oocysts. Appl. Environ. Microbiol. 71(5), 2479–2483. Quintero-Betancourt, W., Peele, E.R. and Rose, J.B. 2002. Cryptosporidium parvum and Cyclospora cayetanensis: a review of laboratory methods for detection of these waterborne parasites. J. Microbiol. Methods 49(3), 209–224. Rahman, M.T., Sobur, M.A., Islam, M.S., Ievy, S., Hossain, M.J., El Zowalaty, M.E., Rahman, A.T. and Ashour, H.M. 2020. Zoonotic Diseases: etiology, Impact, and Control. Microorganisms 8(9), 1405. Ramirez, N.E., Ward, L.A. and Sreevatsan, S. 2004. A review of the biology and epidemiology of cryptosporidiosis in humans and animals. Microbes Infect. 6(8), 773–785. Resnhaleksmana, E., Wijayanti, M.A. and Artama, W.T. 2021. A potential zoonotic parasite: Cryptosporidium parvum transmission in rats, pigs and humans in West Lombok, Indonesia. Afr. J. Infect. Dis. 15(2), 44–51. Rideout, H., Cook, A.J.C. and Whetton, A.D. 2024. Understanding the Cryptosporidium species and their challenges to animal health and livestock species for informed development of new, specific treatment strategies. Front. Parasitol. 3(1), 1448076. Rider, S.D. and Zhu, G. 2010. Cryptosporidium: genomic and biochemical features. Exp. Parasitol. 124(1), 2–9. Riggs, M.W. 2002. Recent advances in cryptosporidiosis: the immune response. Microbes Infect. 4(10), 1067–1080. Riggs, M.W., Stone, A.L., Yount, P.A., Langer, R.C., Arrowood, M.J. and Bentley, D.L. 1997. Protective monoclonal antibody defines a circumsporozoite-like glycoprotein exoantigen of Cryptosporidium parvum sporozoites and merozoites. J. Immunol. 158(4), 1787–1795. Robertson, L.J., Björkman, C., Axén, C. and Fayer, R. 2013. Cryptosporidiosis in Farmed Animals. Cryptosporidium Parasite Dis. 1(1), 149–235. Robinson, G., Wright, S., Elwin, K., Hadfield, S.J., Katzer, F., Bartley, P.M., Hunter, P.R., Nath, M., Innes, E.A. and Chalmers, R.M. 2010. Re-description of Cryptosporidium cuniculus Inman and Takeuchi, 1979 (Apicomplexa: cryptosporidiidae): morphology, biology and phylogeny. Int. J. Parasitol. 40(13), 1539–1548. Roblin, M., Canniere, E., Barbier, A., Daandels, Y., Dellevoet-Groenewegen, M., Pinto, P., Tsaousis, A., Leruste, H., Brainard, J., Hunter, P.R. and Follet, J. 2023. Study of the economic impact of cryptosporidiosis in calves after implementing good practices to manage the disease on dairy farms in Belgium, France, and the Netherlands. Curr. Res. Parasitol. Vector. Borne. Dis. 4(1), 100149. Roche, J.K., Rojo, A.L., Costa, L.B., Smeltz, R., Manque, P., Woehlbier, U., Bartelt, L., Galen, J., Buck, G. and Guerrant, R.L. 2013. Intranasal vaccination in mice with an attenuated Salmonella enterica Serovar 908htr A expressing Cp15 of Cryptosporidium: impact of malnutrition with preservation of cytokine secretion. Vaccine 31(6), 912–918. Rodriguez, J. 2024. One Health Ethics and the Ethics of Zoonoses: a Silent Call for Global Action. Vet. Sci. 11(9), 394. Rossle, N.F. and Latif, B. 2013. Cryptosporidiosis as threatening health problem: a review. Asian Pac. J. Trop. Biomed. 3(11), 916–924. Ruecker, N.J., Braithwaite, S.L., Topp, E., Edge, T., Lapen, D.R., Wilkes, G., Robertson, W., Medeiros, D., Sensen, C.W. and Neumann, N.F. 2007. Tracking host sources of Cryptosporidium spp. in raw water for improved health risk assessment. Appl. Environ. Microbiol. 73(12), 3945–3957. Ryan, U. and Hijjawi, N. 2015. New developments in Cryptosporidium research. Int. J. Parasitol. 45(6), 367–373. Ryan, U., Fayer, R. and Xiao, L. 2014. Cryptosporidium species in humans and animals: current understanding and research needs. Parasitology 141(13), 1667–1685. Ryan, U., Lawler, S. and Reid, S. 2017. Limiting swimming pool outbreaks of cryptosporidiosis – the roles of regulations, staff, patrons and research. J. Water Health 15(1), 1–16. Ryan, U., Zahedi, A. and Paparini, A. 2016. Cryptosporidium in humans and animals-a one health approach to prophylaxis. Parasite Immunol. 38(9), 535–547. Ryan, U., Zahedi, A., Feng, Y. and Xiao, L. 2021. An update on zoonotic Cryptosporidium species and genotypes in humans. Animals 11(11), 3307. Sagodira, S., Buzoni-Gatel, D., Iochmann, S., Naciri, M. and Bout, D. 1999. Protection of kids against Cryptosporidium parvum infection after immunization of dams with CP15-DNA. Vaccine 17(19), 2346–2355. Sanderson, S.J., Xia, D., Prieto, H., Yates, J., Heiges, M., Kissinger, J.C., Bromley, E., Lal, K., Sinden, R.E., Tomley, F. and Wastling, J.M. 2008. Determining the protein repertoire of Cryptosporidium parvum sporozoites. Proteomics 8(7), 1398–1414. Santín, M. 2013. Clinical and subclinical infections with Cryptosporidium in animals. N. Z. Vet. J. 61(1), 1–10. Santin, M. 2020. Cryptosporidium and Giardia in Ruminants. Vet. Clin. North Am. Food Anim. Pract. 36(1), 223–238. Sawant, M., Benamrouz-Vanneste, S., Mouray, A., Bouquet, P., Gantois, N., Creusy, C., Duval, E., Mihalache, A., Gosset, P., Chabé, M., Hot, D., Viscogliosi, E. and Certad, G. 2021. Persistent Cryptosporidium parvum Infection Leads to the Development of the Tumor Microenvironment in an Experimental Mouse Model: results of a Microarray Approach. Microorganisms 9(12), 2569. Scorza, A.V., Tyrrell, P., Wennogle, S., Chandrashekar, R. and Lappin, M.R. 2022. Experimental infection of cats with Cryptosporidium felis. J. Feline Med. Surg. 24(10), 1060–1064. Shaw, H.J., Innes, E.A., Morrison, L.J., Katzer, F. and Wells, B. 2020. Long-term production effects of clinical cryptosporidiosis in neonatal calves. Int. J. Parasitol. 50(5), 371–376. Shehata, A.A., El-Emam, M.M.A., Gouda, H., El-Said, B.M., Salman, M.B. and Abd-Elfatah, E.B. 2024. Molecular characterization of Cryptosporidium parvum infections and analysis of hemological and biochemical changes in diarrheic pre-weaned calves in Egypt. Pak. Vet. J. 44(1), 135–140. Sheoran, A., Carvalho, A., Mimbela, R.P., South, A., Major, S., Ginese, M., Girouard, D. and Tzipori, S. 2022. Pregnant sows immunized with Cryptosporidium parvum significantly reduced infection in newborn piglets challenged with C. parvum but not with C. hominis. PLos Negl. Trop. Dis. 16(7), 10690. Shirley, D.A.T., Moonah, S.N. and Kotloff, K.L. 2012. Burden of disease from cryptosporidiosis. Curr. Opin. Infect. Dis. 25(5), 555–563. Silverlås, C. and Blanco-Penedo, I. 2013. Cryptosporidium spp. in calves and cows from organic and conventional dairy herds. Epidemiol. Infect. 141(3), 529–539. Sinyangwe, N.N., Siwila, J., Muma, J.B., Chola, M. and Michelo, C. 2020. Factors Associated With Cryptosporidium Infection Among Adult HIV Positive Population in Contact With Livestock in Namwala District, Zambia. Front. Public Health 8(1), 74. Šlapeta, J. 2013. Cryptosporidiosis and Cryptosporidium species in animals and humans: a thirty colour rainbow?. Int. J. Parasitol. 43(12–13), 957–970. Slavin, D. 1955. Cryptosporidium meleagridis (sp. nov.). J. Comp. Pathol. 65(3), 262–266. Smith, H.V., Nichols, R.A. and Grimason, A.M. 2005. Cryptosporidium excystation and invasion: getting to the guts of the matter. Trends Parasitol. 21(3), 133–142. Sparks, H., Nair, G., Castellanos-Gonzalez, A. and White, A.C. 2015. Treatment of Cryptosporidium: what We Know, Gaps, and the Way Forward. Curr. Trop. Med. Rep. 2(3), 181–187. Stott, R., May, E., Ramirez, E. and Warren, A. 2003. Predation of Cryptosporidium oocysts by protozoa and rotifers: implications for water quality and public health. Water Sci. Technol. 47(3), 77–83. Stoyanova, K. and Pavlov, S. 2019. Immunity and resistance to cryptosporidiosis: the intricate ways of an enigmatic parasitosis. Biomed. Rev. 30(1), 37–48. Suprihati, E., Puspitasari, H., Indasari, E.N., Galuh, P., Suwanti, L.T., Mufasirin, M., Hastutiek, P. and Setiawan, B. 2024. Molecular detection of Cryptosporidium spp. among wild rats in Surabaya, East Java, Indonesia. Open Vet. J. 14(10), 2687–2692. Tandel, J., English, E.D., Sateriale, A., Gullicksrud, J.A., Beiting, D.P., Sullivan, M.C., Pinkston, B., and Striepen, B. 2019. Life cycle progression and sexual development of the apicomplexan parasite Cryptosporidium parvum. Nat. Microbiol. 4(12), 2226–2236; doi:10.1038/s41564-019-0539-x Takahashi, K., Matsubayashi, M., Ohashi, Y., Naohara, J., Urakami, I., Sasai, K., Kido, Y., Kaneko, A. and Teramoto, I. 2020. Efficacy of ultraviolet light-emitting diodes (UV-LED) at four different peak wavelengths against Cryptosporidium parvum oocysts by inactivation assay using immunodeficient mice. Parasitol. Int. 77(1), 102–108. Tandel, J., English, E.D., Sateriale, A., Gullicksrud, J.A., Beiting, D.P., Sullivan, M.C., Pinkston, B. and Striepen, B. 2019. Life cycle progression and sexual development of the Apicomplexan parasite Cryptosporidium parvum. Nat. Microbiol. 4(12), 2226–2236. Theodos, C.M. 1998. Innate and cell-mediated immune responses to Cryptosporidium parvum. Adv. Parasitol. 40(1), 87–119. Tomley, F.M. and Soldati, D.S. 2001. Mix and match modules: structure and function of microneme proteins in Apicomplexan parasites. Trends Parasitol. 17(2), 81–88. Tosini, F., Drumo, R., Elwin, K., Chalmers, R.M., Pozio, E. and Cacciò, S.M. 2010. The CpA135 gene as a marker to identify Cryptosporidium species infecting humans. Parasitol. Int. 59(4), 606–609. Tyzzer, E.E. 1910. An extracellular Coccidium, Cryptosporidium muris (Gen. Et Sp. Nov.), of the gastric Glands of the Common Mouse. J. Med. Res. 23(3), 487–510. Tyzzer, E.E. 1912. Cryptosporidium parvum (sp. nov.), a coccidium found in the small intestine of the common mouse. Arch. Protistenkd. 26(1), 394–412. Tyzzer. 1907. A sporozoon found in the peptic glands of the common mouse. Proc. Soc. Exp. Biol. Med. 5(1), 12–13. Tzipori, S. and Ward, H. 2002. Cryptosporidiosis: biology, pathogenesis and disease. Microbes. Infect. 4(10), 1047–1058. Tzipori, S. and Widmer, G. 2008. A hundred-year retrospective on cryptosporidiosis. Trends Parasitol. 24(4), 184–189. Ulutaş, B. and Voyvoda, H. 2004. Cryptosporidiosis in Diarrhoeic Lambs on a Sheep Farm. Turkiye. Parazitol. Derg. 28(1), 15–17. Vanathy, K., Parija, S.C., Mandal, J., Hamide, A. and Krishnamurthy, S. 2017. Cryptosporidiosis: a mini review. Trop. Parasitol. 7(2), 72–80. Vermeulen, L.C., Benders, J., Medema, G. and Hofstra, N. 2017. Global Cryptosporidium Loads from Livestock Manure. Environ. Sci. Technol. 51(15), 8663–8671. Veshkini, A., Dengler, F., Bachmann, L., Liermann, W., Helm, C., Ulrich, R., Delling, C., Kühn, C. and Hammon, H.M. 2024. Cryptosporidium parvum infection alters the intestinal mucosa transcriptome in neonatal calves: implications for immune function. Front. Immunol. 15(1), 1351427. Wamsley, M., Wilson, R.T. and Murphy, H.M. 2025. The effects of rain and drought on incidence of enteric disease in Pennsylvania (2010-2019). Environ. Res. 267(1), 120641. Wang, W., Wei, Y., Cao, S., Wu, W., Zhao, W., Guo, Y., Xiao, L., Feng, Y. and Li, N. 2022. Divergent Cryptosporidium species and host-adapted Cryptosporidium canis subtypes in farmed minks, raccoon dogs and foxes in Shandong, China. Front. Cell. Infect. Microbiol. 12(1), 980917. Wanyiri, J. and Ward, H. 2006. Molecular basis of Cryptosporidium-host cell interactions: recent advances and future prospects. Future Microbiol. 1(2), 201–208. Wanyiri, J.W., O’Connor, R., Allison, G., Kim, K., Kane, A., Qiu, J., Plaut, A.G. and Ward, H.D. 2007. Proteolytic processing of the Cryptosporidium glycoprotein gp40/15 by human furin and by a parasite-derived furin-like protease activity. Infect. Immun. 75(1), 184–192. Widmer, G. and Lee, Y. 2010. Comparison of single- and multilocus genetic diversity in the protozoan parasites Cryptosporidium parvum and C. hominis. Appl. Environ. Microbiol. 76(19), 6639–6644. Widmer, G. and Sullivan, S. 2012. Genomics and population biology of Cryptosporidium species. Parasite Immunol. 34(2–3), 61–71. Wilmsmeyer, B., Dopfer, R., Hoppe, J.E. and Niethammer, D. 1993. Kryptosporidienenteritis Cryptosporidium enteritis. Monatsschr. Kinderheilkd. 141(2), 130–132. Winter, G., Gooley, A.A., Williams, K.L. and Slade, M.B. 2000. Characterization of a major sporozoite surface glycoprotein of Cryptosporidium parvum. Funct. Integr. Genomics 1(3), 207–217. Xiao, L., Cama, V.A., Cabrera, L., Ortega, Y., Pearson, J. and Gilman, R.H. 2007. Possible transmission of Cryptosporidium canis among children and a dog in a household. J. Clin. Microbiol. 45(6), 2014–2016. Xiao, L., Fayer, R., Ryan, U. and Upton, S.J. 2004. Cryptosporidium taxonomy: recent advances and implications for public health. Clin. Microbiol. Rev. 17(1), 72–97. Xu, P., Widmer, G., Wang, Y., Ozaki, L.S., Alves, J.M., Serrano, M.G., Puiu, D., Manque, P., Akiyoshi, D., Mackey, A.J., Pearson, W.R., Dear, P.H., Bankier, A.T., Peterson, D.L., Abrahamsen, M.S., Kapur, V., Tzipori, S. and Buck, G.A. 2004. The genome of Cryptosporidium hominis. Nature 431(7012), 1107–1112. Xu, R., Beatty, W.L., Greigert, V., Witola, W.H. and Sibley, L.D. 2024. Multiple pathways for glucose phosphate transport and utilization support growth of Cryptosporidium parvum. Nat. Commun. 15(1), 380. Yang, X.Y., Gong, Q.L., Zhao, B., Cai, Y.N. and Zhao, Q. 2021. Prevalence of Cryptosporidium Infection in Sheep and Goat Flocks in China During 2010-2019: a Systematic Review and Meta-Analysis. Vector Borne Zoonotic Dis. 21(9), 692–706. Yassi, A., Bryce, E.A., Breilh, J., Lavoie, M.C., Ndelu, L., Lockhart, K. and Spiegel, J. 2011. Collaboration between infection control and occupational health in three continents: a success story with international impact. BMC Int. Health Hum. Rights 11(Suppl 2), S8. Zaheer, T., Imran, M., Abbas, R.Z., Zaheer, I. and Malik, M.A. 2021. Avian cryptosporidiosis and its zoonotic significance in Asia. World’s Poult. Sci. J. 77(1), 55–70. Zhang, K., Fu, Y., Li, J. and Zhang, L. 2022. Public health and ecological significance of rodents in Cryptosporidium infections. One Health 14(1), 100364. | ||
| How to Cite this Article |
| Pubmed Style Widodo WT, Khairullah AR, Pratama BP, Ambarika R, Furqoni AH, Kristianto S, Wardhani BWK, Widoretno W, Khariri K, Hermawati L, Suri AA, Moses IB, Putri AK, Kurniasih DAA, Maha MS, Khalisa AT, Ahmad RZ, Wibowo S. Cryptosporidiosis: A global threat to human and animal health. Open Vet. J.. 2025; 15(10): 4814-4833. doi:10.5455/OVJ.2025.v15.i10.3 Web Style Widodo WT, Khairullah AR, Pratama BP, Ambarika R, Furqoni AH, Kristianto S, Wardhani BWK, Widoretno W, Khariri K, Hermawati L, Suri AA, Moses IB, Putri AK, Kurniasih DAA, Maha MS, Khalisa AT, Ahmad RZ, Wibowo S. Cryptosporidiosis: A global threat to human and animal health. https://www.openveterinaryjournal.com/?mno=259470 [Access: January 25, 2026]. doi:10.5455/OVJ.2025.v15.i10.3 AMA (American Medical Association) Style Widodo WT, Khairullah AR, Pratama BP, Ambarika R, Furqoni AH, Kristianto S, Wardhani BWK, Widoretno W, Khariri K, Hermawati L, Suri AA, Moses IB, Putri AK, Kurniasih DAA, Maha MS, Khalisa AT, Ahmad RZ, Wibowo S. Cryptosporidiosis: A global threat to human and animal health. Open Vet. J.. 2025; 15(10): 4814-4833. doi:10.5455/OVJ.2025.v15.i10.3 Vancouver/ICMJE Style Widodo WT, Khairullah AR, Pratama BP, Ambarika R, Furqoni AH, Kristianto S, Wardhani BWK, Widoretno W, Khariri K, Hermawati L, Suri AA, Moses IB, Putri AK, Kurniasih DAA, Maha MS, Khalisa AT, Ahmad RZ, Wibowo S. Cryptosporidiosis: A global threat to human and animal health. Open Vet. J.. (2025), [cited January 25, 2026]; 15(10): 4814-4833. doi:10.5455/OVJ.2025.v15.i10.3 Harvard Style Widodo, W. T., Khairullah, . A. R., Pratama, . B. P., Ambarika, . R., Furqoni, . A. H., Kristianto, . S., Wardhani, . B. W. K., Widoretno, . W., Khariri, . K., Hermawati, . L., Suri, . A. A., Moses, . I. B., Putri, . A. K., Kurniasih, . D. A. A., Maha, . M. S., Khalisa, . A. T., Ahmad, . R. Z. & Wibowo, . S. (2025) Cryptosporidiosis: A global threat to human and animal health. Open Vet. J., 15 (10), 4814-4833. doi:10.5455/OVJ.2025.v15.i10.3 Turabian Style Widodo, Wimbuh Tri, Aswin Rafif Khairullah, Bima Putra Pratama, Rahmania Ambarika, Abdul Hadi Furqoni, Sonny Kristianto, Bantari Wisynu Kusuma Wardhani, Widoretno Widoretno, Khariri Khariri, Luluk Hermawati, Auliyani Andam Suri, Ikechukwu Benjamin Moses, Alifiani Kartika Putri, Dea Anita Ariani Kurniasih, Masri Sembiring Maha, Andi Thafida Khalisa, Riza Zainuddin Ahmad, and Syahputra Wibowo. 2025. Cryptosporidiosis: A global threat to human and animal health. Open Veterinary Journal, 15 (10), 4814-4833. doi:10.5455/OVJ.2025.v15.i10.3 Chicago Style Widodo, Wimbuh Tri, Aswin Rafif Khairullah, Bima Putra Pratama, Rahmania Ambarika, Abdul Hadi Furqoni, Sonny Kristianto, Bantari Wisynu Kusuma Wardhani, Widoretno Widoretno, Khariri Khariri, Luluk Hermawati, Auliyani Andam Suri, Ikechukwu Benjamin Moses, Alifiani Kartika Putri, Dea Anita Ariani Kurniasih, Masri Sembiring Maha, Andi Thafida Khalisa, Riza Zainuddin Ahmad, and Syahputra Wibowo. "Cryptosporidiosis: A global threat to human and animal health." Open Veterinary Journal 15 (2025), 4814-4833. doi:10.5455/OVJ.2025.v15.i10.3 MLA (The Modern Language Association) Style Widodo, Wimbuh Tri, Aswin Rafif Khairullah, Bima Putra Pratama, Rahmania Ambarika, Abdul Hadi Furqoni, Sonny Kristianto, Bantari Wisynu Kusuma Wardhani, Widoretno Widoretno, Khariri Khariri, Luluk Hermawati, Auliyani Andam Suri, Ikechukwu Benjamin Moses, Alifiani Kartika Putri, Dea Anita Ariani Kurniasih, Masri Sembiring Maha, Andi Thafida Khalisa, Riza Zainuddin Ahmad, and Syahputra Wibowo. "Cryptosporidiosis: A global threat to human and animal health." Open Veterinary Journal 15.10 (2025), 4814-4833. Print. doi:10.5455/OVJ.2025.v15.i10.3 APA (American Psychological Association) Style Widodo, W. T., Khairullah, . A. R., Pratama, . B. P., Ambarika, . R., Furqoni, . A. H., Kristianto, . S., Wardhani, . B. W. K., Widoretno, . W., Khariri, . K., Hermawati, . L., Suri, . A. A., Moses, . I. B., Putri, . A. K., Kurniasih, . D. A. A., Maha, . M. S., Khalisa, . A. T., Ahmad, . R. Z. & Wibowo, . S. (2025) Cryptosporidiosis: A global threat to human and animal health. Open Veterinary Journal, 15 (10), 4814-4833. doi:10.5455/OVJ.2025.v15.i10.3 |