| Original Article | ||
Open Vet J. 2020; 10(3): 331-339 doi: 10.4314/ovj.v10i3.12 Open Veterinary Journal, (2020), Vol. 10(3): 331–339 Original Research http://dx.doi.org/10.4314/ovj.v10i3.12 Association of BoLA DRB3 gene polymorphisms with BoHV-1 infection and zootechnical traitsJuan Pablo Arismendy Morales, Albeiro López-Herrera* and Julián Echeverri ZuluagaBiodiversity and Molecular Genetics Research Group “BIOGEM”, Animal Production Department, Universidad Nacional de Colombia, Medellín, Colombia *Corresponding Author: Albeiro López-Herrera. BIOGEM, Animal Production Department, Universidad Nacional de Colombia, Medellín, Colombia. Email: alherrera [at] unal.edu.co Submitted: 19/03/2020 Accepted: 20/08/2020 Published: 25/09/2020 © 2020 Open Veterinary Journal
AbstractBackground: The dairy sector is one of the leading in agricultural production sectors in the world and the bovine herpesvirus 1 (BoHV-1) is an important pathogen that causes great losses in most production systems. Moreover, BoLA DRB3 immunological gene presents different alleles related to protection against many pathogens. Methods: Serological diagnosis was carried out to determine the BoHV-1 infection and through PCR-RFLP 506 Holstein cows from several municipalities of Antioquia were genotyped for BoLA DRB3.2 gene polymorphisms. Results: Alleles 8, 16, 22, and 24 were the most common out of the 42 alleles found. By indirect ELISA technique, a 58.7% prevalence of BoHV-1 infection in this population was diagnosed and Odd ratios for found alleles were calculated by logistic regression; the only significant association was held for allele 37, which showed that it effects confers susceptibility to infection. On the other hand, by using generalized linear models, a significant association between BoLA DRB3.2 gene and milk and fat yield in primiparous and services per conception in multiparous was found, with the most favorable alleles being 11 and 28 in primiparous and 22 and 28 in multiparous; allele 37 was unfavorable only in primiparous. Conclusion: BoLA DRB3.2 gene polymorphisms have shown high variability and significant effects on Holstein cattle and their performance in production systems in Antioquia are at both sanitary or health and productive levels. Keywords: Holstein, Immunity, Prevalence, Production, Reproduction. IntroductionSpecialized dairy is one of the main lines of the agricultural sector, and milk is the main livestock product in most countries. In Colombia, dairy represents 6.4% of the orientation of the herd and the most commonly used breed for specialized dairy is Holstein cows. For many years, the Colombian government has collected information and statistics about productive, reproductive, and milk quality traits of Holstein cows (Jaramillo and Areiza, 2012). Productive and reproductive strategies to optimize system performance are supported by selection, which in recent years has been assisted by the use of molecular markers (Singh et al., 2014). Some of these markers are related to the modulation of immune responses and can affect all aspects of an animal, as production and reproduction. One of these genes is the BoLA DRB3.2, part of the major histocompatibility complex, which has more than 100 polymorphic sites only in exon 2 (Amills et al., 1998). This gene has been widely related to immunological traits (Dietz et al., 1997), resistance or susceptibility to various diseases (Keirn et al., 2001), and production traits and milk quality (Davies et al., 1992; Nascimento et al., 2006; Zambrano et al., 2014). Some diseases affect especially the dairy systems, and bovine herpesvirus 1 (BoHV-1) is an important pathogen that causes great losses in most production systems. The symptoms may be mild and localized or include severe generalized diseases, leading eventually to death. BoHV-1infects the host by viremia and affects different tissues and organs like neuronal cells. In most cases, the virus may replicate inside the neuronal cells and cause neuronal diseases (encephalitis) (Engels and Ackermann, 1996). Due the importance and great effect of this virus over cattle yield, it is necessary to keep researching about it, especially on Holstein cattle population in Antioquia, which is the most important dairy region in Colombia. The aim of this work was to associate BoLA DRB3.2 gene polymorphisms with productive and reproductive traits of Holstein cattle from Antioquia and with BoHV-1 infection, to establish alleles that provide a resistance or susceptibility effect. Material and MethodsPlace and experimental unitsBlood samples from 506 Holstein cows of Belmira, San Pedro de los Milagros, and Entrerríos, municipalities of Antioquia, and the small town of San Felix, in the municipality of Bello, which have not been vaccinated against BoHV-1 were taken. These cows belong to the project “Genetic Evaluation Program and Dairy Control” of the BIOGEM group of the Universidad Nacional de Colombia, Medellín Campus, and Colanta LTDA. The blood samples were taken by coccygeal vein puncture and were collected in BD vacutainer tubes with purple system (EDTA). They were kept in the refrigerator until processing (DNA isolation) in the Laboratory of Basic Animal Science at the Universidad Nacional Colombia, Medellin Campus. Serological diagnosis of infection with BoHV-1Blood plasma was obtained by centrifugation of fresh blood and stored at –80°C until its use in the serological diagnosis of infection with BoHV-1. INgezim IBR R.12.IBR.K1 commercial ELISA kit of INGENASA was used without any modification of the reagents and protocols specified by the manufacturer. Only 450 animals out of the 506 sampled could be analyzed, but it was managed that the samples of all municipalities remained proportionally represented before excluding the 50 animals. The reading of the plates at 450 nm was carried out with a spectrophotometer EL × 800 (BioTek Instruments Inc., Winooski, VT) using Gen5 software (BioTek Instruments Inc., Winooski, VT). DNA isolation and genotyping by PCR-RFLPDNA extraction was carried out by using the salting out procedure (Miller et al., 1988). For semi-nested PCR amplification and genotyping by RFLP, primers and protocols described by van Eijk et al. (1992) for the amplification of a 284 bp final fragment of exon 2 of the BoLA DRB3 gene were used. Primers for the first PCR were HL030: 5′-ATCCTCTCTCTGCAGCACATTTCC-3′ and HL031: 5′-TTTAAATTFYFYCTCACCTFYCFYCT-3′ and it was carried out in a reaction volume of 25 μl which contained 0.4 μM of each dNTP, 0.5 μM of each primer, 2.5 μl of 10× buffer, 2.5 μl of 25 mM MgCl2, ultrapure water, 1U Taq polymerase, and 100–200 ng of genomic DNA. Amplification was conducted with the following thermocycling program: initial denaturation at 94°C for 240 seconds, followed by 35 cycles of denaturation at 94°C for 60 seconds, alignment at 60°C for 120 seconds, extension at 72°C for 60 seconds, and a final extension at 72°C for 300 seconds. Primers for the second PCR were HL030 and HL032: 5′-TFYC FYCTGCACAGTGAAACTCTC-3′ and it was carried out in a reaction volume of 40 μl containing 0.4 mM of each dNTP, 0.6 uM of each primer, 4 μl of 10× buffer, 4 μl of 25 mM MgCl2, ultrapure water, 1U Taq polymerase, and 2.5 μl of first PCR amplicon. The amplification was carried out with the following thermocycling program: initial denaturation at 94°C for 240 seconds, followed by 35 cycles of denaturation at 94°C for 60 seconds, alignment at 67°C for 120 seconds, extension at 72°C for 60 seconds, followed by a final extension of 72°C for 300 seconds. Digestion with restriction enzymes RsaI, BstYI, and HaeIII was conducted in a final volume of 20 μl for each enzyme, which contained 10 μl of the product of the second PCR, 5 U of enzyme and 2 μl of enzyme buffer. Reactions with RsaI and HaeIII were incubated at 37°C and with 6 BstYI at 60ºC for at least 4 hours and visualized on 4% agarose gels stained with EZ-Vision. The strips were read and interpreted according to the nomenclature reported by van Eijk et al. (1992). Odd ratios (OR) calculationTo establish the association between alleles, genotypes, susceptibility, neutrality, or resistance to infection with BoHV-1, odds ratios were calculated using a logistic regression model. If the OR is greater than 1, the relationship of allele or genotype is resistant and there is low risk of being positive for BoHV-1, but if it is less than 1, the relationship is susceptible and there is high risk of being positive for BoHV-1; neutrality is considered when the OR is equal to 1 or when it is not significant. Fisher’s exact test was used to determine the significance of OR (p < 0.05) and, as an additional test, Chi-square (χ2) Mantel–Haenszel test to determine general association between alleles and infection with BoHV-1 was conducted. OR calculations were executed in SAS® software version 9.2 for Windows (SAS Institute Inc, Cary NC) and significance calculations were executed in StatCalc 3.0 (Professor K. Krishnamoorthy, Dept of Mathematics, UL Lafayette, LA, software Lafayette, LA). Evaluated productive and reproductive traitsAll information of the animals was compiled by the “Genetic Evaluation and Dairy Control Program” of the BIOGEM group of the Universidad Nacional de Colombia, Medellin Campus, and Colanta LTDA for 5 years from all lactating animals. The traits evaluated were actual milk yield per lactation (AMYL) in kg, percentages of protein and fat in milk (PP and FP), and protein and fat yield in milk (PY and FY) in kg, calculated with the percentage of each one and the AMYL, calving interval (CI), services per conception (SXC), and somatic cell score (SCS), calculated by logarithmic transformation of somatic cell count in milk (SCC) with the following equation: SCS=(Log2(SCC/100)) + 3. Data were adjusted by eliminating extreme values that could alter the performance of the model; lactations under 250 days and over 450 days, and AMYL under 2,500 kg and over 12,000 kg were removed. Association of BoLA DRB3 gene with productive and reproductive traitsA descriptive analysis of evaluated traits was made and frequency tables were used to characterize the averages of the traits according to the alleles. In order to make the association, primiparous (1 and 2 calving) and multiparous (3–8 calving) were analyzed separately and a generalized linear model was used; the effect of replacing the allele 24 was evaluated through Dunnett’s test. All calculations were made with the SAS® software version 9.2 for Windows (SAS Institute Inc, Cary NC). The model used was: Where:
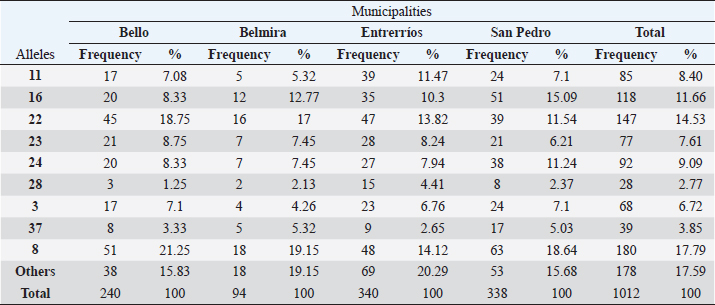
For AMYL trait, was not used. Ethical approval and consent to participateThis study was derived from the project “Efecto de la nutrición sobre la asociación de los polimorfismos de 4 genes (bGH, prolactina, STAT5 e IGF2A) con características productivas y reproductivas en ganado Holstein del departamento de Antioquia”, funded by Colciencias, code No. 111856935121; this project was approved by the Ethics Committee of Agricultural Sciences Faculty of Universidad Nacional de Colombia, Medellín Campus. ResultsGenotyping by PCR-RFLPForty-two alleles were found in the total population (506 animals), with BoLA DRB3.2 *8 (17.79%), *22 (14.53%), *16 (11.66%), and *24 (9.09%) being the most frequent alleles. Alleles that had frequencies lower than 2% were 32 out of the 42 found and were grouped in the category “Other” (*1, *2, *6, *9, *10, *12, *13, *14, *18, *19, *20, *21, *25, *26, *27, *32, *34, *35, *36, *47, *48, *49, *51, *iaa, *gbb, *ibb, *ibe, *kaa, *kha, *dbb, *fdb, *fbb). In Table 1, the allele frequencies of the entire population (506 animals) and by municipality are shown. Allele 8 was the most common in all municipalities, followed by allele 22 in Bello (18.75%), Belmira (17%), and Entrerrios (13.82%), and allele 16 in San Pedro de los Milagros (15.09%). Table 1. Allelic frequencies of BoLA DRB3.2 gene for the total population (506 animals) per municipality.
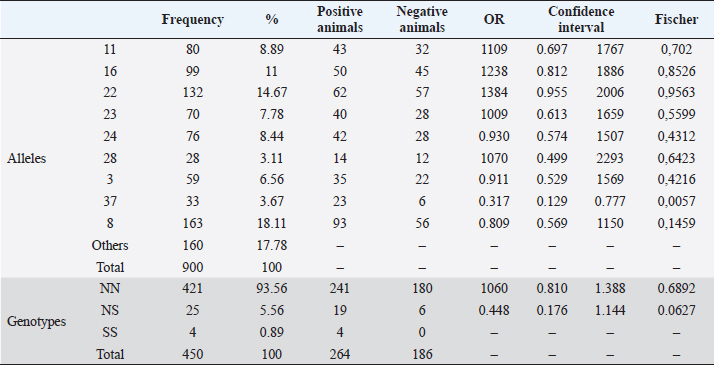
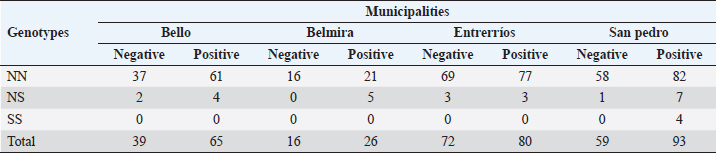
Serological diagnostics and ORIndirect ELISA tests showed a 58.7% prevalence of infection with BoHV-1, having 264 positive animals out of the 450 tested. Chi-square test to establish a relationship between infection with BoHV-1 and the place of origin of the sample (municipality) was 3.49, with p-value=0.32, indicating that in this study there was no association between the municipality and the infection. In Table 2, the OR of each allele and genotype and their descriptive information are shown. Chi-square test showed that there is a significant relationship between DRB3.2 BoLA gene alleles and infection with BoHV-1 (χ2=4.27, p=0.039). By analyzing the relationship of each allele, it was found that the only allele that showed effect on infection was 37 (OR=0.317, p=0.0057), which was considered to be susceptible; all other alleles that showed no significant effect were considered neutral. Twenty nine out of the 450 diagnosed animals had allele 37, and 23 out of 264 animals positive with BoHV-1 had this allele (79.31%), with only 6 negative animals. Table 2 also shows the relationship between genotypes (according to the allele classification made by its OR) and infection with BoHV-1. Alleles of neutrality are denoted by N and susceptibility alleles by S, no resistance (R) alleles were found. As expected, the NN genotype had no significant effect, but NS genotype had an OR <1 (0.448) due to susceptibility allele presence, but the confidence interval contains values above 1 (0.176–1.144). Although the effect of NS genotype was not significant (p=0.0627, p > 0.05), it shows a tendency to be, considering other studies where alleles are considered to have a significant effect with p < 0.1; these results may indicate that the susceptibility allele has no dominance on the allele of neutrality, which does not make the risk of infection increase significantly. Since only 4 animals were found for SS genotype and none of them was negative to infection, it was not possible to properly calculate the OR, since this would be 0 (maximum risk) and the confidence interval would be between 0 and +∞; however, it is clear that there is a relationship of susceptibility to infection with this genotype. Studies with a larger sample number are needed, in which more animals with this genotype are detected and calculating the OR is more reliable. Due to positive or negative animals that have at least one out of the evaluated alleles counted, this might cause each animal to be repeated in different alleles and therefore it is invalid to emit a total count. As shown in Table 1, susceptibility allele 37 was more common in Belmira (5.32%) and San Pedro de los Milagros (5.03%); in parallel, it is in these two municipalities where the prevalence of positive is greater on total animals with this allele, with 100% (5/5) in Belmira and 91.7% (11/12) in San Pedro de los Milagros (Table 3). Table 2. Association between BoLA DRB3 alleles, genotypes, and infection with BoHV-1
Table 3. Distribution of negative and positive BoHV-1 by genotype and by municipality.
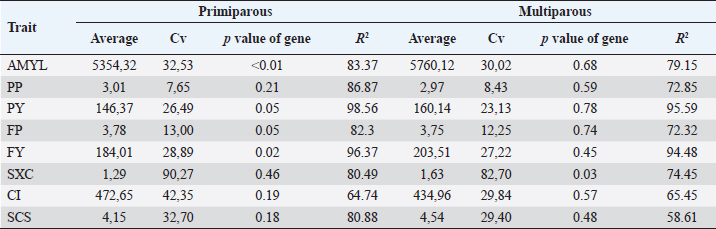
Association with production traitsSignificant association (p < 0.05) of BoLA DRB3.2 gene with milk yield and fat yield in primiparous cows (1 and 2 calving) was found; for protein yield and fat percentage, the p value of the gene in the model was 0.05, which for our study does not indicate an association but a very strong trend to association between the gene and both traits (Table 4). In multiparous cows, the effect of the gene was only significant on SXC. Multiparous cows, compared to primiparous, presented favorable averages in milk yield, protein, and fat yield due to high milk yield and not to percentages, and CI, are unfavorable in SXC and SCS. The traits that presented less variation were protein and fat percentage; and the highest variation was for SXC, which ranged from 1 to 9. The coefficients of determination (R2), in both models primiparous and multiparous cows, were high, with the lowest being 58.61% and the highest above 90%; this indicates that the used models and included variables explain most of the evaluated traits’ variation. Table 4. Descriptive data by calving and p value of BoLA DRB3.2 gene in the model.
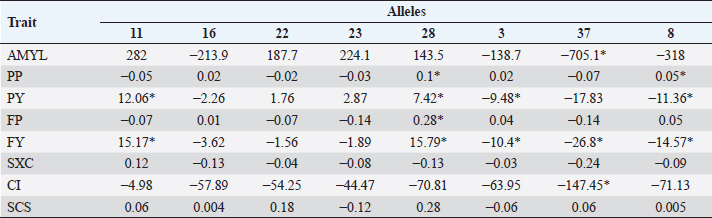
Table 5. Average difference between alleles and allele 24 (substitution allele) in primiparous cows. Differences marked * are significant (p < 0.05).
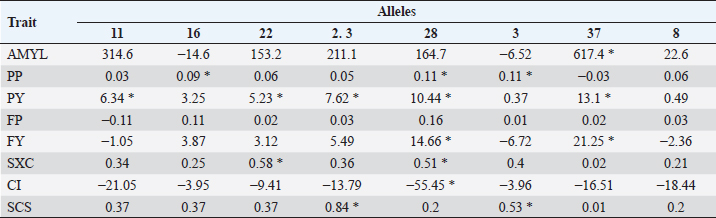
Table 6. Average difference between alleles and allele 24 (substitution allele) in multiparous cows. Differences marked * are significant (p < 0.05).
In Tables 5 and 6, the difference between the averages of each trait by allele and the average of each trait of BoLA DRB3.2 allele *24, which was used as substitution allele, is shown. In primiparous cows, alleles with a significant difference to allele *24 according to Dunnett’s test for one or more traits were *11, *28, *3, *37, and *8. For traits in which the gene had a significant effect on (AMYL and FY), allele *37 was most unfavorable, along with allele *8 (for FY only); for FY, alleles *11 and *28 were favorable. Allele *11 was also favorable for PY, *28 was favorable for PP, PY, and FP, *37 for CI, and *8 for PP; on the other hand, allele *3 was unfavorable for PY and *8 for PY. In multiparous cows, alleles *22 and *28 had a significant favorable effect on SxC (trait on which the gene had a significant effect). In contrast, allele *37 was favorable on the AMYL and FY, and for most traits, contrary to what happened in primiparous cows, in which it had mostly unfavorable effects. For PP, alleles *16, *28, and *3 were favorable; for PY, alleles *11, *22, *23, *28, and *37 were favorable; for FY, allele *28 was also favorable, but unfavorable for CI, and for SCS, alleles *23 and *3 were favorable. Most variables were significant in almost all used models, except days of lactation for SCS (in primiparous cows), FP and CI (in multiparous cows), and AMYL for SxC and CI (in both groups of cows). DiscussionMany studies around the world have genotyped cattle of several breeds for BoLA DRB3.2 polymorphisms, especially Holstein cattle, which have been highly selected for zootechnical traits. Duangjinda et al. (2009) also found in Holstein × Zebu cattle, in addition to alleles 16, 23, 11, and 8, that alleles 52 and 1 were also the most frequent. Gilliespie et al. (1999) found that in Jersey cows, the most frequent alleles are not the same as in the Holstein population, with allele 8 being the only common and most frequent in both breeds. Juliarena et al. (2009) reported alleles 24, 16, and 22 as the most common in the Argentinean Holstein population, along with alleles 8 and 11. Kelm et al. (1997) reported, similar to the above, that the most frequent alleles are 8, 11, 23, 24, 22, and 16 in the US (Iowa) Holstein population. The Canadian Holsteins also presented alleles 8, 22, 24, and 16 as the most common (Rupp et al., 2007). Contrasting to this, populations of Japanese black cattle have allele 10 as the most common, and in Japanese Holstein cattle, alleles 16 and 24 are presented as the most common, revealing that allele frequency depends more on the breed than on the region of origin (Miyasaka et al., 2011). Takeshima et al. (2014) found that in Japanese black cattle the most common allele was 3, while in Holstein and Japanese Shorthorn cattle alleles 22 and 8 were more common, respectively. Starkenburg et al. (1997) found that alleles 22, 23, and 24 were the most common in selected production lines, compared to control lines; these results suggest a relationship of these alleles with performance traits, for which a traditional selection has been made for many years in most dairy systems, which would explain largely why these alleles are so prevalent in Holstein cattle around world. Hernández et al. (2013) found that in Harton del Valle creole breed, the most common allele is 22. Zambrano et al. (2011) also reported alleles 23, 22, 24, 16, 8, and 37 as the most common in a Holstein population of Santa Elena, Medellin, representing more than 50% of the cumulative allele frequency. In the same study, they also found high variability in BON populations and their crossbreeding with Holstein. In several municipalities in northern and other subregions of Antioquia, Ramirez et al. (2014) found that the most common alleles are 8, 24, and 22, with alleles 11 and 16 having greater than 2% prevalence. The serological prevalence of BoHV-1 infection found in this study is similar to other prevalences found in Antioquia and other regions, particularly to areas of the Magdalena; Piedrahita et al. (2010) reported 55.5% prevalences in the middle Magdalena. In Antioquia, BoHV-1 infection has been studied in several towns and has reported prevalences ranging from 85.51% to 4.3% (Ruiz-Sáenz et al., 2010). Several studies have linked BoLA DRB3.2 gene polymorphisms with various animal diseases and alleles that confer resistance and susceptibility to these conditions have been identified. Immunological traits such as lymphocyte response to mitogens, antibody titers, leukocyte count, and neutrophil function have been highly associated with different BoLA DRB3.2 gene alleles (Dietz et al., 1997). Zambrano et al. (2011) found in Holsteins and Holstein x BON cattle that allele 33 offered resistance to subclinical mastitis and allele 8 was associated with susceptibility to it, finding no associations between alleles and clinical mastitis. Ramirez et al. (2014) associated alleles 23 and mbb with increased occurrence of subclinical mastitis and infection with coagulase – negative Staphylococci; they also related alleles jba and 15 with resistance to these last pathogens; this work also showed that allele 23 is related to increased mastitis since it increases the SCS. Although BoLA DRB3.2 gene polymorphisms have not been associated with herpesvirus, they have been associated with another set of viruses with zootechnical importance, particularly with bovine leukemia virus. Juliarena et al. (2009) evaluated BLV viral load with DRB3 alleles and found that alleles 11 and 12 offered resistance (low viral load) and allele 16 generated susceptibility (a high viral load); resistant/susceptible genotypes could not be identified with any trend or particular profile. Concerned about the relationship between low viral load of BLV (resistance) and low antibody titers to BLV, this same group evaluated whether the alleles of resistance to BLV affected the response to other pathogens such as foot-and-mouth bovine virus, virus-linked diarrhea, and BoHV-1, finding no association between these polymorphisms and antibody titers against these pathogens; so, the selection to resistance to BLV does not affect resistance or susceptibility to other evaluated viruses. In general, DRB3.2 gene has been postulated as a great candidate for molecular selection and research on vaccines against viruses such as foot-and-mouth bovine virus, bovine respiratory syncytial virus, and the ones that cause bovine respiratory disease complex (like BoHV-1), given its important role in immune response levels (Glass et al., 2012). Regarding to the association between BoLA DRB3.2 and traits, the different and contrasting effects of the alleles on the traits are due to the different physiological, nutritional, management requirements, uses, and aims of primiparous and multiparous cows. Primiparous cows are in a transition state in which their bodies are adapting to high productive demands that will have the rest of their life in the herd; so, they are particularly sensitive to all factors that influence performance (Wathes et al., 2007). Management and health are some of the factors this will be reflected in production, wherein immune response is essential for the defense of the body and for sustaining the health of the animal (Mehrzad et al., 2001, 2009). It is expected, therefore, that the effect of immunity genes such as BoLA DRB3 on the productive traits is important (significant or high tendency to significance). In the case of allele *37, for example, it can be observed that it considerably reduces the production in primiparous cows, but, as a result of less physiological wear, reproductive processes are facilitated, which is reflected in a considerable reduction of CI. By contrast, in multiparous cows, which have reached or are close to reaching peak production and have already adapted to the production demand of the herd, the reproductive aspect is more sensitive because their reproductive system is under great challenges of performance and efficiency, which only can be accomplish with proper nutrition and reproductive management, especially in heat detection and voluntary waiting period (Wittrock et al., 2011). That is why immunity genes become important in proper recovery and operation of that system; this will be reflected in traits such as SxC. On the other hand, it is known that cows with more lactation usually have higher values of SCS due to wear and sensitivity of the mammary gland (Rodriguez-Zas et al., 2000). Genes that have a favorable or unfavorable effect in primiparous cows may have opposite effects on multiparous cows due to management, demands, and levels of gene expression of their own condition. Relationships between BoLA DRB3.2 gene and animal diseases have been established by other researchers and its effect is also known for various production traits, particularly those related to health quality of milk, as the SCS. Dietz et al. (1997) found that allele *16 of BoLA DRB3 gene is a potential risk factor for high levels of SCS, and *22 decreases it in second lactation. Kelm et al. in (1997) also significantly associated allele *16 with higher levels of SCS; they also associated allele *8 with clinical mastitis, alleles *11 and *23 with lower levels of clinical mastitis, and alleles *3 and *24 with IMI (intramammary infection). This study also recorded allele *3 as an increase factor for SCS. Nascimento et al. (2006) reported an association between decreased protein and fat yield in milk and allele *54, as well as association between decreased milk protein with allele *6, and increase milk protein with allele *7. A trend of increased association between SCS and alleles *3 and *31 was also elucidated. Rupp et al. (2007), contrary to the results of Nascimento et al. (2006) and of this work, reported that allele *3 of DRB3 is associated with low SCS and high clinical mastitis and antibody production. They also found an association between allele *11 with low levels of SCS, *22 and *23 with high levels of SCS, and *8 with high risk of mastitis. Antagonistically, they found that alleles *11 and *23, being the first associated with low levels of SCS and the second with high levels of SCS, were also associated with increased production. Also antagonistically, they found that alleles associated with an increased humoral response reduced cellular response and vice versa. Kulberg et al. (2007) related alleles *3, *9, *11, and *26 with high yields of milk protein, and alleles *22 and *26 were associated with increased clinical mastitis. Starkenburg et al. (1997) found contradictory results; although they found that allele *8 increases acute SCS, they also reported decreases in second lactation mastitis. They reported as well, that allele *16 decreases acute and chronic SCS, that allele *22 increases chronic SCS in first lactation, allele * 7 increases milk yield and udder health problems (but strangely in production systems milk has low frequency), allele *24 increases udder health problems, and allele *26 decreases milk yields. In Colombia, Zambrano et al. (2014), analyzing DRB3 polymorphisms and productive traits of the same type of livestock analyzed for mastitis, determined that allele *36 decreased milk yield adjusted to 305 days, but increased percentages of fat and protein in multiparous and primiparous cows, respectively; likewise, they determined that allele *33 increased percentage of protein and allele *19 increased percentage of fat but only in BON x Holstein cows. In the Holstein cattle population studied, the most common BoLA DRB3.2 alleles were *8, *22, *16, and *24; also, a 58.7% BoHV-1 prevalence was detected. The only allele statistically associated with the BoHV-1 infection was *37, which had a susceptibility effect. Broadly, the most favorable allele, in both primiparous and multiparous cows, is *28, and the less favorable is *37. However, larger studies are needed to continue determining the relationship between these polymorphisms and diseases, and productive and reproductive traits in specialized dairy populations, which are always subject to selection processes that optimize certain traits but hurt others. AcknowledgmentsThe authors are grateful to the Laboratory of Basic Animal Science and the Laboratory of Animal Biotechnology, from the Universidad Nacional de Colombia, Medellín Campus, for providing spaces and services in the development of this work; to the project “Genetic Evaluation and Dairy Control Program” by Biodiversity and Molecular Genetics Research Group “BIOGEM” of the Universidad Nacional de Colombia, Medellin Campus, and Colanta LTDA; and to all producers for their cooperation and provided information. Conflicts of interestThe authors declare that there is no conflict of interest. Authors’ contributionsJPAM carried out all the field and laboratory experiments and analyzed and interpreted the data. ALH guided and supervised all processes, both experimental and analytical, and writing of the manuscript. JEZ was a major contributor in the design and statistical analysis of the data. All authors read and approved the final manuscript. ReferencesAmills, M., Ramiya, V., Nonmine, J. and Lewin, H. 1998. The major histocompatibility complex. Rev. Sci. Tech. Off. Int. Epiz. 17, 108–120. Davies, C.J., Andersson, L., Joosten, I., Mariani, P., Gasbarre, L.C. and Hensen, E.J. 1992. Characterization of bovine MHC class II polymorphism using three typing methods: serology, RFLP and IEF. Int. J. Immuno. 19(5), 253–262. Dietz, A.B., Detilleux, J.C., Freeman, A.E., Kelley, D.H., Stabel, J.R. and Kehrli Jr,M.E. 1997. Genetic association of bovine lymphocyte antigen DRB3 alleles with immunological traits of Holstein cattle. J. Dairy Sci. 80(2), 400–405. Duangjinda, M., Buayai, D., Pattarajinda, V., Phasuk, Y., Katawatin, S., Vongpralub, T. and Chaiyotvittayakul, A. 2009. Detection of bovine leukocyte antigen DRB3 alleles as candidate markers for clinical mastitis resistance in Holstein× Zebu. J. Anim. Sci. 87(2), 469–476. Engels, M. and Ackermann, M. 1996. Pathogenesis of ruminant herpesvirus infections. Vet. Microbiol. 53(1–2), 3–15. Gilliespie, B.E., Jayarao, B.M., Dowlen, H.H. and Oliver, S.P. 1999. Analysis and frequency of bovine lymphocyte antigen DRB3. 2 alleles in Jersey cows. J. Dairy Sci. 82(9), 2049–2053. Glass, E.J., Baxter, R., Leach, R.J. and Jann, O.C. 2012. Genes controlling vaccine responses and disease resistance to respiratory viral pathogens in cattle. Vet. Immunol. Immunopathol. 148(1–2), 90–99. Hernández, D., Posso, A., Muñoz, J., Giovambattista, G. and Álvarez, L. 2013. Polimorfismos del gen BoLA-DRB3. 2* en ganado criollo colombiano. Cordoba, Colombia: Revista MVZ Córdoba vol. 18, pp: 3665–3671. Jaramillo, A. and Areiza, A. 2012. Análisis del mercado de la leche y derivados lácteos en Colombia (2008-2012). Superintendencia de Industria y Comercio, En: Superintendencia de Industria y Comercio, Available via http://www.sic.gov.co/drupal/recursos_user/documentos/promocion_competencia/Estudios_Economicos/Estudios_Economicos/Estudio_Sectorial_Leche1.pdf. Juliarena, M.A., Poli, M., Ceriani, C., Sala, L., Rodriguez, E., Gutierrez, S. and Esteban, E.N. 2009. Antibody response against three widespread bovine viruses is not impaired in Holstein cattle carrying bovine leukocyte antigen DRB3. 2 alleles associated with bovine leukemia virus resistance. J. Dairy Sci. 92(1), 375–381. Keirn, S.C., Freeman, A.E. and Kehrli Jr,M.E. 2001. Genetic control of disease resistance and immunoresponsiveness. Vet. Clin. North Am. Food Anim. Pract. 17(3), 477–493. Kelm, S.C., Dettilleux, J.C., Freeman, A.E., Kehrli Jr,M.E., Dietz, A.B., Fox, L.K. and Kelley, D.H. 1997. Genetic association between parameters of innate immunity and measures of mastitis in periparturient Holstein cattle. J. Dairy Sci. 80(8), 1767–1775. Kulberg, S., Heringstad, B., Guttersrud, O.A. and Olsaker, I. 2007. Study on the association of BoLA-DRB3. 2 alleles with clinical mastitis in Norwegian Red cows. J. Anim. Breed. Genet. 124(4), 201–207. Mehrzad, J., Dosogne, H., Meyer, E., Heyneman, R. and Burvenich, C. 2001. Respiratory burst activity of blood and milk neutrophils in dairy cows during different stages of lactation. J. Dairy Res. 68(3), 399–415. Mehrzad, J., Duchateau, L. and Burvenich, C. 2009. Phagocytic and bactericidal activity of blood and milk-resident neutrophils against Staphylococcus aureus in primiparous and multiparous cows during early lactation. Vet. Microbiol. 134(1–2), 106–112. Miller, S., Dykes, D. and Poletsky, H. 1988. A simple salting out procedure for extracting DNA from human nucleated cells. Nucleic Acids Res. 16(3), 1215; doi: 10.1093/nar/16.3.1215. Miyasaka, T., Takeshima, S.N., Matsumoto, Y., Kobayashi, N., Matsuhashi, T., Miyazaki, Y. and Aida, Y. 2011. The diversity of bovine MHC class II DRB3 and DQA1 alleles in different herds of Japanese Black and Holstein cattle in Japan. Gene 472(1–2), 42–49. Nascimento, C.S.D., Machado, M.A., Martinez, M.L., Silva, M.V.G., Guimarães, M.F.M., Campos, A.L. and Oliveira, D.A. 2006. Association of the bovine major histocompatibility complex (BoLA) BoLA-DRB3 gene with fat and protein production and somatic cell score in Brazilian Gyr dairy cattle (Bos indicus). Genet. Mol. Biol. 29(4), 641–647. Piedrahita, L.E., Montoya, L.M. and Pedraza, F.J. 2010. Herpes Virus Bovino tipo 1 (BoHV-1) como posible causa de encefalitis en bovinos de la región del Magdalena Medio Colombiano. Estudio serológico y análisis epidemiológico. Revista Colombiana de Ciencias Pecuarias 23(2), 191–198. Ramirez, N.F., Montoya, A., Cerón-Muñoz, M.F., Villar, D. and Palacio, L.G. 2014. Association of BoLA-DRB3 and TLR4 alleles with subclinical mastitis in cattle from Colombia. Revista Colombiana de Ciencias Pecuarias, 27(1), 18–28. Rodriguez-Zas, S.L., Gianola, D. and Shook, G.E. 2000. Evaluation of models for somatic cell score lactation patterns in Holsteins. Livestock Prod. Sci. 67(1–2), 19–30. Ruiz-Sáenz, J., Jaime, J. and Vera, V.J. 2010. Prevalencia serológica y aislamiento del Herpesvirus Bovino-1 (BHV-1) en hatos ganaderos de Antioquia y del Valle del Cauca. Revista Colombiana de Ciencias Pecuarias, 23(3), 299–307. Rupp, R., Hernandez, A. and Mallard, B.A. 2007. Association of bovine leukocyte antigen (BoLA) DRB3. 2 with immune response, mastitis, and production and type traits in Canadian Holsteins. J. Dairy Sci. 90(2), 1029–1038. Singh, U., Deb, R., Alyethodi, R.R., Alex, R., Kumar, S., Chakraborty, S. and Sharma, A. 2014. Molecular markers and their applications in cattle genetic research: A review. Biomark. Genom. Med. 6(2), 49–58. Starkenburg, R.J., Hansen, L.B., Kehrli Jr,M.E. and Chester-Jones, H. 1997. Frequencies and effects of alternative DRB3. 2 alleles of bovine lymphocyte antigen for Holsteins in milk selection and control lines. J. Dairy Sci. 80(12), 3411–3419. Takeshima, S.N., Miyasaka, T., Polat, M., Kikuya, M., Matsumoto, Y., Mingala, C.N. and Aida, Y. 2014. The great diversity of major histocompatibility complex class II genes in Philippine native cattle. Meta gene 2, 176–190. van Eijk, M., Stewart-Haynes, J. and Lewin, H. 1992. Extensive polymorphism of the BoLA-DRB3 gene distinguished by PCR-RFLP. Anim. Genet. 23(6), 483–496. Wathes, D.C., Cheng, Z., Bourne, N., Taylor, V.J., Coffey, M.P. and Brotherstone, S. 2007. Differences between primiparous and multiparous dairy cows in the inter-relationships between metabolic traits, milk yield and body condition score in the periparturient period. Domest. Anim. Endocrinol. 33(2), 203–225. Wittrock, J.M., Proudfoot, K.L., Weary, D.M. and Von Keyserlingk, M.A.G. 2011. Metritis affects milk production and cull rate of Holstein multiparous and primiparous dairy cows differently. J. Dairy Sci. 94(5), 2408–2412. Zambrano, A., Echeverri, Z. and López-Herrera, A. 2014. Association of gene BoLA DRB3. 2 with production traits in a dairy herd of Antioquia, Colombia. Revista MVZ Córdoba 19(2), 4116–4129. Zambrano, J.C., Echeverri, J. and López-Herrera, A. 2011. Alelos del gen BoLA DRB3. 2 están asociados con mastitis en vacas lecheras. Revista Colombiana de Ciencias Pecuarias 24(2), 145–156. | ||
| How to Cite this Article |
| Pubmed Style Morales JPA, Herrera AL, Zuluaga JE. Association of BoLA DRB3 gene polymorphisms with BoHV-1 infection and zoothecnical traits. Open Vet J. 2020; 10(3): 331-339. doi:10.4314/ovj.v10i3.12 Web Style Morales JPA, Herrera AL, Zuluaga JE. Association of BoLA DRB3 gene polymorphisms with BoHV-1 infection and zoothecnical traits. https://www.openveterinaryjournal.com/?mno=92566 [Access: June 29, 2025]. doi:10.4314/ovj.v10i3.12 AMA (American Medical Association) Style Morales JPA, Herrera AL, Zuluaga JE. Association of BoLA DRB3 gene polymorphisms with BoHV-1 infection and zoothecnical traits. Open Vet J. 2020; 10(3): 331-339. doi:10.4314/ovj.v10i3.12 Vancouver/ICMJE Style Morales JPA, Herrera AL, Zuluaga JE. Association of BoLA DRB3 gene polymorphisms with BoHV-1 infection and zoothecnical traits. Open Vet J. (2020), [cited June 29, 2025]; 10(3): 331-339. doi:10.4314/ovj.v10i3.12 Harvard Style Morales, J. P. A., Herrera, . A. L. & Zuluaga, . J. E. (2020) Association of BoLA DRB3 gene polymorphisms with BoHV-1 infection and zoothecnical traits. Open Vet J, 10 (3), 331-339. doi:10.4314/ovj.v10i3.12 Turabian Style Morales, Juan Pablo Arismendy, Albeiro Lopez Herrera, and Julian Echeverri Zuluaga. 2020. Association of BoLA DRB3 gene polymorphisms with BoHV-1 infection and zoothecnical traits. Open Veterinary Journal, 10 (3), 331-339. doi:10.4314/ovj.v10i3.12 Chicago Style Morales, Juan Pablo Arismendy, Albeiro Lopez Herrera, and Julian Echeverri Zuluaga. "Association of BoLA DRB3 gene polymorphisms with BoHV-1 infection and zoothecnical traits." Open Veterinary Journal 10 (2020), 331-339. doi:10.4314/ovj.v10i3.12 MLA (The Modern Language Association) Style Morales, Juan Pablo Arismendy, Albeiro Lopez Herrera, and Julian Echeverri Zuluaga. "Association of BoLA DRB3 gene polymorphisms with BoHV-1 infection and zoothecnical traits." Open Veterinary Journal 10.3 (2020), 331-339. Print. doi:10.4314/ovj.v10i3.12 APA (American Psychological Association) Style Morales, J. P. A., Herrera, . A. L. & Zuluaga, . J. E. (2020) Association of BoLA DRB3 gene polymorphisms with BoHV-1 infection and zoothecnical traits. Open Veterinary Journal, 10 (3), 331-339. doi:10.4314/ovj.v10i3.12 |